Abstract
The analysis of grapevine (Vitis vinifera) berries at the transcriptomic, proteomic, and metabolomic levels can provide great insight into the molecular events underlying berry development and postharvest drying (withering). However, the large and very different data sets produced by such investigations are difficult to integrate. Here, we report the identification of putative stage-specific biomarkers for berry development and withering and, to our knowledge, the first integrated systems-level study of these processes. Transcriptomic, proteomic, and metabolomic data were integrated using two different strategies, one hypothesis free and the other hypothesis driven. A multistep hypothesis-free approach was applied to data from four developmental stages and three withering intervals, with integration achieved using a hierarchical clustering strategy based on the multivariate bidirectional orthogonal projections to latent structures technique. This identified stage-specific functional networks of linked transcripts, proteins, and metabolites, providing important insights into the key molecular processes that determine the quality characteristics of wine. The hypothesis-driven approach was used to integrate data from three withering intervals, starting with subdata sets of transcripts, proteins, and metabolites. We identified transcripts and proteins that were modulated during withering as well as specific classes of metabolites that accumulated at the same time and used these to select subdata sets of variables. The multivariate bidirectional orthogonal projections to latent structures technique was then used to integrate the subdata sets, identifying variables representing selected molecular processes that take place specifically during berry withering. The impact of this holistic approach on our knowledge of grapevine berry development and withering is discussed.
Grapevine (Vitis vinifera) is a commercially important fruit crop cultivated for the production of table grapes, juice, wine, distilled liquors, and dry raisins. In addition to its high economic value, wine is now considered a key source of health-promoting secondary metabolites, especially antioxidant polyphenols such as resveratrol (Iriti and Faoro, 2009; Yadav et al., 2009).
The economic importance of grapevine has encouraged many researchers to study the physiological and molecular basis of berry development, particularly those processes that affect wine quality (Conde et al., 2007). The availability of high-throughput analysis methods and a high-quality draft of the grapevine genome sequence (Jaillon et al., 2007) has led to the characterization of berry development at the levels of the transcriptome (Terrier et al., 2005; Waters et al., 2005; Deluc et al., 2007; Pilati et al., 2007), proteome (Giribaldi et al., 2007; Negri et al., 2008; Zhang et al., 2008; Grimplet et al., 2009b), and metabolome (Conde et al., 2007).
Berry development involves two sigmoidal growth periods separated by a transition phase known as veraison (Coombe and McCarthy, 2000; Carmona et al., 2008). The first period (formation) is characterized by rapid cell division and growth, embryo development, and the accumulation of malate and other organic acids in the vacuoles (Coombe and McCarthy, 2000; Sweetman et al., 2009). The second period (ripening) is characterized by sugar accumulation, softening, and changes in color, followed by an increase in pH and the accumulation of polyphenols and flavor compounds (Coombe and McCarthy, 2000). Straw wines require an additional postharvest drying (withering) process to acquire the appropriate must quality characteristics and to increase the concentration of simple sugars. For example, cv Corvina berries undergo a 3-month withering treatment under controlled conditions for the production of straw wines such as Amarone and Recioto. Withering has been investigated at the level of the transcriptome (Zamboni et al., 2008; Rizzini et al., 2009) and through the analysis of certain metabolites (Bellincontro et al., 2004, 2006; Costantini et al., 2006), but no proteomic analysis of withering has previously been reported.
High-throughput analysis methods based on the transcriptome, proteome, and metabolome generate large data sets that must be related to the biological system of interest, in this case berry development. Within this framework, it is necessary to develop analytical tools and strategies that allow relevant biological processes to be described and information to be extracted from parallel analyses carried out with different profiling platforms. Bidirectional orthogonal projections to latent structures (O2PLS; Trygg, 2002; Trygg and Wold, 2003) is a multivariate technique that has been used successfully to integrate such data sets (Bylesjö et al., 2007, 2009). This technique can be applied to systems-level studies carried out using either hypothesis-free (discovery-driven) or hypothesis-driven approaches (Hood et al., 2008). The former involves defining and enumerating the elements of a system, with data analysis leading to a hypothesis (Kell and Oliver, 2004). The latter begins with a hypothesis, which is tested by the capture, analysis, integration, and modeling of global data sets relative to phenotypic responses when the system is perturbed in a defined manner (Hood et al., 2008). The relative merits of the two approaches are debated (Nabel, 2009). The discovery-driven approach can generate new knowledge if important components of the system (and their interactions) can be identified in large omics data sets (Saito and Matsuda, 2010).
We investigated berry development and withering in cv Corvina at the transcriptomic, proteomic, and metabolomic levels. Genome-wide transcriptional analysis was carried out using a microarray containing genomic sequences (Jaillon et al., 2007), proteomic data were obtained by two-dimensional difference gel electrophoresis (2D-DIGE), and metabolomic data were obtained by HPLC coupled to mass spectrometry (MS). The O2PLS-discriminant analysis (DA) technique was used to analyze each data set with respect to the different developmental phases and withering intervals and then to derive putative transcript, protein, and metabolite biomarkers. We used a multistep hypothesis-free approach, in which the first step helped to identify information contained in each data set relevant to the development processes, the second step allowed the identification of subgroups of strongly correlated measured variables characterizing these processes, and the third step involved the application of hierarchical clustering analysis (HCA) and the minimum spanning tree (MST) method to these subgroups of variables in order to build simple correlation networks highlighting the relationships between transcripts, proteins, and metabolites. Each network provides a snapshot of a particular phase of berry development or withering and describes the correlation between variables from different data sets involved in the corresponding underlying molecular events. We used a hypothesis-driven approach to study withering, which is characterized by water stress due to dehydration. Variables from the three withering data sets were selected based on the accumulation of specific metabolites and knowledge of the transcriptional events that characterize withering in Corvina berries (Zamboni et al., 2008). Again, O2PLS allowed the identification of well-correlated transcript, protein, and metabolite variables.
RESULTS
Transcriptomic, Proteomic, and Metabolomic Data
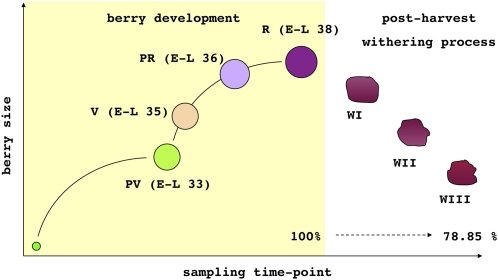
Triplicate samples were taken from four developmental time points and three withering intervals (21 samples in total). Brix values and berry weight increased throughout development, but whereas Brix values continued to increase during withering as sugars concentrated, the total berry weight fell due to dehydration (Fig. 1; Supplemental Table S1).
Figure 1.
The seven sampling time points covering berry development and withering. The first four sampling time points encompass berry development, whereas the last three encompass the 3-month postharvest withering process. The weight loss during withering is indicated by the weight as a percentage of the weight during ripening. PV, Preveraison; V, veraison; PR, preripening; R, ripening; WI, withering I; WII, withering II; WIII, withering III. Each time point is also represented according to the modified E-L system for grapevine growth stages defined by Coombe (1995). [See online article for color version of this figure.]
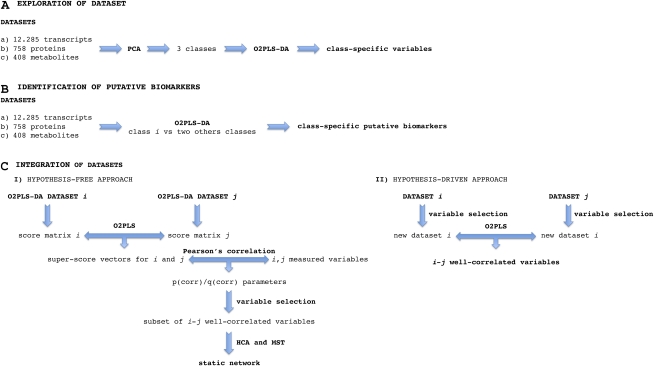
Transcriptome analysis was carried out by interrogating a Combimatrix grapevine genome chip containing 25,471 unique probes, using mRNA prepared from each of the 21 samples. In total, 12,285 transcripts were expressed in at least one of the sampling time points. Proteomic data from the same 21 samples were obtained by 2D-DIGE and subsequent data analysis using the DeCyder software package. This analysis revealed that of the 758 protein spots present in 80% of all protein maps (Supplemental Data Set S8), 68 were differentially expressed (Supplemental Data Set S1). Untargeted metabolomic analysis by HPLC-MS revealed 408 signals (Supplemental Data Set S9) that yielded 220 successful fragmentations and 130 putative identifications, 35 of which were fragments or artifacts (minor isotopes or adducts; Supplemental Table S2). A schematic representation of the data set analysis pipeline is shown in Figure 2.
Figure 2.
Data set analysis strategies. A, Multivariate analysis applied to each data set. The structure of each data set was first defined by unsupervised PCA revealing three main clusters (classes). O2PLS-DA was performed on each data set to identify class-specific variables. B, Identification of putative class-specific markers. Putative markers in classes a, b, and c were identified by a series of three two-class O2PLS-DAs for the transcriptome, proteome, and metabolome data sets. For each series, observations in the class of interest (e.g. a) were separated from those of the other classes (e.g. b and c), the latter identified by two permutations of observations during the definition of first and second classes in the remaining O2PLS-DAs. C, I, Hypothesis-free approach for data integration using a hierarchical analysis strategy. The score vectors arising from the O2PLS-DA models of data sets i and j are combined by O2PLS in order to obtain the superscore vectors to integrate the two data sets. The so-called p(corr) and q(corr) parameters that allowed us to determine which variables were correlated between the two data sets were obtained by calculating the correlation of each measured variable with respect to the superscore vectors of the corresponding block. HCA was performed in order to identify subsets of well-correlated variables, and MST was used to represent graphically the relationships between the variables from each subset. II, Hypothesis-driven approach for data integration. Subdata sets were obtained by selecting variables in relation to known and relevant molecular processes and were integrated using O2PLS models. The so-called p(corr) and q(corr) parameters that allowed us to determine which variables were correlated between the two data sets were obtained by calculating the correlation of each measured variable with respect to the superscore vectors of the corresponding block. [See online article for color version of this figure.]
Data Set Exploration
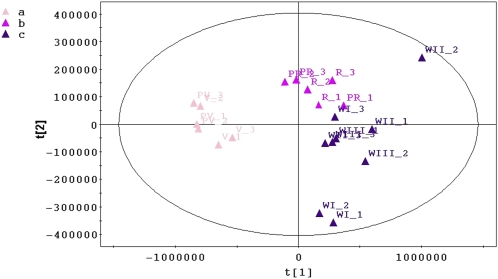
Pattern recognition by principal component analysis (PCA) on each individual data set revealed three clusters (Fig. 2A). Soft independent modeling by class analogy confirmed that each of the three clusters could be considered as a unique class, named a, b, and c (data not shown). The score scatterplot of the PCA model for the metabolome data set is shown in Figure 3, but similar results were obtained for the transcriptome and proteome data sets. The first principal component split the observations of the preveraison and veraison sampling time points (class a) from those of all the other sampling time points, whereas the second principal component split the preripening and ripening observations (class b) from the three withering observations (class c).
Figure 3.
Metabolome PCA scatterplot representing 21 observations. Different colors are used to identify samples in classes a, b, and c. PV, Preveraison; V, veraison; PR, preripening; R, ripening; WI, withering I; WII, withering II; WIII, withering III. [See online article for color version of this figure.]
In order to characterize each class on the basis of class-specific variables, O2PLS-DA was performed on each data set (Fig. 2A). Class-specific variables (more abundant in one class than the others) were selected by inspecting the so-called pq(corr) scatterplot obtained by calculating the Pearson correlation between the measured variables and the two predictive latent components of each model [p(corr)] and that between the dummy variables representing the classes and the predictive components [q(corr)].
