Abstract
Anthocyanins are responsible for the color of many flowers, fruits, and vegetables. An interesting and unique Purple (Pr) gene mutation in cauliflower (Brassica oleracea var botrytis) confers an abnormal pattern of anthocyanin accumulation, giving the striking mutant phenotype of intense purple color in curds and a few other tissues. To unravel the nature of the Pr mutation in cauliflower, we isolated the Pr gene via a combination of candidate gene analysis and fine mapping. Pr encoded a R2R3 MYB transcription factor that exhibited tissue-specific expression, consistent with an abnormal anthocyanin accumulation pattern in the mutant. Transgenic Arabidopsis (Arabidopsis thaliana) and cauliflower plants expressing the Pr-D allele recapitulated the mutant phenotype, confirming the isolation of the Pr gene. Up-regulation of Pr specifically activated a basic helix-loop-helix transcription factor and a subset of anthocyanin structural genes encoding flavonoid 3’-hydroxylase, dihydroflavonol 4-reductase, and leucoanthocyanidin dioxygenase to confer ectopic accumulation of pigments in the purple cauliflower. Our results indicate that the genetic variation including a Harbinger DNA transposon insertion in the upstream regulatory region of the Pr-D allele is responsible for the up-regulation of the Pr gene in inducing phenotypic change in the plant. The successful isolation of Pr provides important information on the regulatory control of anthocyanin biosynthesis in Brassica vegetables, and offers a genetic resource for development of new varieties with enhanced health-promoting properties and visual appeal.
Vegetables and fruits are fundamental components of human diets. They not only are important sources of essential vitamins and minerals, but also contain a wide variety of secondary metabolites important to human health. Colored vegetables and fruits have gained an increasing interest as functional foods, owing to their high levels of plant pigments with potent nutritional and health-promoting effects. Among them, purple cauliflower (Brassica oleracea var botrytis) is a very eye-catching vegetable and available commercially. The purple coloration is due to the accumulation of anthocyanins.
Anthocyanins are a group of flavonoid compounds that fulfill important biological functions in protecting plants against various biotic and abiotic stresses. All of the anthocyanin biosynthetic pathway genes and numerous regulatory factors have been identified from studies of Arabidopsis (Arabidopsis thaliana), maize (Zea mays), petunia (Petunia hybrida), snapdragon (Antirrhinum majus), and other plant species (Broun, 2005; Dixon et al., 2005; Koes et al., 2005; Grotewold, 2006). Transcriptional regulation of structural genes appears to be a major mechanism by which anthocyanin biosynthesis is regulated in plants. R2R3 MYB and basic helix-loop-helix (bHLH) transcription factors as well as WD40 proteins represent the three major families of anthocyanin regulatory proteins (Paz-Ares et al., 1987; Chandler et al., 1989; Ludwig and Wessler 1990; de Vetten et al., 1997; Quattrocchio et al., 1999). They form regulatory complexes to activate expression of anthocyanin structural genes (Goff et al., 1992; Grotewold et al., 2000). In Arabidopsis, several MYB proteins, including PAP1, PAP2, MYB113, MYB114, and MYBL2 (Borevitz et al., 2000; Dubos et al., 2008; Gonzalez et al., 2008; Matsui et al., 2008), three bHLH proteins of TT8, GL3, and EGL3 (Nesi et al., 2000; Payne et al., 2000; Zhang et al., 2003), and a WD40 repeat protein of TTG1 (Walker et al., 1999) are involved in anthocyanin biosynthesis. While the R2R3 MYB proteins of PAP1, PAP2, MYB113, and MYB114 cause tissue-specific anthocyanin accumulation in Arabidopsis (Borevitz et al., 2000; Gonzalez et al., 2008), the R3-MYB protein, MYBL2, acts as an inhibitor of anthocyanin biosynthesis (Dubos et al., 2008; Matsui et al., 2008). TT8 is required for the full transcriptional activation of late anthocyanin pathway genes (Nesi et al., 2000), and is partially functionally redundant with its closest homologs, GL3 and EGL3 (Zhang et al., 2003). The WD40 protein, TTG1, is known to physically interact with the MYB and bHLH transcription factors in controlling anthocyanin biosynthesis (Zhang et al., 2003).
R2R3 MYB transcription factors have been implicated to play an important role for color difference in plant species. Activation of an R2R3 MYB transcription factor confers anthocyanin production in a number of anthocyanin-accumulating plants (Borevitz et al., 2000; Mathews et al., 2003). In contrast, loss of function of a R2R3 MYB results in color loss in the normal anthocyanin-accumulating tissues. For example, a retrotransposon insertion in the promoter of VvMYBA1 and mutations in the adjacent VvMYBA2 inactivate their expression and convert red-skinned grape (Vitis vinifera) into white-skinned one (Walker et al., 2007). In Antirrhinum majus, alteration of MYB-related gene expression controls floral pigmentation intensity and pattern (Schwinn et al., 2006).
Similarly, bHLH transcription factors have also been found to be responsible for color difference in plant species. Alteration of a bHLH transcription factor causes an increase in red pigment production in the aleurone of maize (Burr et al., 1996). Knockout of a bHLH function by a frame shift changes seed pericarp color from red into white in rice (Oryza sativa; Sweeney et al., 2006). An insertion of a DNA transposon into a bHLH regulatory gene alters flower color of the common morning glory (Ipomoea tricolor) into pale pigmented flowers (Park et al., 2007).
