Abstract
ROXY1 and ROXY2 are CC-type floral glutaredoxins with redundant functions in Arabidopsis (Arabidopsis thaliana) anther development. We show here that plants lacking the basic leucine-zipper transcription factors TGA9 and TGA10 have defects in male gametogenesis that are strikingly similar to those in roxy1 roxy2 mutants. In tga9 tga10 mutants, adaxial and abaxial anther lobe development is differentially affected, with early steps in anther development blocked in adaxial lobes and later steps affected in abaxial lobes. Distinct from roxy1 roxy2, microspore development in abaxial anther lobes proceeds to a later stage with the production of inviable pollen grains contained within nondehiscent anthers. Histological analysis shows multiple defects in the anther dehiscence program, including abnormal stability and lignification of the middle layer and defects in septum and stomium function. Compatible with these defects, TGA9 and TGA10 are expressed throughout early anther primordia but resolve to the middle and tapetum layers during meiosis of pollen mother cells. Several lines of evidence suggest that ROXY promotion of anther development is mediated in part by TGA9 and TGA10. First, TGA9 and TGA10 expression overlaps with ROXY1/2 during anther development. Second, TGA9/10 and ROXY1/2 operate downstream of SPOROCYTELESS/NOZZLE, where they positively regulate a common set of genes that contribute to tapetal development. Third, TGA9 and TGA10 directly interact with ROXY proteins in yeast and in plant cell nuclei. These findings suggest that activation of TGA9/10 transcription factors by ROXY-mediated modification of cysteine residues promotes anther development, thus broadening our understanding of how redox-regulated TGA factors function in plants.
TGACG (TGA) motif-binding proteins form a distinct subclade in the basic leucine-zipper (bZIP) superfamily of Arabidopsis (Arabidopsis thaliana) transcription factors (Jakoby et al., 2002). This subclade contains 10 members: TGA1 to TGA7, PERIANTHIA (PAN), and two uncharacterized members, bZIP21/TGA9 (At1g08320) and bZIP65/TGA10 (At5g06839; Jakoby et al., 2002). Past studies have identified overlapping roles for TGA transcription factors in plant disease resistance and stress responses (Després et al., 2003; Zhang et al., 2003; Kesarwani et al., 2007; Fode et al., 2008; Mueller et al., 2008), indicating that these factors operate with a high degree of functional redundancy. To date, developmental roles for TGA factors are known only for PAN, a key regulator of floral patterning. Flowers lacking PAN typically contain an extra sepal and petal and one fewer stamen, resulting in a pentamerous arrangement of floral organs (Running and Meyerowitz, 1996; Chuang et al., 1999).
Recent studies have shed light on how posttranscriptional regulation of TGA transcription factors relays changes in intracellular redox status. A pair of Cys residues in the C terminus of TGA1 or TGA4 form an intermolecular disulfide bridge that is reduced upon exposure to salicylic acid (SA; Després et al., 2003). This modification stimulates DNA binding and permits interaction with the BTB-ankryin protein NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), a transcriptional coactivator (Després et al., 2003; Rochon et al., 2006). PAN similarly partners with BTB-ankryin proteins, BLADE-ON-PETIOLE1 (BOP1) and BOP2, to restrict the number of sepal-whorl organs in flowers (Hepworth et al., 2005). One of two regulatory Cys residues in TGA1 is conserved in PAN and required for activity (Li et al., 2009). Modification of this residue is proposed to depend on the activity of ROXY1, a CC-type glutaredoxin that interacts with PAN in plant cell nuclei (Li et al., 2009). roxy1 mutants initiate fewer petals than the wild type and later petal morphogenesis can be aberrant, indicating that ROXY1 is a regulator of PAN (Xing et al., 2005; Li et al., 2009). ROXY1, together with its closest homolog ROXY2, is also required for anther cell specification and microspore development (Xing and Zachgo, 2008), but TGA partners in this process have yet to be identified.