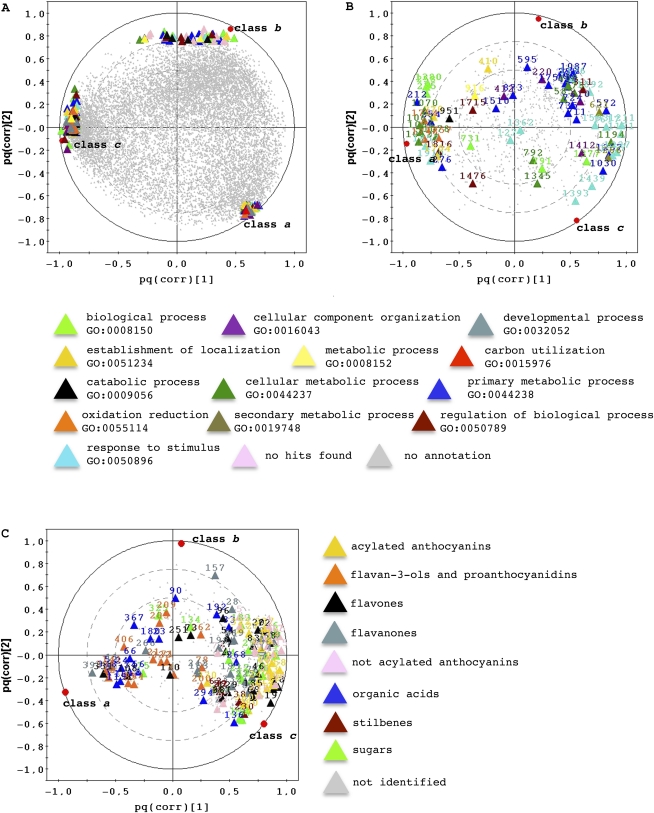
Separate lists were generated from the transcriptome, proteome, and metabolome data sets (Supplemental Data Sets S2–S4). In the transcript data set, each variable was identified by its tentative consensus (TC) number and/or the coding sequence (CDS) identifier code (www.genoscope.cns.fr), and a Gene Ontology biological process annotation was assigned based on BLASTP analysis of the UniProt database (Supplemental Data Sets S2 and S5). Class-specific transcripts were organized into major biological process categories represented by a color code in the scatterplot (Fig. 4A). Although the number of variables characterizing each class was different, the cellular component organization, developmental process, cellular metabolic process, and oxidation-reduction categories were represented predominantly by transcripts specific for classes a and c, catabolic processes by transcripts specific for classes b and c, and the regulation of biological processes by transcripts in all three classes (Fig. 4A).
Figure 4.
O2PLS-DA of each data set. A, O2PLS-DA of the transcriptome data set (UV, 2+2+0, R2Y = 0.97, Q2 = 0.78). B, O2PLS-DA of the proteome data set (UV, 2+3+0, R2Y = 0.98, Q2 = 0.75). C, O2PLS-DA of the metabolome data set (Par, 2+3+0, R2Y = 0.97, Q2 = 0.78). In each case, the pq(corr) scatterplot is obtained by plotting the pq(corr)2 value as a function of the pq(corr)1 value for each variable, and the variable selected on the basis of the best pq(corr)1 and pq(corr)2 for each of the three classes is colored according to the Gene Ontology biological process categories or chemical classes, as appropriate (Supplemental Data Sets S1 and S2). [See online article for color version of this figure.]
For proteins and metabolites, the color code in the scatterplots was used only for variables regardless of each class (Fig. 4, B and C). The protein color code was assigned according to the Gene Ontology terms revealed by the BLASTP results (Supplemental Data Set S1), whereas for metabolites the code was assigned according to their grouping in major chemical families (Supplemental Table S2). The response to stimulus and secondary metabolic process categories were represented predominantly by proteins specific for classes b and c, oxidation reduction by proteins specific for class a, and primary metabolic processes by proteins in all three classes (Fig. 4B). Few metabolites were specifically characteristic of class a, whereas a larger number of metabolites were associated with classes b and c (Fig. 4C). Class a mainly comprised flavan-3-ols, proanthocyanidins, and organic acids, whereas classes b and c included sugars, flavones, flavanones, and acetylated and nonacetylated anthocyanins. Stilbenes were exclusively present in class c (Fig. 4C).
Identification of Putative Biomarkers
Three distinct two-class O2PLS-DA models were built to identify putative biomarkers for each of the three classes (Fig. 2B). For example, to identify putative class a biomarkers, class a observations were used as a reference whereas class b and c observations were associated in a unique distinct class. The other two models were similarly designed, using b and c for the reference classes, respectively. The so called S-plot [i.e. cov(x,tp) versus corr(x,tp)] was then used to select putative biomarkers (Wiklund et al., 2008).
Putative biomarkers were defined as molecules indicating or correlating with the physiological changes that occur during berry development or withering. These were further described as “increasing” if they were more abundant in the class being considered relative to the other two classes or as “decreasing” if they were less abundant in the class being considered relative to the other two classes.
By applying three consecutive O2PLS-DAs for each data set (Fig. 2B), putative biomarkers specific for all the three classes were identified (Table I). In the transcriptome data set, we identified six increasing and four decreasing biomarkers in class a, three increasing and three decreasing in class b, and six increasing and two decreasing in class c. In the proteome data set, we identified two increasing and three decreasing biomarkers in class a, two increasing and two decreasing in class b, and four increasing and four decreasing in class c. In the metabolome data set, we identified four decreasing biomarkers but no increasing ones in class a, whereas classes b and c were characterized by six and four increasing biomarkers, respectively, and no decreasing ones (Table I). Some putative biomarkers yielded no information: that is, transcripts with no sequence homology to known proteins (“no hits found”), proteins that were not sequenced (“not sequenced”), and metabolites that were not identified (“not identified”).
Table I. Putative class-specific biomarkers for transcript, protein, and metabolite data sets.
| IDa | Descriptionb | p(corr)c | Type of Putative Biomarkerd |
| Transcripts | |||
| Class a (O2PLS-DA, UVe, 1+2+0, R2Y = 0.97 Q2 = 0.91)f | |||
| TC56559 | Endo-1,3-1,4-β-d-glucanase, putative | 0.98 | ↑ |
| TC58936 | 11-β-Hydroxysteroid dehydrogenase-like | 0.97 | ↑ |
| TC59336 | Putative Pro-rich protein | 0.97 | ↑ |
| TC61079 | Tubulin β-chain, putative | 0.97 | ↑ |
| TC62885 | Putative Pro-rich cell wall protein | 0.97 | ↑ |
| TC67142 | Peptide transporter-like protein | 0.97 | ↑ |
| TC65181 | Laccase, putative | −0.91 | ↓ |
| TC68733 | Hypothetical binding protein | −0.87 | ↓ |
| TC70396 | LYK10 | −0.86 | ↓ |
| TC70352 | Ser/Thr-protein kinase BRI1-like3 | −0.86 | ↓ |
| Class b (O2PLS-DA, UVe, 1+3+0, R2Y = 0.98 Q2 = 0.83)f | |||
| TC62912 | No hits found | 0.90 | ↑ |
| TC60484 | Putative uncharacterized protein | 0.87 | ↑ |
| GSVIVT00011164001 | Uncharacterized protein At5g10470.2 | 0.87 | ↑ |
| TC67036 | Putative uncharacterized protein | −0.84 | ↓ |
| TC66870 | No hits found | −0.81 | ↓ |
| TC70381 | Expansin | −0.81 | ↓ |
| Class c (O2PLS-DA, UVe, 1+2+0, R2Y = 0.97 Q2 = 0.87)f | |||
| TC55160 | Putative uncharacterized protein | 0.97 | ↑ |
| TC62844 | Pentatricopeptide repeat-containing protein At1g80270, mitochondrial | 0.95 | ↑ |
| TC67468 | Histone H3 | 0.95 | ↑ |
| TC63796 | Histone H2A | 0.94 | ↑ |
| TC61232 | F-box protein At1g23780 | 0.94 | ↑ |
| TC53432 | GTP-binding nuclear protein Ran-3 | 0.96 | ↑ |
| TC56308 | Cytochrome P450 | −0.93 | ↓ |
| TC55457 | No hits found | −0.92 | ↓ |
| Proteins | |||
| Class a (O2PLS-DA, UVe, 1+1+0, R2Y = 0.95 Q2 = 0.83)f | |||
| 1211 | Class IV chitinase | 0.95 | ↓ |
| 1171 | Class IV chitinase | 0.95 | ↓ |
| 1199 | Not sequenced | 0.95 | ↓ |
| 210 | Not sequenced | −0.87 | ↑ |
| 1062 | Not sequenced | −0.88 | ↑ |
| Class b (O2PLS-DA, UVe, 1+2+0, R2Y= 0.95 Q2 = 0.71)f | |||
| 1917 | Not sequenced | 0.81 | ↓ |
| 1861 | Not sequenced | 0.77 | ↓ |
| 860 | Not sequenced | −0.73 | ↑ |
| 421 | Not sequenced | −0.72 | ↑ |
| Class c (O2PLS-DA, UVe, 1+2+0, R2Y = 0.97 Q2 = 0.69)f | |||
| 1393 | Osmotin-like protein | 0.87 | ↑ |
| 1420 | Not sequenced | 0.87 | ↑ |
| 1542 | Not sequenced | 0.82 | ↑ |
| 1439 | Thaumatin-like protein TLP | 0.81 | ↑ |
| 258 | OSJNBa0006A01.15 protein | −0.71 | ↓ |
| 1156 | Not sequenced | −0.70 | ↓ |
| Metabolites | |||
| Class a (O2PLS-DA, Pare, 1+2+0, R2Y = 0.98 Q2 = 0.89)f | |||
| 15 | Peonidin 3-O-glucoside (chlorine adduct) | 0.92 | ↓ |
| 17 | Malvidin O-glucoside (chlorine adduct) | 0.91 | ↓ |
| 65 | Malvidin 3-O-glucoside derivative | 0.93 | ↓ |
| 6 | Dihexose derivative | 0.94 | ↓ |
| Class b (O2PLS-DA, Pare, 1+2+0, R2Y = 0.96 Q2 = 0.79)f | |||
| 157 | Not identified | 0.74 | ↑ |
| 197 | Not identified | 0.63 | ↑ |
| 249 | Not identified | 0.68 | ↑ |
| 258 | Not identified | 0.65 | ↑ |
| 312 | Not identified | 0.79 | ↑ |
| Class c (O2PLS-DA, Pare, 1+3+0, R2Y = 0.99 Q2 = 0.87)f | |||
| 7 | Taxifolin deoxyhexoside | 0.89 | ↑ |
| 150 | Not identified | 0.90 | ↑ |
| 18 | Taxifolin hexoside (chlorine adduct) | 0.91 | ↑ |
| 324 | Not identified | 0.92 | ↑ |
| 19 | Tetrahydroxyflavanone O-deoxyhexoside | 0.94 | ↑ |
Identifier number of putative transcript, protein, and metabolite biomarkers.
Description of putative transcript, protein, and metabolite biomarkers.
p(corr) value of putative transcript, protein, and metabolite biomarkers.
Increasing (↑) or decreasing (↓)putative transcript, protein, and metabolite biomarkers.
Scaling method: UV, unit variance; Par, pareto.
Parameter of each O2PLS-DA model (scaling, components, R2, and Q2 values).
Integration of Data Sets (Hypothesis-Free Approach)
Data integration in the hypothesis-free approach was carried out using a hierarchical multivariate data analysis strategy (Fig. 2C). The predictive scores obtained for each data set in the class characterization step described above were used as supervariables to represent the three data sets. This overcame the dominance effect in data integration caused by the different number of variables in each data set (12,285 transcripts, 758 proteins, and 408 metabolites). When integrating a pair of data sets, O2PLS identified the joint covariation between the two blocks. The supervariable arising from each data set was rotated, yielding a useful basis to project the measured variables. Indeed, for each block, we generated two orthogonal superscore vectors closely correlated to those of the other block. By calculating the correlation of each measured variable with respect to the superscore vectors of the corresponding block, we obtained the so-called p(corr) and q(corr) parameters that allowed us to evaluate which variables were correlated between the two data sets and which were not (Bylesjö et al., 2007). Three different kinds of variables were defined: those correlating well with the first superscore vector, those correlating well with the second superscore vector, and those showing negligible correlation with either vector. Data integration involved only those variables in each block showing good correlation with one of the two superscore vectors. A permutation test was used to establish useful thresholds for correlation, with a significance level of 0.95.