Mutant analyses have facilitated gene discovery and elucidation of the regulatory control of anthocyanin biosynthesis. Although there are large numbers of researches on the underlying mechanisms controlling anthocyanin production in flowers, fruits, and model plants, only few have focused on vegetables. Purple cauliflower mutant represents an interesting mutation that confers profound anthocyanin production in the otherwise low-pigmented curds and seeds—the two tissues of agricultural importance. Thus, this mutant provides an excellent opportunity to reveal regulatory control of anthocyanin biosynthesis in vegetable crops.
The cauliflower purple mutation was found to be controlled by a single, semidominant gene. We designate the symbol Pr-D for the Purple allele, and pr for its wild-type counterpart. Through a combination of candidate gene analysis and fine mapping, we isolated the Pr gene and found that it encoded a R2R3 MYB transcription factor. Comparison of Pr sequences from wild type and the mutant allele revealed allelic variation including a Harbinger DNA transposon insertion in the upstream regulatory region of the Pr-D allele. Such an alteration caused an increased Pr gene transcription, which in turn up-regulated anthocyanin structural gene expression to produce the striking purple phenotype. The activation of the Pr gene, probably by introduction of new regulatory motifs in the promoter region, provides a way that anthocyanin transcriptional regulation can be switched on differently in plants.
RESULTS
Phenotypic Characterization of the Purple Cauliflower Mutant
The cauliflower purple mutant plants grew and developed normally when compared to white cauliflower under normal growth conditions in field and in greenhouse. The mutant exhibited a tissue-specific pattern of anthocyanin accumulation. Intense purple coloration was observed in curd and a few other tissues of the plant (Fig. 1, A–D, F, and G), including young seedlings, very young leaves, and very young flower buds and siliques, as well as the endosperm of seeds, a tissue that is rarely reported to accumulate pigments in anthocyanin-accumulating mutants, e.g. in red cabbage (Brassica oleracea var capitata; Yuan et al., 2009). The purple color was not observed in older leaves, stems, flower petals, and older siliques (Fig. 1, B, E, and F). The purple phenotype appears to be associated primarily with very young tissues, curds, and seeds, rather than in flower petals, the most common anthocyanin-accumulating tissue. Under the same growth conditions, the wild-type cauliflower plants exhibited no purple hue in these tissues (Fig. 1, A–G).
Figure 1.
Phenotypic comparison between wild type and the cauliflower Pr-D mutant. A, Young seedlings of 5-d-old plants. B, Young plants of 3-week-old. C, Curds of cauliflower plants grown in field. D, Young flower buds. E, Flowers. F, Young (inner) and old siliques of wild type (left) and mutant (right). G, Seed endosperms.
Purple Mutant Predominantly Accumulates Cyanidin Glucosides
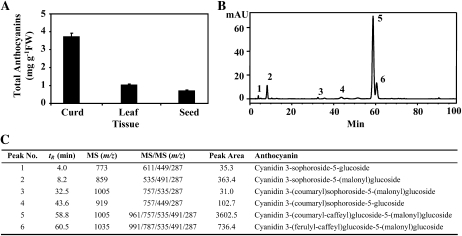
To examine the composition and content of anthocyanins accumulated in young leaves, curds, and seeds of the purple mutant, we performed HPLC analysis and found high levels of anthocyanins in the purple tissues (Fig. 2A). The curds accumulated approximately 3.75 mg cyanidin diglucoside equivalent g−1 fresh weight, a level that was comparable with that found in blueberries (Vaccinium myrtillus; Gao and Mazza, 1994). In contrast, the samples of the wild-type control plants contained undetectable amounts of anthocyanins (data not shown), indicating that the anthocyanin pathway was biochemically quiescent under normal growth conditions.
Figure 2.
Analysis of anthocyanin content and composition in the Pr-D mutant. A, Total anthocyanin levels as cyanidin diglucoside equivalent in curds, leaves, and seeds. FW, Fresh weight. B, HPLC elution profile of anthocyanins accumulated in curds. The absorbance was monitored at A520 nm. C, Major anthocyanins identified by HPLC-ESI-MS/MS in curds.
Several different groups of anthocyanins (e.g. delphinidin, pelargonidin, and cyanidin) exist in the plant kingdom (Tanaka et al., 2008). To identify anthocyanin composition in the mutant, anthocyanins in curd tissue were separated and analyzed using HPLC-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS; Wu and Prior, 2005b). The purple mutant contained one major peak (Fig. 2B), which was identified as cyanidin 3-(coumaryl-caffeyl) glucoside-5-(malonyl)glucoside (Fig. 2C). The other minor peaks were also identified as different forms of cyanidin glycosides based on the dominant ion pairs observed in the mass spectrometry data and anthocyanin structures (Mazza and Miniati, 1993; Wu and Prior, 2005a).
The Purple Mutation Is Controlled by a Single Semidominant Gene
To examine whether the purple mutant was controlled by one or multiple genes, we crossed the purple mutant Graffiti to an inbred white cultivar Stovepipe, selfed the heterozygous F1 plants, and generated a large F2 population. A subpopulation of 102 individuals was germinated in a greenhouse. Genotyping of these F2 individuals was completed by visually examining the absence and presence of light or dark purple color in very young leaves and curds of the progeny, followed by further confirmation of the individual genotype in some cases by visual examination of the color with 16 F3 individuals. The segregation ratio of these F2 plants for white:light purple:dark purple was 31:53:18. χ2 test showed a good fit with a 1:2:1 ratio (P > 0.05), which was consistent with the ratio expected for the progeny derived from selfing a parent heterozygous at one locus. This result suggests that the purple phenotype is controlled by a single, semidominant gene, Pr.