The Arabidopsis anther is a bilaterally symmetrical four-lobed structure in which pollen develops. The mitotic division of archesporial cells specified in the interior of each lobe generates pollen mother cells surrounded by three morphologically distinct cell layers: the tapetum, middle layer, and endothecium, underlying the epidermis (Sanders et al., 1999). Pollen mother cells divide meiotically to generate tetrads of haploid microspores that separate and mature into pollen grains. The tapetum contributes to microspore development by secreting enzymes necessary for microspore release and by providing nutrients and structural components essential for the production of viable pollen (Scott et al., 2004). Pollen grains are released from the anthers by dehiscence, a process whose terminal step is breakage of the stomium connecting the abaxial and adaxial anther lobes (Sanders et al., 1999; Ma, 2005). Interestingly, abaxial and adaxial anther lobe development is differentially dependent on ROXY activity. Loss of ROXY1/2 blocks sporogenous cell formation only in adaxial anther lobes. In abaxial lobes, anther cell layers are specified but development of pollen mother cells and the tapetum is abnormal, leading to microspore degradation (Xing and Zachgo, 2008).
We show here that mutations in TGA9 and TGA10 lead to male sterility and differential defects in abaxial and adaxial anther lobe development similar to roxy1 roxy2 mutants. Additional defects not seen in roxy1 roxy2 mutants include abnormal stability of the middle layer and lack of septum and stomium degeneration, resulting in nondehiscent anthers. We show that TGA9 and TGA10 are expressed in an overlapping fashion with ROXY1 and ROXY2 and regulate an overlapping set of genes with tapetum functions. In addition, ROXY proteins interact directly with TGA9 and TGA10 in the nuclei of yeast and plant cells, suggesting that activation of TGA9 and TGA10 transcription factors by the modification of redox-sensitive Cys residues is needed for normal anther development. These data reveal a new role for TGA factors in anther patterning and dehiscence, thus broadening our understanding of how redox-regulated TGA factors contribute to plant development.
RESULTS
Mutant Isolation and Characterization
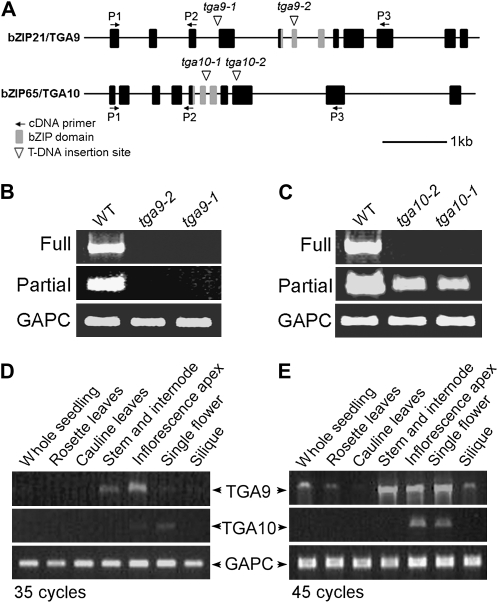
TGA9 and TGA10 form a distinct subclade in the phylogenetic tree of Arabidopsis TGA bZIP family members (Supplemental Fig. S1), but their functions have yet to be elucidated. Homologs of TGA9 and TGA10 in other plant species have developmental roles. In tobacco (Nicotiana tabacum), overexpression of NtTGA10 causes derepression of lateral branching and defects in plant defense signaling (Schiermeyer et al., 2003). In maize (Zea mays), mutation of the TGA9 homolog LIGULELESS2 causes defects in the formation of the blade-sheath boundary in leaves and delayed flowering (Walsh et al., 1998; Walsh and Freeling, 1999). To examine the functions of Arabidopsis TGA9 and TGA10, we obtained insertion lines from the Salk T-DNA collection (Fig. 1A). Lines designated tga9-1 and tga9-2 contain T-DNA inserts at nucleotides 2,023 and 3,325 of the coding sequence, respectively. No transcripts were produced in these lines (Fig. 1B). Lines designated tga10-1 and tga10-2 contain T-DNA inserts at nucleotides 1,826 and 2,284 of the coding sequence, respectively. Partial transcripts were produced in both lines, but the insertion in tga10-1 disrupts the bZIP domain (Fig. 1, A and C) so as to encode a nonfunctional protein. No single mutants showed an obvious phenotype. Analysis of TGA9 and TGA10 transcript by reverse transcription (RT)-PCR revealed an overlapping expression pattern in flowers, suggesting that their activities may overlap in floral development (Fig. 1, D and E). To examine this possibility, we constructed tga9 tga10 double mutants.
Figure 1.