HCA was performed in order to identify subsets of well-correlated variables, and MST was used to represent graphically the relationships between the variables from each subset. The inverse of the pair-wise correlation was used to determine the edge distance. In order to integrate the transcriptome and proteome data sets, transcript variables were selected by setting p(corr)1 ≥ 0.83 and ≤ –0.83 and setting p(corr)2 ≥ 0.80 and ≤ –0.82. The protein variables were selected by setting q(corr)1 ≥ 0.73 and ≤ –0.71 and setting q(corr)2 ≥ 0.79 and ≤ –0.75. HCA was carried out using the selected transcript and protein variables, resulting in 15 clusters, only two of which contained both transcripts and proteins (Fig. 5). Similar analysis was carried out to integrate the metabolome and transcriptome data sets and the proteome and metabolome data sets. In the former case, metabolite variables were selected by setting p(corr)1 ≥ 0.82 and ≤ –0.71 and setting p(corr)2 ≥ 0.73 and ≤ −0.68, whereas transcript variables were selected by setting q(corr)1 ≥ 0.81 and ≤ –0.82 and setting q(corr)2 ≥ 0.78 and ≤ –0.76. In the latter case, metabolite variables are selected by setting p(corr)1 ≥ 0.83 and ≤ –0.72 and setting p(corr)2 ≥ 0.71 and ≤ –0.68, whereas protein variables were selected by setting q(corr)1 ≥ 0.90 and ≤ –0.86 and setting q(corr)2 ≥ 0.64 and ≤ –0.63. In both cases, two HCAs were performed, resulting in 21 clusters for the transcript/metabolite integration, one of which contained both transcripts and metabolites, as represented by the network shown in Figure 6. Thirteen clusters were identified for the protein/metabolite integration, two of which contained both proteins and metabolites, as represented by the network shown in Figure 7.
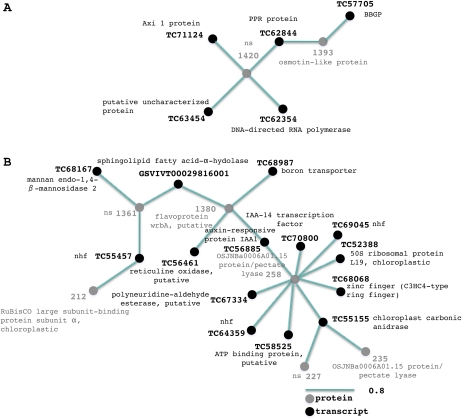
Figure 5.
Transcript-protein networks. A network representation of the two transcript-protein clusters generated by HCA performed on selected transcript and protein variables from O2PLS-DA data is shown. The length of the variable interconnection is inversely proportional to the pair-wise correlation between the two variables. Black, Transcript; gray, protein; nhf, no hits found; ns, not sequenced. [See online article for color version of this figure.]
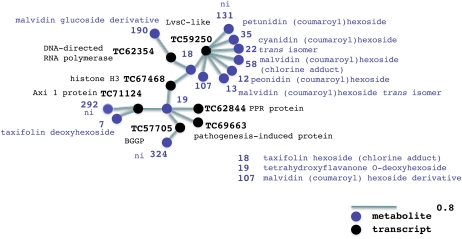
Figure 6.
Transcript-metabolite network. A network representation of the transcript-metabolite cluster generated by HCA performed on selected transcript and metabolite variables from O2PLS-DA data is shown. The length of the variable interconnection is inversely proportional to the pair-wise correlation between the two variables. Black, Transcript; violet, metabolite; nhf, no hits found; ni, not identified. [See online article for color version of this figure.]
Figure 7.
Protein-metabolite network. A network representation of the protein-metabolite cluster generated by HCA performed on selected metabolite and protein variables from O2PLS-DA data is shown. The length of the variable interconnection is inversely proportional to the pair-wise correlation between the two variables. Gray, Protein; violet, metabolite; ni, not identified; ns, not sequenced. [See online article for color version of this figure.]
To integrate all three data sets, O2PLS was carried out to identify the joint covariation structures between the metabolome data set and the joint covariation structures from the transcriptome and proteome data sets. By following the same approach proposed for two-block integration, the two orthogonal pairs of superscore vectors were used to calculate the p(corr) and q(corr) parameters. On the basis of these parameters, we selected the sets of correlated variables between the three data sets. Using the permutation test, thresholds were defined for the p(corr) and q(corr) values with a significance of 0.95. Transcript variables were selected by setting p(corr)1 ≥ 0.83 and ≤ –0.83 and setting p(corr)2 ≥ 0.85 and ≤ –0.95. Protein variables were selected by setting p(corr)1 ≥ 0.60 and ≤ –0.60 and setting p(corr)2 ≥ 0.77 and ≤ –0.78. Finally, the metabolite variables were selected by setting q(corr)1 ≥ 0.60 and ≤ –0.61 and setting p(corr)2 ≥ 0.75 and ≤ –0.60. HCA identified 58 clusters, two of which contained transcripts, proteins, and metabolites, as represented by the networks in Figure 8. All networks showed only positive correlations between transcripts, proteins, and metabolites (Supplemental Data Set S1, Supplemental Table S2, and Supplemental Data Set S5, respectively).
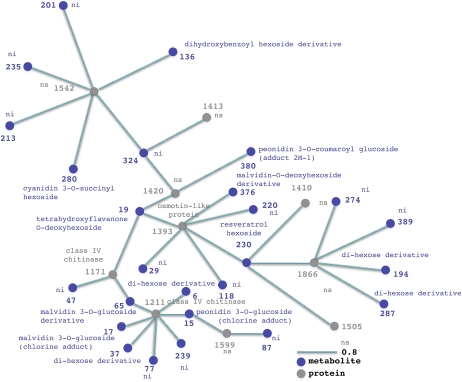
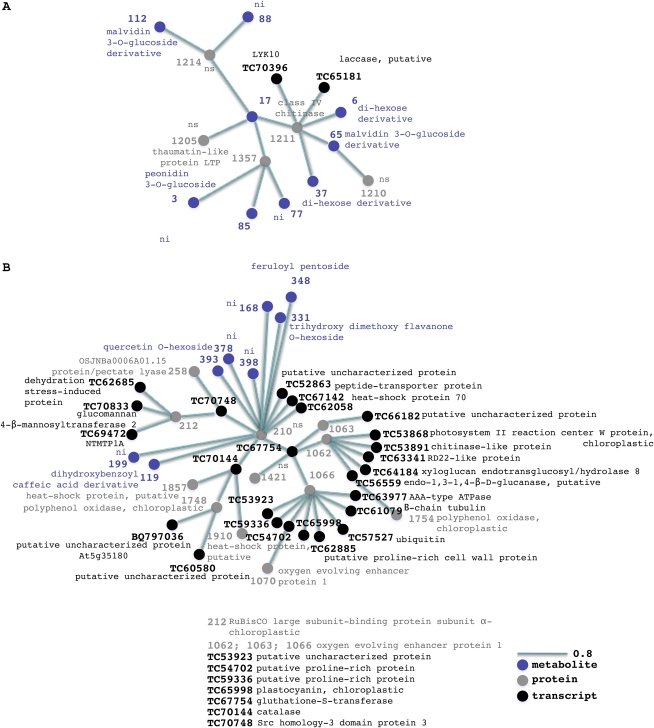
Figure 8.
Transcript-protein-metabolite networks. A network representation of the two transcript-protein-metabolite clusters generated by HCA performed on selected transcript, protein, and metabolite variables from O2PLS-DA data is shown. The length of the variable interconnection is inversely proportional to the pair-wise correlation between the two variables. Black, transcript; gray, protein; violet, metabolite; nhf, no hits found; ni, not identified; ns, not sequenced. [See online article for color version of this figure.]
Integration of Data Sets (Hypothesis-Driven Approach)
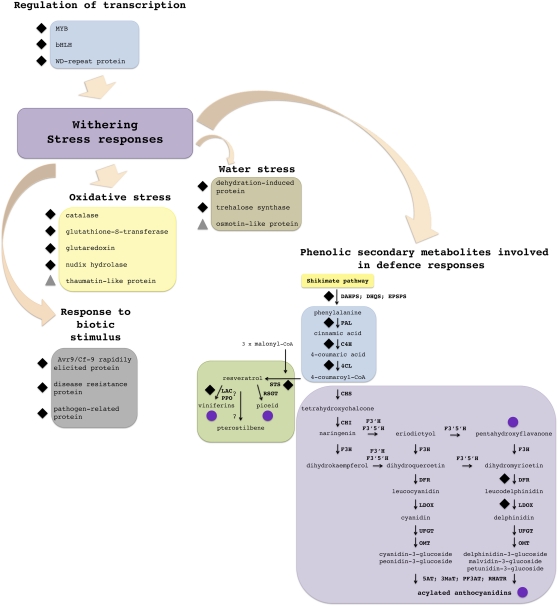
Withering was analyzed separately using a hypothesis-driven approach to integrate data sets selected on the basis of secondary metabolites accumulating specifically during withering and withering-specific changes in gene expression described in a previous investigation (Zamboni et al., 2008). Specific transcriptome, proteome, and metabolome subdata sets were selected accordingly. A 45-metabolite subdata set was obtained by selecting all secondary metabolites belonging to the chemical groups clearly associated with the withering process (Fig. 4C) such as stilbenes, flavanones, and acylated anthocyanins (Supplemental Data Set S4; Supplemental Table S2). We also selected a 134-transcript subdata set (Supplemental Data Set S6) including transcripts involved in Phe synthesis (shikimate pathway and terminal reactions of Phe synthesis), in the synthesis of secondary metabolites (general phenylpropanoid, stilbene, and flavonoid pathways), and those encoding MYB, bHLH, and WD-repeat proteins. O2PLS (UV, 2+3+0, R2X = 0.74 R2Y = 0.62) identified 57 well-correlated variables: 27 transcripts and 30 metabolites (Supplemental Data S6). We also selected a 169-transcript and 11-protein subdata set reflecting the stress response (to biotic stimuli, oxidative stress, and dehydration) as well as WRKY transcription factors (Supplemental Data Set S7). O2PLS (UV, 2+3+0, R2X = 0.70 R2Y = 0.69) identified 15 well-correlated variables: 13 transcripts and two proteins (Supplemental Data Set S7). A schematic representation of well-correlated transcript, protein, and metabolite variables representing phenolic compound synthesis and the stress responses during withering is shown in Figure 9.
Figure 9.
Schematic representation of the molecular events characterizing grapevine berry withering determined by hypothesis-driven data integration. The well-correlated variables involved in these molecular events, resulting from two O2PLS models performed with the new transcript, protein, and metabolite data sets, were tagged with different symbols (black diamonds, transcript; gray triangles, protein; violet circles, metabolite), and identifier numbers are shown in parentheses. Regulation of transcription: MYB protein (TC53952, TC58746, TC60089, TC60338, TC61058, and TC65609); bHLH protein (TC52373); WD-repeat protein (TC52291, TC55356, TC57076, TC61305, TC63183, TC67542, and GSVIVT00027473001). Phenolic secondary metabolite biosynthesis: DHAPS, 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (TC57642); DHQS, 3-dehydroquinate synthase (TC56854); EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase (GSVIVT00026406001); PAL, Phe ammonia lyase (TC69585); C4H, cinnamate 4-hydroxylase (TC70715); 4CL, 4-coumarate-CoA ligase (TC57438 and TC67505); CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; DFR, dihydroflavonol-4-reductase (TC51699); LDOX, leucoanthocyanidin dioxygenase (TC54858); UFGT, UDP-Glc:flavonoid 3-O-glucosyltransferase; OMT, O-methyltransferase; 5AT, anthocyanidin-5-aromatic acyltransferase; 3MaT, anthocyanidin 3-O-glucoside-6′′-O-malonyltransferase; PF3AT, hydroxycinnamoyl-CoA:anthocyanin 3-O-glucoside-6′′-O-hydroxycinnamoyltransferase; RHATR, anthocyanidin-3-glucoside rhamnosyltransferase; STS, stilbene synthase (TC61642 and GSVIVT00009242001); LAC, laccase (TC65181); PPO, polyphenol oxidase (TC58764); flavanone (7, 18, 19, 20, 46, 68, 69, 72, 83, and 185); acylated anthocyanidins (12, 13, 22, 35, 36, 58, 60, 81, 107, 112, 115, 127, 215, 237, 247, 280, and 380); stilbenes (129 and 230); viniferins (38). Response to biotic stimulus: Avr9/Cf-9 rapidly elicited protein (TC58894); disease resistance protein (NP596488 and TC67507); pathogen-related protein (TC51862). Oxidative stress: catalase (TC53791); glutathione-S-transferase (TC53088, TC55724, TC56532, and TC66064); glutaredoxin (TC65527); nudix hydrolase (TC52130); thaumatin-like protein (1439). Water stress: dehydration-induced protein (TC59129); trehalose synthase (TC60540); osmotin-like protein (1393).