Expression of Anthocyanin Biosynthetic and Regulatory Genes
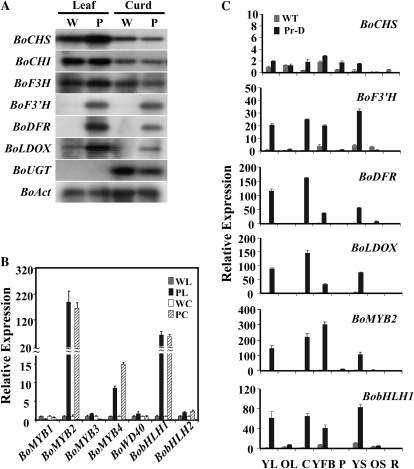
To investigate whether Pr represented one of the biosynthetic genes that was significantly up-regulated in the Pr-D mutant, we examined the expression of anthocyanin pathway genes in curds and leaves of wild type and the Pr-D mutant by northern-blot analysis. The cDNAs coding chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucosyltransferase (UGT) were used as probes. While the early pathway genes were expressed at similar levels between wild type and the Pr-D mutant in both leaves and curds, the late pathway genes, BoF3’H, BoDFR, and BoLDOX, were dramatically up-regulated in the mutant (Fig. 3A). The co-up-regulation of three structural genes suggests that Pr is unlikely a mutation of one specific anthocyanin pathway gene.
Figure 3.
Expression of anthocyanin structural and regulatory genes in wild type and the Pr-D mutant. A, northern-blot analysis of transcript levels of anthocyanin pathway genes in curds and leaves. BoAct serves as equal loading control. P, Pr-D mutant; W, wild-type control. B, qRT-PCR analysis of anthocyanin regulatory genes in curds and leaves. The identities of the amplified products were confirmed by sequencing. Expression of genes in wild-type leaves and curds were set to 1. WL, Wild-type leaves; PL, Pr-D leaves; WC, wild-type curds; PC, Pr-D curds. BoWD40, BobHLH1, and BobHLH2 are cauliflower homologs of Arabidopsis TTG1, TT8, and EGL3, respectively. C, qRT-PCR analysis of transcript levels of BoCHS and the differentially expressed genes during different developmental stages. Expression of genes in wild-type (WT) young leaves was set to 1. YL, Young leaves; OL, old leaves; C, curds; YFB, young flower buds; P, petals; YS, young siliques; OS, old siliques; R, roots. qRT-PCR results represent mean values + sd from three biological replicates with three technical replicates for each.
Many regulatory genes that control anthocyanin biosynthesis have been isolated from plant species (Paz-Ares et al., 1987; Chandler et al., 1989; Ludwig and Wessler 1990; de Vetten et al., 1997; Quattrocchio et al., 1999). Since cauliflower genes typically share high coding sequence identity with Arabidopsis genes, the available sequences of anthocyanin regulatory genes from Arabidopsis and the significant amounts of Brassica sequence information in the public domains provide direct resources for designing the gene-specific primers for some cauliflower homologs. In Arabidopsis, the R2R3 MYB family proteins of PAP1, PAP2, MYB113, and MYB114, bHLH proteins of TT8 and EGL3, and WD40 protein of TTG1 are known to regulate anthocyanin biosynthesis. To investigate whether the Pr gene represented a mutation of one of the known regulatory genes, transcript levels of these homologous genes in wild type and the Pr-D mutant were examined by quantitative reverse transcription (qRT)-PCR using gene-specific primers (Yuan et al., 2009; Supplemental Table S1). As shown in Figure 3B, PAP-like MYB family genes, i.e. BoMYB2 and BoMYB4, as well as BobHLH1, a homologous gene of Arabidopsis TT8 (Nesi et al., 2000) exhibited differential expression in both leaves and curds between wild type and the Pr-D mutant.
Anthocyanins in the Pr-D mutant exhibited tissue- and development-specific accumulation (Fig. 1). To examine whether the specific anthocyanin accumulation pattern was correlated with gene expression pattern, we analyzed transcript levels of BoCHS and the differentially expressed genes in different tissues of wild-type and the Pr-D plants. Similar level of BoCHS transcript was observed in tissues between wild type and the Pr-D mutant. In contrast, the transcripts of BoF3’H, BoDFR, and BoLDOX, as well as BoMYB2 and BobHLH1 accumulated highly in very young leaves, curds, very young flower buds, and young siliques in the mutant. The expression patterns of these genes were consistent with the tissue-specific anthocyanin accumulation pattern in the Pr-D mutant.
Identification of a MYB Gene That Cosegregates with Pr-D
To test whether BobHLH1 or one of the PAP-like MYB family genes represented Pr, association mapping was carried out to investigate whether any of them cosegregated with the Pr-D locus. To develop markers for association mapping, full genomic fragments of BobHLH1 and four BoMYB genes including promoter sequences were isolated through genomic DNA walking from both wild type and the Pr-D mutant. Comparison of the DNA sequences between wild type and mutant allele revealed that there were no polymorphisms for BobHLH1 and BoMYB1, suggesting that they were unlikely the gene representing Pr. Based on different insertion/deletion or single nucleotide polymorphisms in BoMYB2, BoMYB3, and BoMYB4, PCR-based markers were developed (Supplemental Table S1). These three MYB genes were found to cosegregate with the Pr-D locus in a mapping population of 102 F2 plants.