Characterization of tga9 and tga10 mutant alleles, and tissue expression of TGA9 and TGA10. A, Scale diagrams of TGA9 and TGA10 genomic sequences showing the positions of features as indicated in the key at bottom left. Black boxes indicate exons, and horizontal arrows represent annealing positions of primers used for transcript analysis. B, RT-PCR analysis of TGA9 transcripts in the wild type (WT) and tga9 mutants (40 cycles). C, RT-PCR analysis of TGA10 transcripts in the wild type and tga10 mutants (40 cycles). Full, Product obtained using P1/P3 primer combination; Partial, product obtained using P1/P2 primer combination. D and E, RT-PCR analysis of TGA9 and TGA10 transcripts in wild-type plant tissues (35 and 45 cycles). In B to E, GAPC control transcript used 23 cycles.
Male-Sterile Phenotype in tga9 tga10 Double Mutants
Strikingly, four of 72 F2 plants derived from a cross between tga9-1 and tga10-1 failed to set seed. Genotyping confirmed that sterility was restricted to double mutants (Fig. 2). Examination of tga9 tga10 flowers showed no changes in floral patterning, but anther morphology was abnormal (Fig. 3). Stage 12 anthers in the tga9 tga10 mutant were only partially filled with pollen and failed to dehisce at stage 13 (Fig. 3). Hand pollination of tga9 tga10 carpels with wild-type pollen yielded a similar number of viable seeds (1.29% aborted seeds; n = 465) as a wild-type control cross (1.30% aborted seeds; n = 461), confirming that female fertility was normal. Unfortunately, expression of TGA9 and TGA10 coding sequences under the control of either a cauliflower mosaic virus 35S or native promoter failed to complement the tga9 tga10 mutant phenotype. However, an identical male-sterility phenotype was seen in tga9-1 tga10-2 double mutants (data not shown), providing further evidence that the anther defect in tga9 tga10 mutants is indeed due to a loss of TGA9 and TGA10 function.
Figure 2.
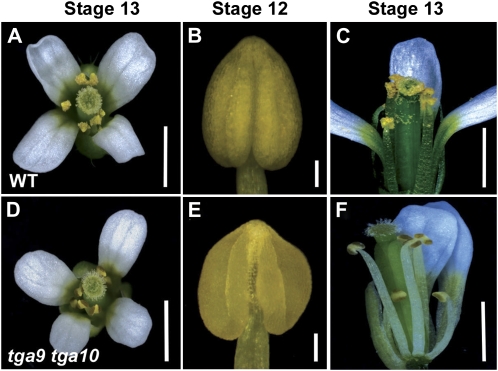
Sterility phenotype in tga9 tga10 double mutants. A and B, Comparison of wild-type (WT) and tga9 tga10 inflorescences. Arrows indicate empty siliques. C, Wild-type and tga9 tga10 siliques. D and E, Wild-type and tga9 tga10 dissected siliques. Bars = 1 mm. [See online article for color version of this figure.]
Figure 3.
Flower and anther morphology in the wild type and tga9 tga10 mutants. Floral stages are as indicated. A, Wild-type (WT) flower. B, Wild-type anther, adaxial view. C, Wild-type dissected flower. D, tga9 tga10 flower. E, tga9 tga10 anther, adaxial view. F, tga9 tga10 dissected flower; anthers are nondehiscent. Bars = 1 mm, except 100 μm in B and E. [See online article for color version of this figure.]
To further examine the male-sterility phenotype in tga9 tga10 mutants, Alexander’s stain was used to assess pollen viability (Alexander, 1969). The anthers of late stage 12 flowers in the mutant contained pollen that were abnormally aggregated but most stained purple-red, suggesting that they were potentially viable (Supplemental Fig. S2, A, B, D, and E). However, pollen grains that were excised from indehiscent anthers failed to germinate on agar medium, revealing them to be unviable (Supplemental Fig. S2, C and F; 0% germination for tga9 tga10 pollen versus 70.5% germination for wild-type pollen [n = 200]). Scanning electron micrographs of mutant pollen showed that grains were small and irregular, lacking the characteristic oblong shape of wild-type pollen grains. The exine structure was also abnormal, with highly compressed columella, indicating defects in pollen wall maturation (Supplemental Fig. S2, G–J).