DISCUSSION
Overview
In order to achieve what is, to our knowledge, the first holistic, systems-level analysis of grapevine development and withering by integrating transcriptomic, proteomic, and metabolomic data sets, we used the same 21 samples representing four developmental time points and three withering intervals to improve the consistency of microarray, 2D-DIGE, and untargeted large-scale metabolomic analysis by HPLC-MS. The inherent difference in data set complexity (12,285 transcripts, 758 proteins, 408 metabolites) reflects both the limitations of the technologies and the hierarchical organization of biological information (Oltvai and Barabási, 2002).
Data Set Exploration
The variability within each data set was examined by unsupervised PCA, which showed that the first principal component split the preveraison and veraison observations from those of the other sampling time points, and the second principal component split the preripening and ripening observations from those of the three withering time points. PCA thus grouped the observations from the seven sampling time points into three major biological classes representing preveraison/veraison (a), preripening/ripening (b), and withering (c; Fig. 3). Although the preveraison and veraison stages were described as physiologically different (Deluc et al., 2007; Pilati et al., 2007), when studied together with ripening and withering they are not discriminated in PCA analysis.
O2PLS-DA was used to determine the variability of each data set with respect to the three biological classes, and this showed that the transcriptome data set was unique because of the clear separation between class c and the other two classes in terms of shared variables (Fig. 4A). In contrast, there was a more distinct separation between class a and the other two classes in the proteome and metabolome data sets (Fig. 4, B and C), probably reflecting the temporal differences between transcription, protein synthesis, and metabolite synthesis.
The transcriptome O2PLS-DA data generally supported previous microarray studies (Terrier et al., 2005; Waters et al., 2005, Deluc et al., 2007; Pilati et al., 2007) but also identified novel transcripts involved in berry development and withering (Supplemental Text S1; Supplemental Data Set S2). Class a (preveraison/veraison) transcripts were predominantly involved in photosynthesis and carbon assimilation (Terrier et al., 2005; Waters et al., 2005, Pilati et al., 2007), protein synthesis (Pilati et al., 2007), and sugar catabolism (Deluc et al., 2007; Pilati et al., 2007), all of which are known to decline in ripening fruit. Additional class a transcripts were related to the oxidative burst that occurs at veraison (Pilati et al., 2007). Transcripts in the response-to-stimulus category were found in both classes a and b, including those involved in ethylene (class a) and gibberellin (class b) signaling, whereas several transcripts encoding transcription factors were found in classes b and c. Class c (withering-specific) transcripts included those involved in cell wall metabolism, stress responses, aerobic fermentation, volatile compound synthesis, and cell death, as suggested by previous genomic studies (Zamboni et al., 2008; Rizzini et al., 2009) and physiological investigations (Bellincontro et al., 2004; Costantini et al., 2006; Chkaiban et al., 2007).
The proteome O2PLS-DA data showed that proteins involved in photosynthesis and primary/cellular metabolic processes were mainly associated with class a, and their abundance declined after veraison as reported previously (Giribaldi et al., 2007; Negri et al., 2008). The second latent component allowed the identification of class b and c proteins, although some annotated proteins lay between the classes, indicating their involvement in both ripening and withering (Fig. 4B). Proteins involved in primary/cellular metabolic processes, sugar catabolism, cellular component organization, and biogenesis were more likely to group in class b, reflecting the activation of glycolysis and gluconeogenesis during ripening (Sarry et al., 2004; Negri et al., 2008). Proteins involved in the switch to aerobic fermentation during ripening (Sarry et al., 2004) and withering (Bellincontro et al., 2006; Costantini et al., 2006; Chkaiban et al., 2007) were found between classes b and c along with those involved in the production of flavonoids (Coombe and McCarthy, 2000).
The metabolome data set included large numbers of semipolar compounds such as organic acids, sugars, and phenols (Fig. 4C). The O2PLS-DA data indicated that class a is associated with the initial synthesis of organic acids, flavan-3-ols, and proanthocyanins. This is in line with the accumulation of monomeric catechins and tannins in berry skin during the formation phase (Conde et al., 2007), whereas classes b and c contained predominantly sugars, organic acids, flavones, and flavanones, together with acylated and nonacylated anthocyanins that are responsible for the onset of color changes at veraison (Conde et al., 2007; Braidot et al., 2008). Stilbenes clustered as class c-specific compounds. The coumaroylation and succinylation of anthocyanins occurred during withering along with the synthesis of stress-related secondary metabolites such as resveratrol and viniferin (Dercks and Creasy, 1989; Adrian et al., 1997; Zamboni et al., 2008), as reported previously in Corvina berry skin during wilting (Versari et al., 2001). Taxifolin, originally identified in pine (Pinus sp.) because of its antifungal activity (Bonello and Blodgett, 2003), was also found in class c.
Putative Biomarkers
Putative biomarkers were identified in each data set by two-class O2PLS-DA (Table I). They comprise transcripts, proteins, and metabolites whose relative abundance identifies the preveraison/veraison phases (class a), the preripening/ripening phases (class b), and berries that have undergone withering (class c). The putative biomarkers were observed to either increase or decrease in abundance at a specific phase.
In class a, we identified eight increasing putative biomarkers (six transcripts and two proteins) and 11 decreasing ones (four transcripts, three proteins, and four metabolites; Table I). The increasing transcripts included two encoding Pro-rich proteins (TC59336 and TC62885), one tubulin β-chain (TC61079), and one endo-1,3-1,4-β-d-glucanase (TC56559), all of which have been implicated in cell growth (Josè-Estanyol and Puigdoménech, 1998; Mayer and Jürgens, 2002; Glissant et al., 2008). The expression of certain Pro-rich protein and tubulin genes is known to be restricted to the formation phase of berry development (Waters et al., 2005, Grimplet et al., 2007). The two remaining increasing transcripts encoded a peptide transporter (TC67142) and an 11-β-hydroxysteroid dehydrogenase (TC58936). Although 11-hydroxysteroids have yet to be isolated from plants, an Arabidopsis (Arabidopsis thaliana) 11-β-hydroxysteroid dehydrogenase is thought to be involved in the synthesis of brassinosteroids (Li et al., 2007), which are also required for cell growth and division (Belkhadir et al., 2006). The two putative increasing protein biomarkers were not sequenced.
The four decreasing transcripts encoded a laccase (TC65181), a LYK10 (Lys motif receptor-like kinase 10) homolog (TC70396), and a Ser/Thr kinase (TC70352) as well as an unknown protein (TC68733). Plant laccases are involved in phenylpropanoid metabolism (e.g. polymerization of lignin and resveratrol; Mayer and Staples, 2002) as part of the stress responses characteristic of ripening and withering (Deluc et al., 2007; Pilati et al., 2007; Zamboni et al., 2008). LYK kinases recognize peptidoglycans and chito-oligosaccharides, and they facilitate defense responses against fungal pathogens (Zhang et al., 2009). The Ser/Thr kinase (TC70352) is homologous to Arabidopsis BRI1-like3, which is involved in brassinosteroid perception and signal transduction (Oh et al., 2009), so the grapevine transcript may respond to brassinosteroid signaling after veraison, even though the components of the signaling pathway are expressed and assembled prior to veraison (Symons et al., 2006). Two of the three putative decreasing protein biomarkers correspond to different isoforms of a class IV chitinase (1171 and 1211) that are also involved in the ripening/withering stress response (Deluc et al., 2007; Pilati et al., 2007; Negri et al., 2008; Zamboni et al., 2008; Rizzini et al., 2009). All the putative metabolic biomarkers in class a were shown to decrease in abundance. They comprised a dihexose derivative (6) and three modified anthocyanidins (15, 17, and 65). The latter accumulate during ripening and withering, particularly in red cultivars (Conde et al., 2007).
In class b, we identified 10 putative increasing biomarkers (three transcripts, two proteins, and five metabolites) and five decreasing ones (three transcripts and two proteins). Only one of the six transcripts matched an annotated sequence, and this putative decreasing biomarker corresponded to an expansin (TC70381). Expansins are involved in cell wall metabolism and remodeling and therefore could conceivably be involved in both the growth that occurs in early berry development (low-level expression) and the cell wall reconstruction involved in withering (high-level expression). None of the putative protein biomarkers were sequenced, and none of the putative metabolite biomarkers were identified.
In class c, we identified 15 putative increasing biomarkers (six transcripts, four proteins, and five metabolites) and four decreasing ones (two transcripts and two proteins). Four of the increasing transcripts may have roles in gene regulation during withering. One matched the Arabidopsis mitochondrial pentatricopeptide repeat (PPR) protein (TC62884), which is known to regulate organellar RNA processing (Saha et al., 2007). Another matched the GTP-binding nuclear protein Ran-3 (TC53432), which regulates RNA and protein transport through nuclear pores and the assembly of the mitotic spindle (Yudin and Fainzilber, 2009). Finally, two corresponded to histones (TC63796 and TC67468) and therefore could regulate gene expression directly by controlling chromatin structure (Pandey et al., 2008). A further increasing transcript matched the Arabidopsis F-box protein At1g23780 (TC61232), a component of the ASK-Cullin-F-box E3 ubiquitin ligase complex involved in proteasomal degradation (Risseeuw et al., 2003), indicating that protein degradation could play an important role in withering. Two of the putative increasing protein biomarkers were identified, one representing an osmotin-like protein (1393) and the other a thaumatin-like protein (1439), both of which are known to be involved in stress responses. The three putative increasing metabolic biomarkers included two taxifolins (7 and 18) and a tetrahydroxyflavanone-O-deoxyhexoside (19), which may be involved in the withering stress response. To our knowledge, taxifolin has never previously been proposed as a withering biomarker and is more representative of the process than stilbenes, which are induced during withering (Versari et al., 2001). Two putative increasing metabolic biomarkers were not identified. The two decreasing transcripts encoded a cytochrome P450 and a protein of unknown function. The two increasing and the two decreasing proteins were not sequenced.
Integration of Data Sets (Hypothesis-Free Approach)
Hypothesis-free data integration was achieved by performing hierarchical multivariate data modeling combined with HCA. The predictive latent components arising from the class characterization step were combined by O2PLS in the data integration process to obtain a new base set representing the measured variables, while a permutation procedure allowed us to select variables from different data sets with significant pair-wise correlations.
We first integrated the transcriptome and proteome data sets. The interaction between transcripts and proteins whose abundance increases during ripening and withering (Supplemental Data Sets S1 and S5, respectively) is shown in Figure 5A, highlighting the role of stress response genes and proteins as described previously (Arnholdt-Schmitt, 2004; Sarnowski et al., 2005). The network links the expression of genes for a DNA-directed RNA polymerase (TC62354), a mitochondrial PPR protein (TC62844), an auxin response gene (TC71124), and a β-1-3-galactosyl-O-glycosyl-glycoprotein (TC57705) to an unidentified protein (1420) and an osmotin-like protein (1393). PPR proteins in plants play a role in organelle biogenesis and posttranscriptional regulation (Lurin et al., 2004), and an Arabidopsis PPR protein connects abiotic stress responses to mitochondrial electron transport (Zsigmond et al., 2008). The grapevine mitochondrial PPR protein transcript, therefore, could have a similar role in the stress response during ripening and withering, along with the osmotin-like protein, which may respond to osmotic stress caused by increased sugar accumulation (Qureshi et al., 2007). The β-1-3-galactosyl-O-glycosyl-glycoprotein may modify the putative glycosylation site present in osmotin-like proteins, as reported for tobacco (Nicotiana tabacum; Takeda et al., 1991).