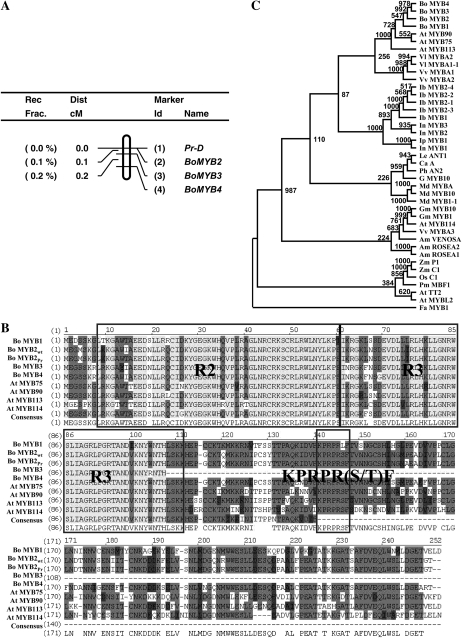
To define the gene that represented Pr, the three BoMYB genes were mapped in a large mapping population of 1,898 F2 individuals. Two and seven recombinant events were detected for BoMYB3 and BoMYB4 marker, respectively. In contrast, no recombinant event was observed for BoMYB2 marker, indicating that BoMYB2 cosegregated with the Pr-D locus and most likely the Pr gene. A high-resolution genetic map for this region is shown in Figure 4A.
Figure 4.
High-resolution genetic map of the Pr region, MYB sequence alignment, and phylogenetic tree of BoMYB homologs. A, Linkage map of BoMYB2, BoMYB3, BoMYB4, and the Pr-D locus in a mapping population of 1,898 F2 individuals. B, Sequence alignment of cauliflower and Arabidopsis R2R3 MYB proteins. R2 and R3 repeat domains as well as the conserved KPRPR[S/T]F motif are indicated. C, Phylogenetic tree of BoMYBs and R2R3 MYBs from other plant species. Numbers along branches indicate bootstrap support determined from 1,000 trials. The length of the branch lines indicates the extent of divergence. The GenBank accession numbers of these proteins are provided in Supplemental Table S2.
Structural Analysis of Pr
The Pr gene from both wild-type and mutant plants contained an open reading frame of 744 bp that encoded a protein of 247 amino acids with an estimated molecular mass of 27.99 kD. Pr encoded BoMYB2 that was predicted to be a nuclear protein (WoLF PSORT, wolfpsort.org) and contained R2 and R3 MYB repeat domains with the signature motif of KPRPR[S/T]F, specific for MYB proteins that activate anthocyanin biosynthesis (Stracke et al., 2001; Fig. 4B). Noticeably, the KPRPR[S/T]F motif overlaps with a TAS4-siR81 target site identified in Arabidopsis PAP1 and PAP2 (Rajagopalan et al., 2006), suggesting a possible addition level of BoMYB2 regulation. BoMYB2 shared 79.8% to 89.9% amino acid sequence identity to the other BoMYB proteins, and 69.1% to 86.3% to the Arabidopsis PAP-like proteins (Fig. 4B). Alignment of the coding sequences of wild type and the mutant gene showed that there were only two single nucleotide differences, which resulted in two amino acid changes from Ile to Thr and Pro to Ala at position 14 and 159, respectively (Fig. 4B). Phylogenetic analysis indicates that BoMYBs are most closely related to Arabidopsis PAP-like proteins and clustered with R2R3 MYB transcription factors involved in regulating anthocyanin biosynthesis from other plant species (Fig. 4C).
Functional Complementation of the Purple Mutant Phenotype in Arabidopsis and Cauliflower
To confirm the function of Pr, genomic fragments of the Pr gene from both wild-type and mutant plants were introduced into wild-type Arabidopsis and cauliflower. Over 40 independent transgenic Arabidopsis lines were generated for each construct. Like the vector-only control (Fig. 5A, 1–6), the transgenic Arabidopsis expressing the wild-type pr allele under the control of endogenous promoter (prpro:pr) exhibited no or very low levels of pigmentation (Fig. 5C, 1–6). In contrast, the Pr-D (Prpro:Pr-D) transformants showed tissue-specific anthocyanin accumulation (Fig. 5B, 1–6). Purple pigments accumulated mainly in young tissues of Arabidopsis, such as young leaves and young flower buds, and in seeds, but not in old leaves, flower petals, or mature siliques. A similar pattern of anthocyanin accumulation was also observed in transgenic cauliflower plants expressing the Pr-D allele (Fig. 5G, 1–6). Ectopic expression of the Pr-D allele under the control of endogenous promoter in both transgenic Arabidopsis and cauliflower induced tissue-specific anthocyanin accumulation, which showed the same anthocyanin accumulation pattern as in the Pr-D mutant. Thus, these results further confirm the isolation of the Pr gene.
Figure 5.