The interaction between transcripts and proteins (Supplemental Data Sets S1 and S5, respectively) that are abundant during preveraison and veraison is shown in a second network and primarily involves cell wall and chloroplast metabolism (Fig. 5B). Two isoforms of pectate lyase (235 and 258) and a gene encoding a mannan-endo-1,4-β-mannosidase (TC68167) are associated with cell wall metabolism during berry growth (Nunan et al., 2001; Glissant et al., 2008). The network also includes a Rubisco large subunit-binding protein subunit α (212), which is involved in Rubisco oligomer assembly (Viitanen et al., 1995), flavoprotein WrbA (1380), which is related to flavodoxins (Zurbriggen et al., 2007), and a transcript encoding plastid carbonic anhydrase (TC55155; Majeau and Coleman, 1991), all of which support photosynthesis during berry formation. Transcripts for the 50S ribosomal protein L19 (TC52388) and a boron transporter (TC68987) are also present, the latter likely involved in the transport of HCO3−which is a potential supplier of CO2 to Rubisco. A transcript encoding a sphingolipid fatty acid α-hydroxylase (GSVIVT00029816001) is also found in this network. Sphingolipids in the plasma membrane and tonoplast play a signaling role in guard cell closure and cell death (Chen et al., 2008). The presence in the network of transcripts encoding an auxin-responsive protein (TC56885) and an indole-3-acetic-acid 14 transcription factor (TC70800) suggests a role for auxin signaling in the repression of ripening (Davies and Bottcher, 2009). Auxin levels are high during berry formation, and auxin treatment delays berry ripening (Deytieux-Belleau et al., 2007; Davies and Bottcher, 2009).
Figure 6 shows the network representing the integrated transcriptome and metabolome data sets, describing the molecular events taking place during ripening and withering (Supplemental Table S2 and Supplemental Data Set S5, respectively). Most of the transcripts belong also to the network shown in Figure 5A (see above), but the transcript-metabolite network adds further information. The presence of a transcript encoding histone H3 (TC67468) suggests transcriptional regulation in response to stress (Pandey et al., 2008), supported by the presence of another transcript encoding a pathogenesis-induced protein (TC69633). Metabolites (7 and 19) linked to these processes include flavanones (Bonello and Blodgett, 2003) and coumaroyl-anthocyanins (13, 12, 22, 35, 58, and 107). A LysC-like BEACH protein transcript (TC59250) provides possible links to cytokinesis, contractile vacuole function during osmoregulation (Gerald et al., 2002; De Lozanne, 2003), endosome and lysosome activity (Kypri et al., 2007), vesicle and membrane fusion (De Lozanne, 2003; Saedler et al., 2009), and membrane trafficking (Khodosh et al., 2006). The grapevine LysC-like BEACH protein could facilitate the compartmentalization of coumaroyl-anthocyanins through membrane trafficking.
The integrated proteome and metabolome data sets (Supplemental Data Set S1 and Supplemental Table S2, respectively) are represented by the network shown in Figure 7, and this also describes the molecular events taking place during ripening and withering, highlighting the involvement of a stress response (Deluc et al., 2007; Pilati et al., 2007; Zamboni et al., 2008). In fact, this network includes an osmotin-like protein (1393), two isoforms of a class IV chitinase (1171 and 1211), and two stress-related metabolites: tetrahydroxyflavanone-O-hexoside (19) and resveratrol hexoside (230). As discussed above, osmotin-like proteins respond to osmotic stress, presumably due to sugar accumulation (Qureshi et al., 2007), and chitinases are induced by dehydration (Grimplet et al., 2009b). Flavanone compounds such as tetrahydroxyflavanone-O-hexoside could fulfill a similar role to stilbenes in response to stress (Langcake and Pryce, 1976; Dercks and Creasy, 1989). The two chitinases are positively correlated with glycosylated anthocyanidins such as malvidin-O-glucoside (65 and 17) and a peonidin-O-glucoside (15), which are known to accumulate during ripening and withering (Coombe and McCarthy, 2000; Conde et al., 2007). The network also contains additional modified anthocyanidins (280, 376, and 380) as well as dihexose derivatives (6, 37, 194, and 287).
Figure 8 presents the networks derived by integrating all three data sets (Supplemental Data Sets S1 and S5; Supplemental Table S2). Figure 8A shows the network of variables whose abundance increases during ripening and withering. Many of the variables in this network are also featured in the protein-metabolite network shown in Figure 7 and are not discussed further here. Additional variables unique to this network include a malvidin-coumaroyl-hexoside derivative (112), a peonidin-3-O-glucoside (3), and a malvidin-3-O-glucoside (17), which is positively linked to the class IV chitinase (1211) mentioned above but also to a thaumatin-like protein (1357) known to be involved in biotic stress responses (Ng, 2004). The class IV chitinase (1211) is also positively correlated with two stress-related transcripts encoding a laccase (TC65181) and a LYK10 kinase (TC70396), which may transduce stress responses during ripening and withering.
Figure 8B shows the network of variables that are abundant during the preveraison and veraison phases. Many of the components are also present in the transcript-protein network shown in Figure 5B and will not be discussed further here. Like the network in Figure 5B, they predominantly reflect cell wall and chloroplast metabolism. Additional variables involved in cell wall metabolism include transcripts encoding glucomannan 4-β-mannosyltransferase 2 (TC70833), xyloglucan endotransglucosylase/hydrolase 8 (TC64184), endo-1,3-1,4-β-d-glucanase (TC56559; Liepman et al., 2005; Glissant et al., 2008), and three Pro-rich proteins (TC59336, TC54702, and TC62885). The genes encoding these proteins are expressed predominantly during berry formation (Glissant et al., 2008). There are indications that cell wall metabolism during berry formation involves pectin degradation by pectate lyase (258) followed by hemicellulose reassembly mediated by xyloglucan endotransglucosylase and β-1,4-glucanase (Glissant et al., 2008). The expression of Pro-rich proteins before veraison could boost the levels of these proteins during the subsequent ripening phase, reinforcing the cell wall while it undergoes extensive remodeling (Nunan et al., 2001). Cellulose synthesis is likely to take place during this phase of cell wall remodeling, as shown by the presence of transcripts encoding glucomannan 4-β-mannosyltransferase 2 and chitinase, the latter required for primary and secondary wall synthesis (Zhang et al., 2004). The cellulose content of cell walls may increase during ripening in some fruit (Glissant et al., 2008), although in grapevine the cellulose content of the berries increases after veraison but declines thereafter (Vicens et al., 2009). The network presented in Figure 8B, therefore, reveals a very dynamic system including molecular events linked to the first phase of berry growth (e.g. photosynthesis) as well as the induction of molecular changes that will have a physiological effect later in development (e.g. the oxidative burst marking veraison).
Photosynthetic variables include the above-mentioned Rubisco large subunit-binding protein subunit α (212) but also four isoforms of oxygen-evolving enhancer protein 1 (1062, 1063, 1066, and 1070) and a transcript encoding PSII reaction center W protein (TC53868), which may be essential for the stability of PSII (Sugihara et al., 2000). In addition, it includes two isoforms of a plastid polyphenol oxidase (1748 and 1754) that potentially regulates photosynthesis (Dry and Robinson, 1994; Kuwabara, 1995). Another transcript linked to chloroplast metabolism encodes a plastid plastocyanin (TC65998) whose role may be to facilitate electron transport between the cytochrome b6f complex and PSI (Sugawara et al., 1999). A further linked transcript encodes a protein homologous to tobacco NTMTP1Aa, which confers zinc and cobalt tolerance in yeast mutants, sequestering metals into vacuoles (Shingu et al., 2005). Heavy metals such as zinc, cadmium, cobalt, nickel, and mercury interact with PSII and inhibit its activity (Ghirardi et al., 1996). The transporter encoded by this grapevine transcript, therefore, could protect PSII by sequestering heavy metal ions in the vacuole. Two heat shock proteins (1857 and 1910) and a transcript encoding a cucumber (Cucumis sativus) heat shock protein 70 (TC62058) could help to stabilize old and newly synthesized proteins at veraison (Negri et al., 2008).
Additional transcripts in the network encode catalase (TC70144), a detoxifying enzyme, and glutathione-S-transferase (TC67754), which balances the oxidized and reduced forms of glutathione (Pilati et al., 2007). This suggests that the forming berry begins to adapt to the anticipated oxidative burst that occurs during the ripening phase. The expression of two transcripts that encode dehydration-induced proteins (TC62685 and TC63341) may indicate that berries activate other stress response mechanisms at this stage. TC63341 corresponds to Vvdr22, a dehydration-responsive gene involved in abiotic stress tolerance that is expressed in grapevine cell suspension cultures in response to abscisic acid particularly in combination with Suc (Hanana et al., 2008). The tubulin β-chain 6 transcript (TC61079) indicates that the cell division and enlargement in berry formation may involve cytoskeleton modifications. The transcripts encoding a ubiquitin carrier protein (TC57527) and an AAA-type ATPase (TC63977) indicate that proteolysis or its regulation may be an important component of berry formation (Kedzierska, 2006). The metabolites in the network reflect the genes and proteins that are active during berry growth. Two modified hydroxycinnamic acid derivatives, a dihydroxybenzoyl-caffeic acid derivative (119) and a feruloyl pentoside (348), are present in the network, reflecting the accumulation of hydroxycinnamic acids in skin and flesh during berry formation as precursors of volatile phenols (Conde et al., 2007). A trihydroxy-dimethoxy-flavanone-O-hexoside (331) and flavonol quercetin-O-hexoside (393) are also present in the network, the latter reflecting the biphasic nature of flavonol biosynthesis in grapevine berries (one during flowering, another 3–4 weeks after veraison; Mattivi et al., 2006).
Integration of Data Sets (Hypothesis-Driven Approach)
Hypothesis-driven data integration (Fig. 9) indicated that withering involves transcriptomic, proteomic, and metabolomic changes representing a stress response to dehydration and eventual pathogen attack. The metabolome data set showed that stilbenes (38, 128, and 230), flavanones (7, 18, 19, 20, 46, 68, 69, 72, 83, and 185), and acylated anthocyanins (12, 13, 22, 35, 36, 58, 60, 81, 107, 112, 115, 127, 215, 237, 247, 280, and 380) are the predominant metabolites that accumulate during withering, with stilbenes and flavonones already implicated in stress responses (Dercks and Creasy, 1989; Adrian et al., 1997; Versari et al., 2001; Bonello and Blodgett, 2003) and a similar role likely for acetylated anthocyanins. The treatment of Merolt grapevine with benzothiadiazole increases the synthesis of trans-resveratrol and anthocyanins, particularly malvidine-3-glucoside and its acylated derivates, and thus enhances resistance to Botrytis cinerea (Iriti et al., 2004). The acetylated anthocyanins that accumulate during withering are mostly coumarate derivates that are important in winemaking because they influence pigmentation and increase light absorption, thus affecting red wine color (Iriti et al., 2004). Acylation can also increase anthocyanin activity (Yoshimoto et al., 2001). The accumulation of nutritionally valuable and health-promoting secondary metabolites such as stilbenes and modified anthocyanins in berries in response to a technical process such as postharvest withering has a positive impact on wine quality.