Complementation of the purple phenotype in wild-type Arabidopsis and cauliflower. Top images: Arabidopsis transgenic lines containing empty vector (VC), the Pr-D allele with endogenous promoter (Prpro:Pr-D), the wild-type pr allele with endogenous promoter (prpro:pr), the Pr-D allele with CaMV 35S promoter (35Spro:Pr-D), and the wild-type pr allele with CaMV 35S promoter (35Spro:pr). The sections from top represent 7-d-old seedlings, 3-week-old plants, young flower buds, flowers, older siliques, and seeds. Scale bars for seed images: 0.1 mm. Bottom images: Cauliflower transgenic lines containing the same five constructs as top images. The different sections from top represent T0 young plants, young curds, flowers, part of more mature curds, siliques, and seeds.
Both wild-type and mutant Pr genes under the control of cauliflower mosaic virus (CaMV) 35S (35Spro:pr and 35Spro:Pr-D) were also introduced into wild-type Arabidopsis and cauliflower. Overexpression of either wild-type or mutant gene resulted in the production of dark-purple transgenic plants with high levels of anthocyanin accumulation in Arabidopsis (Fig. 5, D and E, 1–6). Pigments accumulated in the entire transgenic plants that included leaves, roots, flower buds, flower petals, siliques, and seeds. Similarly, when these gene constructs were introduced into wild-type cauliflower plants, anthocyanins appeared in the entire transgenic plants (Fig. 5I, 1–6 and J, 1 and 2). Interestingly, the young top curd tissues of some overexpression lines were white (Fig. 5, I and J, 2) with the below curd tissues purple (Fig. 5I, 4). These results suggest that both wild-type and mutant genes encode functional proteins.
Pr Specifically Regulates a bHLH Transcription Factor and a Subset of Genes Involved in Anthocyanin Production
A number of late anthocyanin pathway genes and a transcription factor BobHLH1 along with Pr were expressed highly in the Pr-D mutant (Fig. 3). To investigate whether expression of Pr in transgenic cauliflower caused such specific activation of gene expression, transcript levels of anthocyanin biosynthetic genes and a number of regulatory genes in various transgenic lines were analyzed. The late pathway genes, BoF3’H, BoDFR, BoLDOX, and the transcription factor BobHLH1 were expressed highly in the Prpro:Pr-D transformants as well as in 35Spro:Pr-D and 35Spro:pr transgenic lines in comparison with vector-only control and wild-type prpro:pr lines (Fig. 6). The early pathway genes showed no dramatic up-regulation. These results clearly demonstrate that Pr specifically activates BobHLH1 and the late pathway genes in controlling anthocyanin biosynthesis in cauliflower.
Figure 6.
Expression of anthocyanin structural and regulatory genes in transgenic cauliflower. qRT-PCR analysis of transcript levels of selected genes in young leaves of cauliflower transformants containing different Pr gene constructs. The transcript levels of genes in vector control lines were set to 1. Results reported represent mean values ± sd from three biological replicate lines with three technical replicates for each.
Allelic Variation at Upstream Regulatory Region of the Pr-D Allele Enhances Promoter Activity
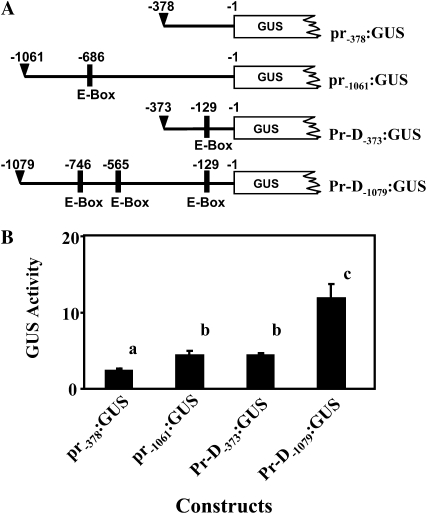
Pr was expressed highly in the mutant and shared 99.2% nucleotide sequence identity in the coding region with the wild-type gene. Thus, we hypothesized that the mutation in the promoter region controlled Pr expression in regulating anthocyanin accumulation. Analysis of the promoter sequences between the wild-type and Pr-D alleles revealed that the first −370-bp sequences were nearly identical except five nucleotide differences and one extra TATA box at the wild-type promoter (Supplemental Fig. S1). A Harbinger DNA transposon insertion (giri: www.girinst.org) was found at −373 bp of the Pr-D mutant allele. To test whether the sequence rearrangement due to the mutation altered promoter activity, comparable lengths of promoter sequences from wild-type and Pr-D allele were fused to GUS gene (Fig. 7A) and transformed into Arabidopsis. Interestingly, although the promoter sequences between the −378/−373 regions of Pr-D and pr were nearly identical, the mutant Pr-D−373:GUS transformants exhibited significantly higher GUS activity than the wild-type pr−378:GUS transformants (P < 0.01). Similarly, transformants expressing Pr-D−1079:GUS that included part of the transposon sequence also yielded significantly higher GUS activity than transgenic lines containing wild-type pr−1061:GUS construct (Fig. 7B). These results indicate that the allelic alteration at the upstream regulatory region of the mutant allele enhances the promoter activity of the Pr gene in activating its expression.
Figure 7.
Analysis of promoter activity of the Pr gene. A, GUS constructs containing different lengths of promoter sequences. The promoter regions were chosen based on transposon insertion site at −373 bp and the transposon transposase site at −1,079 bp. B, GUS activity of 4-week-old stable Arabidopsis transgenic lines containing different GUS constructs. GUS activity was measured from at least four independent transgenic lines with three repeats and expressed as μmol 4-methylumbelliferone min−1 mg−1 protein. Error bars indicate sd (n > 4 plants). The letters above the bars indicate significant differences (P < 0.01).