Figure 9 also shows that the accumulation of stilbenes, flavanones, and acylated anthocyanins correlates with the accumulation of certain transcripts encoding enzymes involved in their synthesis, such as three enzymes in the shikimate pathway (3-deoxy-d-arabino-heptulosonate-7-phosphate synthase [TC57642], 3-dehydroquinate synthase [TC56854], and 5-enolpyruvylshikimate-3-phosphate synthase [GSVIVT00026406001]), and others involved in general phenylpropanoid metabolism (Phe ammonia lyase [TC69585], cinnamate 4-hydroxylase [TC70715], and 4-coumarate-CoA ligase [TC57438 and TC67505]). The phenylpropanoid pathway provides coumaroyl-CoA, a precursor for stilbene and flavonoid synthesis. The synthesis of stilbenes such as resveratrol (129 and 230) and ε-viniferin (38) correlates with the expression of stilbene synthase (TC61642 and GSVIVT00009242001), laccase (TC65181), and polyphenol oxidase (TC58764), the latter possibly involved in the oxidative dimerization of resveratrol to viniferins (Breuil et al., 1999; Pezet et al., 2003). No transcript or protein variables related to flavanone synthesis increased in abundance during withering, although anthocyanin biosynthesis genes such as dihydroflavonol-4-reductase (TC51699) and leucoanthocyanidin dioxygenase (TC54858) could promote the accumulation of acylated anthocyanins during the same process even if transcripts involved in the anthocyanin acylation were not modulated (i.e. anthocyanidin-5-aromatic acyltransferase, anthocyanidin 3-O-glucoside-6′′-O-malonyltransferase, hydroxycinnamoyl-CoA:anthocyanin 3-O-glucoside-6′′-O-hydroxycinnamoyltransferase, and anthocyanidin-3-glucoside rhamnosyltransferase). However, the accumulation of flavanones and coumarated anthocyanins was clearly shown not only by HPLC-electrospray ionization-MS but also by quantitative HPLC-diode array (data not shown). The production of these classes of secondary compounds, therefore, may involve enzymes that are already active during ripening and do not need to be induced specifically during withering.
The hypothesis-driven approach also identified four well-correlated transcripts characteristic of the withering process, encoding an Avr9/Cf-9 rapidly elicited protein (TC58894), two disease resistance proteins (NP596488 and TC67507), and a pathogen-related protein (TC51862). The up-regulation of similar transcripts was also reported in the previous amplified fragment length polymorphism-transcriptional profiling analysis of withering (Zamboni et al., 2008) and in ripening berries under water-deficit conditions (Grimplet et al., 2007). Further variables associated with stress responses included a dehydration-induced transcript (TC59129), a trehalose synthase transcript (TC60540), and an osmotin-like protein (1393). The osmotin-like protein (1393), already identified as a putative class c biomarker and as a component of a network describing withering and the last phase of ripening, was also identified using our hypothesis-free approach (Fig. 5A) and is probably a response to the increased sugar concentration (Qureshi et al., 2007). The transcript encoding a trehalose synthase (TC60540) is probably involved in this process too (Garg et al., 2002). The induction of transcripts encoding catalase (TC53791), glutathione-S-transferase (TC53088, TC55724, TC56532, and TC66064), glutaredoxin (TC65527), nudix hydrolase (TC52130), and a thaumatin-like protein (1439) is likely to be a response to the oxidative stress burst that occurs during withering (Zamboni et al., 2008). This involves a different catalase and different glutathione-S-transferases to those identified during berry formation using the hypothesis-free approach (Fig. 8B).
The hypothesis-driven approach also identified transcripts encoding new transcription factors of the MYB (TC53952, TC58746, TC60089, TC60338, TC61058, and TC65609), bHLH (TC52373), and WD-repeat families (GSVIVT00027473001, TC52291, TC55356, TC57076, TC61305, TC63183, and TC67542), which have previously been implicated in the transcriptional regulation of genes involved in the synthesis of flavonoids (Matus et al., 2008, 2010; Czemmel et al., 2009; Mahjoub et al., 2009; Terrier et al., 2009; Hichri et al., 2010). These transcripts could be candidates for functional analysis, with the aim of dissecting the regulation of these classes of secondary compounds in grapevine. However, the hypothesis-driven approach did not identify transcripts encoding WRKY proteins, which are also involved in coordinating responses to abiotic and biotic stresses (Pandey and Somssich, 2009).
Comparison of Hypothesis-Free and Hypothesis-Driven Approaches
The hypothesis-free and hypothesis-driven strategies for data integration highlighted similar molecular mechanisms underlying the withering process and identified similar variables predominantly among the metabolic data set. The correlation between metabolites that accumulate during withering and transcripts encoding enzymes involved in their synthesis was observed only with the hypothesis-driven approach, suggesting that such relationships may be masked in the hypothesis-free approach by the larger, unselected data sets. The hypothesis-free approach cannot only identify correlations between variables involved in a same molecular process but also those acting at the same stage of development. The hypothesis-free approach can provide the greatest insight when there is a large number of variables and therefore a high density of information (Nabel, 2009). Therefore, these approaches should be regarded as complementary rather than alternatives, and for the optimum results they should be applied iteratively (Kell and Oliver, 2004).
CONCLUSION
The transcriptome, proteome, and metabolome of grapevine berries were analyzed during development and withering, and three data sets were obtained. Class-specific variables and putative increasing and decreasing biomarkers were identified for each data set using the O2PLS-DA approach.
Our results revealed that the multivariate O2PLS technique, which has previously been applied in a limited context known as OPLS to analyze the structure of metabolomic and metabonomic data sets and to identify putative biomarkers in such studies, can also be a useful approach for the analysis of transcriptomic and proteomic data sets, which have distinct characteristics. This technique allowed us to identify not only metabolites but also transcripts and proteins as putative biomarkers, which were then analyzed in terms of the molecular events characterizing grapevine berry development and withering. O2PLS is an efficient analysis technique for the identification of known and putative transcript, protein, and metabolite biomarkers of different biological systems linked to developmental processes or environmental interactions.
In the hypothesis-free strategy, we applied a hierarchical approach to data integration based on the O2PLS technique and composed of three levels. The first level identifies the information contained in each data set corresponding to the grapevine berry development and withering processes by using O2PLS-DA and three classes recognized by PCA. By this approach, we obtained a subspace spanned by two latent variables for each data set containing only the information related to grapevine berry development and withering. In the second level, the reduced data sets were compared by O2PLS and a permutation test was applied to select significant, well-correlated subsets of measured variables. These two steps were used to reduce the space to be analyzed in the third level, in which networks were built by applying HCA and MST on the subsets of variables previously selected. HCA was used to construct networks of correlated transcripts, proteins, and metabolites, and a thorough analysis of the results (focusing on the biological function of each of the variables) produced a holistic view of each network. These networks provide, to our knowledge, the first comprehensive systems-level picture of berry development and withering and have allowed us to identify major features of each phase by correlating gene expression, protein activity, and the resulting metabolic profiles. Such “static” networks, grouping transcripts, proteins, and metabolites on the basis of profile correlation, are the starting points for the construction of “dynamic” networks, where specific perturbations allow the identification of linked elements that take part in the same molecular process.
We used a hypothesis-driven approach to identify transcript, protein, and metabolite variables involved in the molecular events underpinning withering, which predominantly reflected a general stress response. This revealed the correlation between variables in different data sets related to the same process (i.e. secondary metabolite accumulation and genes encoding enzymes involved their biosynthesis).
Our results have confirmed the molecular events that are known to characterize grapevine berry development and withering, but the application of powerful systems biology approaches has also provided new insights into these processes. We can confirm that berry formation involves active cell wall metabolism and photosynthesis, whereas ripening and withering are characterized by the induction of stress responses. Novel findings include the discovery that carbonic acid acts as a putative supplier of CO2 to Rubisco and that sphingolipid fatty acids act as signals during the first berry growth phase. Berry ripening and withering are characterized by the accumulation of secondary metabolites such as acylated anthocyanins, whose compartmentalization correlates with the expression of a BEACH transcript, but we also found that withering involves the activation of specific osmotic and oxidative stress response genes and the specific production of acylated anthocyanins, stilbenes, and taxifolin. Our identification of stage-specific functional networks of linked transcripts, proteins, and metabolites, therefore, has provided important insights into the key molecular processes that determine the quality characteristics of wine.
MATERIALS AND METHODS
Plant Material and Sampling
Grapevine (Vitis vinifera cv Corvina, clone 48) berries were sampled during the 2006 growing season at four developmental time points and at three additional time points during the 91-d postharvest withering process (Fig. 1). The four developmental time points were 59 (E-L 33), 71 (E-L 35), 98 (E-L 36), and 112 (E-L 38) d after fruit set, corresponding to preveraison, veraison, early ripening, and late ripening, and the three withering time points (WI, WII, and WIII) were 35, 56, and 91 d after harvest (Supplemental Table S1). Three biological replicates were taken at each time point, resulting in a total of 21 observations. Each biological replicate comprised a pool of deseeded berries from five clusters collected from five different plants. Mean Brix degree values (Supplemental Table S1) were recorded at each sampling stage using a PR-32 bench refractometer (Atago). The postharvest withering phase was analyzed by collecting berry clusters and storing them in the special room (fruttaio) normally used for this process at room temperature and a relative humidity not exceeding 65%. Berry weight was determined by averaging the weight of 400 berries, and percentage weight loss in withering berries was calculated using ripening berries as a reference (Fig. 1). The pools of deseeded berries from each of the 21 samples were used for RNA, protein, and metabolite extraction.
Transcriptome Analysis
Transcriptome analysis was carried out using a Combimatrix Vitis vinifera chip, produced by the Plant Functional Genomics Center, University of Verona. The chip carries 24,571 nonredundant probes randomly distributed in triplicate across the array, each comprising a 35- to 40-mer oligonucleotide designed using the program Oligoarray 2.1 (Rouillard et al., 2003). The source of sequence information included TC sequences derived from the Dana-Farber Cancer Institute (DFCI) Grape Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=grape) release 5.0 (19,062 probes), singletons based on 3′ poly(A) tail sequences (1,904 probes), ESTs (55 probes), and genomic sequences produced by the International Grape Genome Project (Jaillon et al., 2007) with no TC matches (3,490 probes). Nine bacterial oligonucleotide sequences provided by Combimatrix, 40 probes designed on seven Ambion spikes, and 11 probes based on Bacillus anthracis, Haemophilus ducreyi, and Alteromonas phage sequences were used as negative controls.
RNA was isolated according to Rezaian and Krake (1987) and quantified by spectrophotometry (ATI Unicam). An aliquot of each RNA sample was also analyzed using an Agilent 2100 Bioanalyzer. Total RNA (1 μg) was amplified using the SuperScript Indirect RNA Amplification System (Invitrogen) to incorporate amino-allyl UTP (aRNA) and was fluorescently labeled with Alexa Fluor 647. The purified labeled aRNA was quantified by spectrophotometry as above. A 3-μg sample of labeled RNA was hybridized to the array according to the manufacturer’s recommendations (http://www.combimatrix.com/support_docs.htm). The array was scanned with a ScanArray 4000XL (Perkin-Elmer). TIFF images were exported to the Microarray Imager 5.8 (Combimatrix) for densitometric analysis. A scale normalization was applied to raw data (Smyth and Speed, 2003). Probe signals were then filtered on median values calculated on the negative probes, considering only those probes with signal higher than median negative values for at least one of the seven sampling time points and for all three biological replicates taken at each sampling time point. Probe signals were then filtered according to the coefficient of variation calculated using the signals of the replicates of each probe, setting a threshold of 0.5. All microarray expression data are available at the Gene Expression Omnibus under the series entry GSE20511 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rjetpeisaacakji&acc=GSE20511).
Proteome Analysis
Samples were prepared according to Tsugita and Kamo (1999) with some modifications (Di Carli et al., 2009). Extracted proteins were purified using the Clean-Up kit (GE Healthcare) and quantified using the DC Protein Assay (Bio-Rad). Two-dimensional gel electrophoresis was carried out using the GE Healthcare DIGE system. Purified proteomes were covalently labeled using the CyDye DIGE Fluor (Cy2, Cy3, and Cy5) according to the manufacturer’s instructions and analysis carried out as described previously (Di Carli et al., 2009). A random design with 12 gels and a dye-swap approach was used to avoid labeling artifacts and ensure statistical significance. For the first-dimensional separation, immobilized pH gradient strips (pH 4–7/18 cm) containing solubilized protein samples were passively rehydrated as recommended, and isoelectric focusing was performed on an IPGphor unit (GE Healthcare) at 20°C with a 50-μA current limit per strip and a program setting of 3 h at 300 V, 1 h at 500 V, 6 h at 1,000 V, and 5 h at 8,000 V (analytical gels) or a 60-μA current limit per strip and a program setting of 10 h at 200 V, 3 h at 300 V, 1 h at 500 V, 6 h at 1,000 V, and 5 h at 8,000 V (preparative gels). After focusing, proteins were reduced, alkylated, and separated in the second dimension by 12.5% SDS-PAGE using 18-cm × 20-cm × 1-mm gels on the Ettan DALT-12 system (GE Healthcare). Separated CyDye-labeled proteins were visualized by scanning with a Typhoon 9410 Imager (GE Healthcare). To compare protein spots across gels, a match set was created from the images of all 12 gels. The statistical analysis of protein level changes between different maturation stages was performed using the DeCyder-BVA (for Biological Variation Analysis, version 6.5; GE Healthcare) software module. A one way-ANOVA was used for univariate analysis. Protein spots with a statistically significant variation (P ≤ 0.05) showing a greater than 1.5-fold difference in volume between samples were considered to be differentially expressed and were selected for further analysis by MS. In-gel reduction using iodoacetamide and digestion with trypsin was followed by MS/MS analysis. All liquid chromatography(LC)-MS/MS experiments were performed using an Eksigent NanoLC-1D Plus (Eksigent Technologies) HPLC system and an LTQ Orbitrap mass spectrometer (ThermoFisher) as described by Deery et al. (2009). MS data were then submitted to the Mascot search algorithm (Matrix Science) and searched against the National Center for Biotechnology Information Viridiplantae (Green Plants) database (419,956 sequences) using a fixed modification of carbamidomethyl and a variable modification of oxidation.
Metabolome Analysis
Powdered samples were extracted with three volumes of ice-cold methanol containing 0.1% formic acid. An HPLC system (Beckman Coulter System Gold 127, Solvent Module) with a Rheodyne sample injector, coupled online with a Bruker ion-trap mass spectrometer (Esquire 6000) equipped with an electrospray ionization source, was used for LC-MS analysis. The system was fitted with a 150- × 2.1-mm Alltima HP C18 3-μm column and a 7.5- × 2.1-mm guard column (Alltech Associates). Two solvents were used for separation: 5% (v/v) formic acid/5% (v/v) acetonitrile in water (solvent A) and 100% acetonitrile (solvent B). After injecting 5 μL of sample at a flow rate of 200 μL min−1, a solvent gradient was established from 0% to 10% B in 5 min, from 10% to 20% B in 20 min, from 20% to 25% B in 5 min, and from 25% to 70% B in 15 min; a 20-min equilibration followed each analysis, which was carried out in duplicate (for detailed procedures and technical data, see Supplemental Text S1). The column, the flow rate, the solvents, and the gradient solvent for HPLC-diode array analysis were the same as used for LC-MS analysis above, although with an injection volume of 20 μL. Chromatography data were extracted using metAlign software (http://metAlign.nl; RIKILT Institute of Food Safety). The data matrix was processed (Supplemental Text S1) to provide 408 molecular parent ion signals. For compound identification, mass-to-charge ratio values and fragmentation patterns (MS/MS and MS3) were compared with those of appropriate commercial standards. When direct comparison was impossible due to the lack of appropriate commercial standards, fragmentation patterns were compared with those reported in the literature (Supplemental Text S1). Matrix effects for all compounds were evaluated as described by Cavaliere et al. (2008).
Statistical Methods
O2PLS is a multivariate projection method that is able to find relationships between two data blocks (X and Y). O2PLS decomposes the systematic variation in the X-block (or Y-block) into two model parts: a predictive part, which models the joint X-Y correlated variation, and an orthogonal part, which is not related to Y (or X). The latent structures of the joint X-Y correlated variation can be used to identify small groups of correlated variables belonging to the two different blocks. This was achieved by evaluating the similarity between each variable and the predictive latent components of the X-Y O2PLS model by means of their correlation. In order to set the significance threshold for the similarity, a permutation test was carried out, and data integration was performed on each small group of X-Y variables with significant correlation. O2PLS was also used to perform DA. O2PLS-DA allowed the identification of latent variables that were able to yield a parsimonious and very efficient representation of the process. A hierarchical strategy of analysis based on these latent variables allowed us to integrate the data sets. In order to define the number of latent components for OPLS(-DA) models, we applied partial cross-validation and a permutation test to highlight overfitting. Multivariate data analysis was performed by using SIMCA P+ (Umetrics).
Transcript Annotation
Class-specific transcripts, putative transcript biomarkers, and transcripts identified using the hypothesis-free data integration approach were analyzed using BLASTN (Altschul et al., 1997) to link corresponding TCs in the DFCI Grape Gene Index Releases 5.0 and 6.0 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=grape). For Release 6.0 TCs and 8.4× CDS that did not match Release 5.0, BLASTP analysis (Altschul et al., 1997) was performed against the UniProt database (http://www.uniprot.org/) using the 8.4× CDS predicted protein sequences and an E-value cutoff of 10−8. Biological process Gene Ontology terms (http://www.geneontology.org/) were assigned to each probe using the BLASTP results.
Manual gene annotation, correspondences between the DFCI Grape Gene Index Release 5.0 and the predicted 8.4× genome CDS, and grapevine molecular networks described by Grimplet et al. (2009a) were used to select transcripts for the hypothesis-driven data integration approach.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Sampling time points during berry development and postharvest withering.
Supplemental Table S2. Identified metabolites.
Supplemental Data Set S1. Sequenced and identified protein spots.
Supplemental Data Set S2. Class-specific transcripts.
Supplemental Data Set S3. Class-specific proteins.
Supplemental Data Set S4. Class-specific metabolites.
Supplemental Data Set S5. Annotated transcripts.
Supplemental Data Set S6. The 134-transcript and 45-metabolite data sets related to secondary metabolism generated using the hypothesis-driven approach, showing well-correlated variables identified by O2PLS.
Supplemental Data Set S7. The 169-transcript and 13-protein data sets related to defense responses generated using the hypothesis-driven approach, showing well-correlated variables identified by O2PLS.
Supplemental Data Set S8. Proteome data set.
Supplemental Data Set S9. Metabolome data set.
Supplemental Text S1. Supplemental Discussion and Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Pasqua Vini e Cantine (Verona, Italy) for allowing us to sample material from its vineyard.
References
- Adrian M, Jeandet P, Veneau J, Weston LA, Bessis R. (1997) Biological activity of resveratrol, stilbenic compound from grapevine, against Botrytis cinerea, the causal agent for gray mold. J Chem Ecol 23: 1689–1702 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnholdt-Schmitt B. (2004) Stress-induced cell reprogramming: a role for global genome regulation? Plant Physiol 136: 2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Wang X, Chory J. (2006) Arabidopsis brassinosteroid signaling pathway. Sci STKE 2006: cm5. [DOI] [PubMed] [Google Scholar]
- Bellincontro A, De Santis D, Botondi R, Villa I, Mencarelli F. (2004) Different post-harvest dehydration rates affect quality characteristics and volatile compounds of Malvasia, Trebbiano and Sangiovese grape for wine production. J Sci Food Agric 84: 1791–1800 [Google Scholar]
- Bellincontro A, Fardelli A, De Santis D, Botondi R, Mencarelli F. (2006) Postharvest ethylene and 1-MCP treatments both affect phenols, anthocyanins, and aromatic quality of Aleatico grapes and wine. Aust J Grape Wine Res 12: 141–149 [Google Scholar]
- Bonello P, Blodgett J. (2003) Pinus nigra-Sphaeropsis sapinea as a model pathosystem to investigate local and systemic effects of fungal infection of pines. Physiol Mol Plant Pathol 63: 249–261 [Google Scholar]
- Braidot E, Zancani M, Petrussa E, Peresson C, Bertolini A, Patui S, Macrì F, Vianello A. (2008) Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal Behav 3: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuil AC, Jeandet P, Adrian M, Chopin F, Pirio N, Meunier P, Bessis R. (1999) Characterization of pterostilbene dehydrodimer produced by laccase of Botrytis cinerea. Phytopathology 89: 298–302 [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Eriksson D, Kusano M, Moritz T, Trygg J. (2007) Data integration in plant biology: the O2PLS method for combined modeling of transcript and metabolite data. Plant J 52: 1181–1191 [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Nilsson R, Srivastava V, Grönlund A, Johansson AI, Jansson S, Karlsson J, Moritz T, Wingsle G, Trygg J. (2009) Integrated analysis of transcript, protein and metabolite data to study lignin biosynthesis in hybrid aspen. J Proteome Res 8: 199–210 [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Chaïb J, Martínez-Zapater JM, Thomas MR. (2008) A molecular genetic perspective of reproductive development in grapevine. J Exp Bot 59: 2579–2596 [DOI] [PubMed] [Google Scholar]
- Cavaliere C, Foglia P, Gubbiotti R, Sacchetti P, Samperi R, Laganà A. (2008) Rapid-resolution liquid chromatography/mass spectrometry for determination and quantitation of polyphenols in grape berries. Rapid Commun Mass Spectrom 22: 3089–3099 [DOI] [PubMed] [Google Scholar]
- Chen M, Markham JE, Dietrich CR, Jaworski JG, Cahoon EB. (2008) Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20: 1862–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkaiban L, Botondi R, Bellincontro A, De Santis D, Kefalas P, Mencarelli F. (2007) Influence of postharvest water stress on lipoxygenase and alcohol dehydrogenase activities, and on the composition of some volatile compounds of Gewürztraminer grapes dehydrated under controlled and uncontrolled thermohygrometric conditions. Aust J Grape Wine Res 13: 142–147 [Google Scholar]
- Conde C, Silva P, Fontes N, Dias ACP, Tavares RM, Sousa MJ, Agasse A, Delrot S, Geros H. (2007) Biochemical changes throughout grape berry development and fruit and wine quality. Food 1: 1–22 [Google Scholar]
- Coombe BG. (1995) Adoption of a system for identifying grapevine growth stages. Aust J Grape Wine Res 1: 100–110 [Google Scholar]
- Coombe BG, McCarthy MG. (2000) Dynamics of grape berry growth and physiology of ripening. Aust J Grape Wine Res 6: 131–135 [Google Scholar]
- Costantini V, Bellincontro A, De Santis D, Botondi R, Mencarelli F. (2006) Metabolic changes of Malvasia grapes for wine production during postharvest drying. J Agric Food Chem 54: 3334–3340 [DOI] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J. (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Bottcher C. (2009) Hormonal control of grapevine berry ripening. In KA Roubelakis-Angelakis, ed, Grapevine Molecular Physiology and Biotechnology, Ed 2. Springer, Dordrecht, The Netherlands, pp 229–261 [Google Scholar]
- Deery MJ, Maywood ES, Chesham JE, Sládek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS.et al (2009) Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol 19: 2031–2036 [DOI] [PubMed] [Google Scholar]
- De Lozanne A. (2003) The role of BEACH proteins in Dictyostelium. Traffic 4: 6–12 [DOI] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dercks W, Creasy LL. (1989) The significance of stilbene phytoalexins in the Plasmopara viticola grapevine interaction. Physiol Mol Plant Pathol 34: 189–202 [Google Scholar]
- Deytieux-Belleau C, Gagne S, L’Hyvernay A, Doneche B, Geny L. (2007) Possible roles of both abscisic acid and indol-acetic acid in controlling grape berry ripening process. J Int Sci Vigne Vin 41: 141–148 [Google Scholar]