DISCUSSION
The Pr gene of cauliflower confers anthocyanin production in otherwise anthocyanin nonaccumulating tissues, such as curds and seed endosperms, turning them purple. Here we report the successful isolation of the Pr gene via a combination of candidate gene analysis and fine mapping. The Pr gene was found to encode a R2R3 MYB protein. R2R3 MYB transcription factors belong to a large protein family (Stracke et al., 2001; Allan et al., 2008). In Arabidopsis, MYB transcription factor subgroup that activates anthocyanin biosynthesis includes four genes, PAP1, MYB113, MYB114, and PAP2, with the last three localized in tandem on chromosome one, an arrangement also seen in grape (Walker et al., 2007). A total of four different MYB-like genes, BoMYB1, BoMYB2, BoMYB3, and BoMYB4, were isolated from the cauliflower genome. Three of them, i.e. BoMYB2, BoMYB3, and BoMYB4, appear to be located next to each other. High-resolution mapping and functional complementation in both cauliflower and Arabidopsis clearly confirm the identification of BoMYB2 as the Pr gene. BoMYB2 appears to be an important MYB transcription factor in regulating anthocyanin biosynthesis in Brassica genomes. A previous study of red cabbage varieties also reveals that the constitutive anthocyanin production is associated with an increased expression of only BoMYB2 among the four BoMYBs (Yuan et al., 2009). BoMYB2 from cauliflower shares 99.6% nucleotide sequence identity with that from cabbage.
Pr and BobHLH1 as well as a number of late pathway genes including BoF3’H, BoDFR, and BoLDOX, were expressed highly in the Pr-D mutant. Introduction of the Pr-D allele into wild-type cauliflower specifically increased the transcript levels of BobHLH1 and the same set late structural genes in the transformants. The up-regulation of BobHLH1 in both the Pr-D mutant and the Pr-D transformants strongly suggests that Pr regulates the expression of BobHLH1. Although expression of MdMYB10 does not elevate a bHLH transcription factor expression in apple (Malus domestica; Espley et al., 2007), PAP1 has been shown to regulate the expression of TT8 in Arabidopsis (Baudry et al., 2006). Similarly, AN2 and AN4, the genes encoding MYB transcription factors, activate the bHLH transcription factor of AN1 in petunia (Spelt et al., 2000). Pr and BobHLH1 likely work together to coordinately regulate several transcripts of anthocyanin late pathway genes in conferring anthocyanin accumulation in the Pr-D mutant and transformants.
Despite the fact that MYB and bHLH transcription factors share similar functions among plants, they exhibit species-specific differences in activating part or the entire set of anthocyanin pathway genes. For example, in maize the C1/P1 family of MYB proteins and the R/B family of bHLH proteins activate the entire set of anthocyanin structural genes (Chandler et al., 1989; Ludwig and Wessler 1990; Cone et al., 1993; Grotewold et al., 1994). In petunia and Arabidopsis, these two families of proteins control a subset of structural genes (Quattrocchio et al., 1993; Spelt et al., 2000; Gonzalez et al., 2008). Like the later cases, BoMYB2 and BobHLH1 regulated a subset of anthocyanin structural genes in inducing tissue-specific expression in the Pr-D mutant. However, how Pr exhibits its unique tissue-specific regulation of anthocyanin accumulation remains to be determined.
The activity of MYB-like genes has been suggested to be the primary cause of natural variation in anthocyanin pigmentation in plants (Quattrocchio et al., 1999; Schwinn et al., 2006). Increased expression of R2R3 MYB transcription factors was found to be responsible for anthocyanin production in a number of anthocyanin-accumulating mutants. For example, the constitutive up-regulation of PAP1, ANT1, and MdMYB10 causes anthocyanin accumulation throughout the plant in pap1-D Arabidopsis (Borevitz et al., 2000), antl tomato (Solanum lycopersicum; Mathews et al., 2003), and red-fleshed apple (Espley et al., 2007). While overexpression of PAP1 and ANT1 MYB transcription factors is due to activation-tagged insertions in their promoter sequences (Borevitz et al., 2000; Mathews et al., 2003), the high transcript level of MdMYB10 was recently found to be due to the formation of a minisatellite-like structure comprising multiple repeats of a promoter segment that generates a novel autoregulatory motif (Espley et al., 2009). Unlike these reported anthocyanin-accumulating mutants, the Pr-D mutant appears to display a different mechanism in activating Pr expression.