- Di Carli M, Villani ME, Renzone G, Nardi L, Pasquo A, Franconi R, Scaloni A, Benvenuto E, Desiderio A. (2009) Leaf proteome analysis of transgenic plants expressing antiviral antibodies. J Proteome Res 8: 838–848 [DOI] [PubMed] [Google Scholar]
- Dry IB, Robinson SP. (1994) Molecular cloning and characterisation of grape berry polyphenol oxidase. Plant Mol Biol 26: 495–502 [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald NJ, Siano M, De Lozanne A. (2002) The Dictyostelium LvsA protein is localized on the contractile vacuole and is required for osmoregulation. Traffic 3: 50–60 [DOI] [PubMed] [Google Scholar]
- Ghirardi ML, Lutton TW, Seibert M. (1996) Interactions between diphenylcarbazide, zinc, cobalt, and manganese on the oxidizing side of photosystem II. Biochemistry 35: 1820–1828 [DOI] [PubMed] [Google Scholar]
- Giribaldi M, Perugini I, Sauvage FX, Schubert A. (2007) Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 7: 3154–3170 [DOI] [PubMed] [Google Scholar]
- Glissant D, Dedaldechamp F, Delrot S. (2008) Transcriptomic analysis of grape berry softening during ripening. J Int Sci Vigne Vin 42: 1–13 [Google Scholar]
- Grimplet J, Cramer GR, Dickerson JA, Mathiason K, Van Hemert J, Fennell AY. (2009a) VitisNet: “omics” integration through grapevine molecular networks. PLoS ONE 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC. (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Wheatley MD, Jouira HB, Deluc LG, Cramer GR, Cushman JC. (2009b) Proteomic and selected metabolite analysis of grape berry tissues under well-watered and water-deficit stress conditions. Proteomics 9: 2503–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanana M, Deluc L, Fouquet R, Daldoul S, Léon C, Barrieu F, Ghorbel A, Mliki A, Hamdi S. (2008) [Identification and characterization of “rd22” dehydration responsive gene in grapevine (Vitis vinifera L.).] C R Biol 331: 569–578 [DOI] [PubMed] [Google Scholar]
- Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. (2010) The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant 3: 509–523 [DOI] [PubMed] [Google Scholar]
- Hood L, Rowen L, Galas DJ, Aitchison JD. (2008) Systems biology at the Institute for Systems Biology. Brief Funct Genomics Proteomics 7: 239–248 [DOI] [PubMed] [Google Scholar]
- Iriti M, Faoro F. (2009) Bioactivity of grape chemicals for human health. Nat Prod Commun 4: 611–634 [PubMed] [Google Scholar]
- Iriti M, Rossoni M, Borgo M, Faoro F. (2004) Benzothiadiazole enhances resveratrol and anthocyanin biosynthesis in grapevine, meanwhile improving resistance to Botrytis cinerea. J Agric Food Chem 52: 4406–4413 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Josè-Estanyol M, Puigdoménech P. (1998) Rapid changes induced in developmental programmes of the maize embryo detected by analysis of the expression of genes encoding proline-rich proteins. FEBS Lett 422: 400–402 [DOI] [PubMed] [Google Scholar]
- Kedzierska S. (2006) [Structure, function and mechanisms of action of ATPases from the AAA superfamily of proteins]. Postepy Biochem 52: 330–338 [PubMed] [Google Scholar]
- Kell DB, Oliver SG. (2004) Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays 26: 99–105 [DOI] [PubMed] [Google Scholar]
- Khodosh R, Augsburger A, Schwarz TL, Garrity PA. (2006) Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133: 4655–4665 [DOI] [PubMed] [Google Scholar]
- Kuwabara T. (1995) The 60-kDa precursor to the dithiothreitol-sensitive tetrameric protease of spinach thylakoids: structural similarities between the protease and polyphenol oxidase. FEBS Lett 371: 195–198 [DOI] [PubMed] [Google Scholar]
- Kypri E, Schmauch C, Maniak M, De Lozanne A. (2007) The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic 8: 774–783 [DOI] [PubMed] [Google Scholar]
- Langcake P, Pryce RJ. (1976) Production of resveratrol by Vitis vinifera and other members of Vitaceae as a response to infection or injury. Physiol Plant Pathol 9: 77–86 [Google Scholar]
- Li F, Asami T, Wu X, Tsang EW, Cutler AJ. (2007) A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol 145: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub A, Hernould M, Joubès J, Decendit A, Mars M, Barrieu F, Hamdi S, Delrot S. (2009) Overexpression of a grapevine R2R3-MYB factor in tomato affects vegetative development, flower morphology and flavonoid and terpenoid metabolism. Plant Physiol Biochem 47: 551–561 [DOI] [PubMed] [Google Scholar]
- Majeau N, Coleman JR. (1991) Isolation and characterization of a cDNA coding for pea chloroplastic carbonic anhydrase. Plant Physiol 95: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. (2006) Metabolite profiling of grape: flavonols and anthocyanins. J Agric Food Chem 54: 7692–7702 [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Poupin MJ, Cañón P, Bordeu E, Alcalde JA, Arce-Johnson P. (2010) Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol Biol 72: 607–620 [DOI] [PubMed] [Google Scholar]
- Mayer AM, Staples RC. (2002) Laccase: new functions for an old enzyme. Phytochemistry 60: 551–565 [DOI] [PubMed] [Google Scholar]
- Mayer U, Jürgens G. (2002) Microtubule cytoskeleton: a track record. Curr Opin Plant Biol 5: 494–501 [DOI] [PubMed] [Google Scholar]
- Nabel GJ. (2009) Philosophy of science: the coordinates of truth. Science 326: 53–54 [DOI] [PubMed] [Google Scholar]
- Negri AS, Prinsi B, Rossoni M, Failla O, Scienza A, Cocucci M, Espen L. (2008) Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TB. (2004) Antifungal proteins and peptides of leguminous and non-leguminous origins. Peptides 25: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Nunan KJ, Davies C, Robinson SP, Fincher GB. (2001) Expression patterns of cell wall-modifying enzymes during grape berry development. Planta 214: 257–264 [DOI] [PubMed] [Google Scholar]
- Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA 106: 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltvai ZN, Barabási AL. (2002) Systems biology: life’s complexity pyramid. Science 298: 763–764 [DOI] [PubMed] [Google Scholar]
- Pandey A, Chakraborty S, Datta A, Chakraborty N. (2008) Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol Cell Proteomics 7: 88–107 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet R, Perret C, Jean-Denis JB, Tabacchi R, Gindro K, Viret O. (2003) δ-Viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. J Agric Food Chem 51: 5488–5492 [DOI] [PubMed] [Google Scholar]
- Pilati S, Perazzolli M, Malossini A, Cestaro A, Demattè L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C. (2007) Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at vèraison. BMC Genomics 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MI, Qadir S, Zolla L. (2007) Proteomics-based dissection of stress-responsive pathways in plants. J Plant Physiol 164: 1239–1260 [DOI] [PubMed] [Google Scholar]
- Rezaian MA, Krake LR. (1987) Nucleic acid extraction and virus detection in grapevine. J Virol Methods 17: 277–285 [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Rizzini FM, Bonghi C, Tonutti P. (2009) Postharvest water loss induced marked changes in transcript profiling in skin of wine grape berry. Postharvest Biol Technol 52: 247–253 [Google Scholar]
- Rouillard JM, Zuker M, Gulari E. (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 31: 3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Jakoby M, Marin B, Galiana-Jaime E, Hülskamp M. (2009) The cell morphogenesis gene SPIRRIG in Arabidopsis encodes a WD/BEACH domain protein. Plant J 59: 612–621 [DOI] [PubMed] [Google Scholar]
- Saha D, Prasad AM, Srinivasan R. (2007) Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Biochem 45: 521–534 [DOI] [PubMed] [Google Scholar]
- Saito K, Matsuda F. (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Sarnowski TJ, Ríos G, Jásik J, Swiezewski S, Kaczanowski S, Li Y, Kwiatkowska A, Pawlikowska K, Koźbiał M, Koźbiał P, et al. (2005) SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarry JE, Sommerer N, Sauvage FX, Bergoin A, Rossignol M, Albagnac G, Romieu C. (2004) Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 4: 201–215 [DOI] [PubMed] [Google Scholar]
- Shingu Y, Kudo T, Ohsato S, Kimura M, Ono Y, Yamaguchi I, Hamamoto H. (2005) Characterization of genes encoding metal tolerance proteins isolated from Nicotiana glauca and Nicotiana tabacum. Biochem Biophys Res Commun 331: 675–680 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP. (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Inoue T, Li C, Gotowda M, Hibino T, Takabe T, Kai Y. (1999) Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene. J Biochem 125: 899–903 [DOI] [PubMed] [Google Scholar]
- Sugihara K, Hanagata N, Dubinsky Z, Baba S, Karube I. (2000) Molecular characterization of cDNA encoding oxygen evolving enhancer protein 1 increased by salt treatment in the mangrove Bruguiera gymnorrhiza. Plant Cell Physiol 41: 1279–1285 [DOI] [PubMed] [Google Scholar]
- Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. (2009) Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70: 1329–1344 [DOI] [PubMed] [Google Scholar]
- Symons GM, Davies C, Shavrukov Y, Dry IB, Reid JB, Thomas MR. (2006) Grapes on steroids: brassinosteroids are involved in grape berry ripening. Plant Physiol 140: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Sato F, Ida K, Yamada Y. (1991) Nucleotide sequence of a cDNA for osmotin-like protein from cultured tobacco cells. Plant Physiol 97: 844–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Glissant D, Grimplet J, Barrieu F, Abbal P, Couture C, Ageorges A, Atanassova R, Léon C, Renaudin JP, et al. (2005) Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development. Planta 222: 832–847 [DOI] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C. (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J. (2002) O2-PLS for qualitative and quantitative analysis in multivariate calibration. J Chemometr 16: 283–293 [Google Scholar]
- Trygg J, Wold S. (2003) O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J Chemometr 17: 53–64 [Google Scholar]
- Tsugita A, Kamo M. (1999) 2-D electrophoresis of plant proteins. Methods Mol Biol 112: 95–97 [DOI] [PubMed] [Google Scholar]
- Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C. (2001) Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J Agric Food Chem 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Vicens A, Fournand D, Williams P, Sidhoum L, Moutounet M, Doco T. (2009) Changes in polysaccharide and protein composition of cell walls in grape berry skin (cv. Shiraz) during ripening and over-ripening. J Agric Food Chem 57: 2955–2960 [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Schmidt M, Buchner J, Suzuki T, Vierling E, Dickson R, Lorimer GH, Gatenby A, Soll J. (1995) Functional characterization of the higher plant chloroplast chaperonins. J Biol Chem 270: 18158–18164 [DOI] [PubMed] [Google Scholar]
- Waters DLE, Holton TA, Ablett EM, Lee LS, Henry RJ. (2005) cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct Integr Genomics 5: 40–58 [DOI] [PubMed] [Google Scholar]
- Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. (2008) Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem 80: 115–122 [DOI] [PubMed] [Google Scholar]
- Yadav M, Jain S, Bhardwaj A, Nagpal R, Puniya M, Tomar R, Singh V, Parkash O, Prasad GB, Marotta F, et al. (2009) Biological and medicinal properties of grapes and their bioactive constituents: an update. J Med Food 12: 473–484 [DOI] [PubMed] [Google Scholar]
- Yoshimoto M, Okuno S, Yamaguchi M, Yamakawa O. (2001) Antimutagenicity of deacylated anthocyanins in purple-fleshed sweetpotato. Biosci Biotechnol Biochem 65: 1652–1655 [DOI] [PubMed] [Google Scholar]
- Yudin D, Fainzilber M. (2009) Ran on tracks: cytoplasmic roles for a nuclear regulator. J Cell Sci 122: 587–593 [DOI] [PubMed] [Google Scholar]
- Zamboni A, Minoia L, Ferrarini A, Tornielli GB, Zago E, Delledonne M, Pezzotti M. (2008) Molecular analysis of post-harvest withering in grape by AFLP transcriptional profiling. J Exp Bot 59: 4145–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Hrmova M, Wan CH, Wu C, Balzen J, Cai W, Wang J, Densmore LD, Fincher GB, Zhang H, et al. (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54: 353–372 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ma H, Feng J, Zeng L, Wang Z, Chen S. (2008) Grape berry plasma membrane proteome analysis and its differential expression during ripening. J Exp Bot 59: 2979–2990 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G. (2009) Evolutionary genomics of LysM genes in land plants. BMC Evol Biol 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Otvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. (2008) Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol 146: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen MD, Tognetti VB, Carrillo N. (2007) Stress-inducible flavodoxin from photosynthetic microorganisms: the mystery of flavodoxin loss from the plant genome. IUBMB Life 59: 355–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.