The coding regions of Pr-D and pr shared 99.2% sequence identity and both of them encoded functional proteins. Thus, the sequence variation in the promoter region of Pr is likely responsible for the activation of Pr in controlling anthocyanin biosynthesis. Indeed, the promoter from wild type and the Pr-D alleles showed different capacity in activating GUS expression. For example, although the promoter of the −378/−373 regions of Pr-D and pr shared nearly identical sequences, significantly higher GUS activity was observed in the transformants expressing the mutant Pr-D−373:GUS than the wild-type pr−373:GUS. Detailed examination of the Pr-D−373/pr−373 sequences revealed that an insertion of two nucleotides in Pr-D−373 range generated a new regulatory motif, the E-box (5′-CANNTG-3′), which is a cis-acting element with binding consensus site for bHLH proteins (Toledo-Ortiz et al., 2003; Supplemental Fig. S1). Further, both Pr-D−373 and wild-type pr−1061 sequences contained one E-box and the transformants expressing these constructs exhibited similar promoter activity. The Pr-D−1079 sequence harbored two additional E-boxes and gave significantly higher promoter activity than wild-type pr−1061 sequence (Fig. 7). The transposon insertion in the Pr-D allele introduced additional E-box cis-acting elements, which likely provide more binding sites for bHLH transcription factors to activate Pr expression, as suggested in a recent study of the DNA transposon mPing in rice (Naito et al., 2009). E-boxes are identified in the promoter region of anthocyanin structural genes and have been shown to be required for bHLH transcription factor binding in enhancing gene expression (Shirley et al., 1992; Hartmann et al., 2005). Future experiments will help clarify the involvement of E-box amplification in activating Pr expression. Nevertheless, our results reported here clearly indicate that the mutation at the upstream promoter region in the Pr-D allele resulted in activating Pr gene transcription to confer the phenotypic change in cauliflower. Pr exerts profound effect on anthocyanin production in plants. The successful cloning of Pr and the discovery of it as primary determinant of color in cauliflower have implications for breeding vegetable crops with enhanced health-promoting properties and visual appeal.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Purple cauliflower (Brassica oleracea var botrytis) arose from a spontaneous mutation found in a cauliflower field about 20 years ago. A commercial purple cauliflower cultivar Graffiti (Harris Seeds) and a white cultivar Stovepipe as wild-type control were used in this study. Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for genetic transformation. Cauliflower plants were grown either in a greenhouse under 14-h-light/10-h-dark photoperiod at 23°C or in a field. Arabidopsis plants were grown in a growth room under 14-h-light/10-h-dark photoperiod at 23°C.
HPLC-ESI-MS/MS Analysis of Anthocyanins
Anthocyanins in Graffiti was extracted and analyzed following the method as described by Wu and Prior (2005b). Freeze-dried curd sample (0.5 g) was ground into powder and extracted twice in total 25 mL methanol/water/acetic acid (85:15:0.5). The extract was diluted 2-fold. Aliquot (10 μL) was injected into a Zorbax stablebond analytical SB-C18 column (4.6 × 250 mm, 5 μm, Agilent Technologies) and separated using 5% formic acid (A) and 100% methanol (B) as mobile phases on an HP 1100 series HPLC. Low-resolution electrospray mass spectrometry as described (Wu and Prior, 2005b) was performed to identify the major anthocyanin peaks in the purple curd sample. Quantification was carried out based on peak areas and a calibration curve generated with a commercial standard of cyanidin 3,5-diglucoside chloride (INDOFINE Chemical Company).
Nucleic Acid Analysis
Genomic DNA was isolated as described previously (Lu et al., 2006). Total RNA from purple and white cauliflower tissues was extracted using Trizol reagent following the manufacturer’s instruction (Invitrogen). mRNA was isolated from total RNA using PolyATtract mRNA isolation system IV (Promega).
For northern-blot analysis, mRNA (2 μg) samples were used. Prehybridization and hybridization with P32-labeled probes were performed as described previously (Li et al., 2001). Probes used included CHS (U21762), CHI (U20894), F3H (U14735), and LDOX (YAY780) from Arabidopsis, as well as F3’H, DFR, UGT, and Actin amplified from white cauliflower DNA using primers as listed in Supplemental Table S1.
qRT-PCR analysis was conducted according to the requirements and guidelines described by Udvardi et al. (2008). The cDNAs were synthesized from DNase I treated total RNA (5 μg) using Superscript III reverse transcriptase (Invitrogen). qRT-PCR was carried out using SYBR Green PCR master mix following the manufacturer’s instruction (Applied Biosystems) in an Applied Biosystems 7900HT fast real-time PCR system. Gene-specific primers, which were confirmed to produce specific gene products by sequencing (Yuan et al., 2009), are listed in Supplemental Table S1. The relative transcript levels were calculated as described previously (Lyi et al., 2007).
PCR walking on genomic DNA was performed using the Universal GenomeWalker kit following the manufacturer’s instruction as described previously (Li and Garvin, 2003). Wild-type and mutant cauliflower genomic DNA (2.5 μg) were digested with HincII, RsaI, SmaI, StuI, SwaI, DraI, EcoRV, PvuII, ScaI, and SspI, respectively, and ligated to the GenomeWalker adaptors to produce Genome walking libraries. PCR products of secondary nested reactions were cloned into pCR2.1 vector (Invitrogen) and sequenced.
Association Mapping of the Candidate Genes
DNA of the parents and 1,898 F2 individuals of the mapping population were extracted (Lu et al., 2006). To design PCR-based markers, the candidate genes from both mutant and wild-type plants were isolated. Based on insertions and deletions or single nucleotide polymorphisms in the two alleles of these genes, primer sets for BoMYB2, BoMYB3, and BoMYB4 markers were developed (Supplemental Table S1). BoMYB2m and BoMYB4m were codominant markers and BoMYB3m was dominant marker. Genetic linkage map was generated using MapMaker (www.broadinstitute.org/ftp/distribution/software/ mapmaker3).
Sequence Analysis
DNA and protein sequences were analyzed using the program of DNAStar (Lasergene). Multiple sequence alignments were produced by Vector NTI (Invitrogen) or CLC sequence viewer using default setting. The phylogenic tree was calculated by DNAStar and visualized by Treeview version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/ treeview.html).
Plasmid Construction and Plant Transformation
To create the constructs for phenotypic complementation, genomic DNA of Pr including 1,755 or 1,061 bp of promoter sequences for the mutant and wild-type allele, respectively, were amplified. Despite of extensive efforts, the upstream above 1,061 bp promoter sequence for wild-type allele could not be obtained due to highly repeated sequence and was not found from public sequence databases. The amplified products were cloned into pCAMBIA 1300 vector (CAMBIA) with a nopaline synthase terminator to produce prpro:pr and Prpro:Pr-D constructs. To generate the overexpression constructs, comparable genomic DNA from start to stop codon of both Pr alleles were amplified and inserted into pCAMBIA 1300S containing CaMV 35S promoter (Zhou et al., 2009) to produce 35Spro:pr and 35Spro:Pr-D. To create the GUS constructs, region of wild-type pr promoter (−378 and −1061 bp), and Pr-D promoter (−373 and −1,079) were amplified and inserted into pSG506 vector (kindly provided by Dr. Susheng Gan, Cornell University). The promoter fragments were selected based on transposable element insertion site. The fragments were then subcloned into pCAMBIA 1300 to produce various promoter-GUS constructs. All constructs were verified by sequencing. The constructs and vector-only controls were electroporated into Agrobacterium tumefaciens strain GV3101, and transformed into wild-type Arabidopsis using a flower-dipping method (Clough and Bent, 1998), and into cauliflower using A. tumefaciens-mediated tissue culture method (Lu et al., 2006).
GUS Activity Analysis
Quantitative analysis of GUS activity in transformants expressing different GUS constructs was carried out using fluorometric assay (Blazquez, 2007) with slight modification. Four-week-old Arabidopsis leaf tissues (25 mg) from at least four independent transgenic lines for each construct were extracted independently in 100 μL GUS extraction buffer. Total proteins in the crude extract were quantified using the RC DC protein assay kit (Bio-Rad). Extract (5 μL) was then added to 450 μL GUS extraction buffer containing 1 mm 4-methylumbelliferyl β-d-glucuronide and incubated at 37°C. Aliquot of the reaction mixture (20 μL) was added into 180 μL stop solution (1 m sodium carbonate) every 10 min. Fluorescence was then measured using fluorometer Fluorolite 1000 (DYNEX technologies) at excitation wavelength of 365 nm and emission wavelength of 450 nm. The analysis was repeated three times. The mean values for each construct were compared.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU219986 (BoMYBPr-D), GU219987 (BoMYB2pr), GU219985 (BoMYB1), GU219988 (BoMYB3), GU219989 (BoMYB4), and GU219990 (BobHLH1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence comparison of the promoter region at −378/−373 bp of the wild type and mutant Pr gene.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. List of the GenBank accession numbers of the proteins used in the phylogenic tree of Figure 4C.
Supplementary Material
Acknowledgments
We are especially grateful to Leon Kochian and Jim Giovannoni for valuable comments. We thank the Li lab members for helpful discussion and technical advice. We also thank Susheng Gan for providing the pSG506 vector, Don Reed for providing field space and assistance in growing cauliflower, and Chrissa McFarlane for her excellent technical assistance.
References
- Allan AC, Hellens RP, Laing WA. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13: 99–102 [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Blazquez M. (2007) Quantitative GUS activity assay of plant extracts. Cold Spring Harb Protoc 2007: doi/10.1101/pdb.prot4690 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Burr FA, Burr B, Scheffler BE, Blewitt M, Wienand U, Matz EC. (1996) The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D. (1989) Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B. (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R. (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11: 1422–1434 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165: 9–28 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Mazza G. (1994) Quantitation and distribution of simple and acylated anthocyanins and other phenolics in blueberries. J Food Sci 59: 1057–1059 [Google Scholar]
- Goff SA, Cone KC, Chandler VL. (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6: 864–875 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97: 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Li L, Garvin DF. (2003) Molecular mapping of Or, a gene inducing beta-carotene accumulation in cauliflower (Brassica oleracea L. var. botrytis). Genome 46: 588–594 [DOI] [PubMed] [Google Scholar]
- Li L, Paolillo DJ, Parthasarathy MV, Dimuzio EM, Garvin DF. (2001) A novel gene mutation that confers abnormal patterns of beta-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J 26: 59–67 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Wessler SR. (1990) Maize R gene family: tissue-specific helix-loop-helix proteins. Cell 62: 849–851 [DOI] [PubMed] [Google Scholar]
- Lyi SM, Zhou X, Kochian LV, Li L. (2007) Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 68: 1112–1119 [DOI] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al. (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Mazza G, Miniati E. (1993) Anthocyanins in Fruits, Vegetables, and Grains. CRC Press, Boca Raton, FL [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S. (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49: 641–654 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6: 3553–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. (2000) anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP. (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wu X, Prior RL. (2005a) Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: fruits and berries. J Agric Food Chem 53: 2589–2599 [DOI] [PubMed] [Google Scholar]
- Wu X, Prior RL. (2005b) Identification and characterization of anthocyanins by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts, and grains. J Agric Food Chem 53: 3101–3113 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chiu LW, Li L. (2009) Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhou X, Yuan Y, Yang Y, Rutzke M, Thannhauser TW, Kochian LV, Li L. (2009) Involvement of a broccoli COQ5 methyltransferase in the production of volatile selenium compounds. Plant Physiol 151: 528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.