Differential Effect of tga9 tga10 Mutation on Abaxial and Adaxial Anther Development
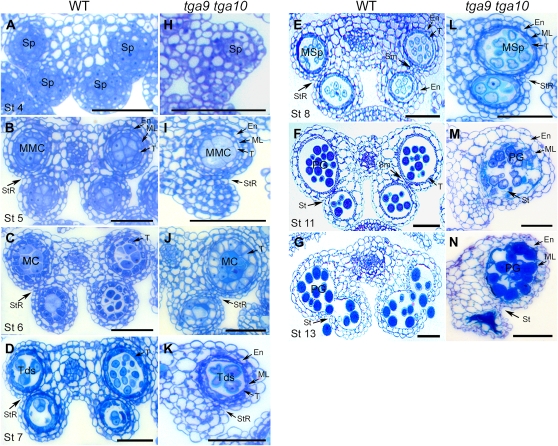
Anther cross-sections were used to examine the detailed morphology of wild-type and tga9 tga10 anthers during development (Fig. 4) Morphological abnormalities in the mutant were apparent from stage 4 onward. In wild-type anthers, primary parietal cells and sporogenous cells were specified in all four anther lobes and underwent cell division to form microspore mother cells surrounded by four morphologically distinct cell layers: the tapetum, middle layer, endothecium, and epidermis (Fig. 4, A and B; Sanders et al., 1999). In the double mutant, sporogenous cells formed reliably only in abaxial anther lobes (Fig. 4H). In adaxial lobes with sporogenous cells, cell divisions were often variable or disorganized, resulting in developmental arrest. Cell division in abaxial lobes followed a pattern similar to the wild type, resulting in microspore mother cells surrounded by tapetum, middle, and endothecium cell layers at stage 5 (Fig. 4, B and I). During stages 6 and 7, pollen mother cells in abaxial lobes underwent meiosis to form tetrads encapsulated by callose (Fig. 4, C, D, J, and K; Supplemental Fig. S3). However, cells in the tapetum layer of the mutant at late stage 6 were expanded, with large vacuoles compared with the wild type (Fig. 4, C and J), suggesting abnormalities in tapetum function.
Figure 4.
Comparison of wild-type and tga9 tga10 anther development. Cross-sections of wild-type (WT; A–G) and tga9 tga10 (H–N) anthers were stained with toluidine blue. A, Stage 4. Sporogenous cells in all four lobes. B, Stage 5. Characteristic layered structure of anther lobes is resolved. C, Stage 6. Meiosis of microspore mother cells. D, Stage 7. Tetrads form, and the middle layer is crushed. E, Stage 8. Developing microspores; tapetum degeneration is under way. F, Stage 11. Developing microspores; septum degeneration begins and endothecium expands. G, Stage 13. Anthers are dehisced; ruptured stomium. H, Stage 4. Sporogenous cells are often missing in adaxial lobes. I, Stage 5. Sporogenous cells proliferate in abaxial lobes; adaxial lobes are underdeveloped. J, Stage 6. Microspore mother cells in abaxial lobes undergo meiosis, and tapetal cells are abnormally vacuolated; development is variable in adaxial lobes. K, Stage 7. Tetrads appear in abaxial lobes. L, Stage 8. Microspores are released in abaxial lobes; the middle layer fails to degrade. M, Stage 11. Microspore development is delayed; breakdown of the middle layer is incomplete, but endothecium cells are expanded. N, Stage 13. Anthers are indehiscent; pollen is clumped or degraded. En, Endothecium; MC, meiotic cells; ML, middle layer; MMC, microspore mother cells; MSp, microspores; PG, pollen grains; Sm, septum; Sp, sporogenous cells; St, stomium; StR, stomium region; T, tapetum; Tds, tetrads. Bars = 50 μm. [See online article for color version of this figure.]
In the wild type, following the release of microspores at stage 8, the middle layer is crushed and the tapetum compacts as programmed cell death of these layers takes place (Fig. 4, D and E; Sanders et al., 1999). In the mutant, the middle layer was only partly degraded, and by stages 10 and 11, microspore development was visibly impaired (Fig. 4, L and M; Supplemental Fig. S4). Pollen grains in stage 13 anthers of the mutant were clumped together and irregular in appearance (Fig. 4N). Even in anthers with pollen in adaxial lobes, the septum and stomium were nonfunctional, blocking dehiscence (Fig. 4, K and L; Supplemental Fig. S4). Staining of stage 11 anthers with phloroglucinol indicated that the endothecium in both the wild type and mutant is lignified to the same degree, suggesting that reinforcement of this cell layer, essential for dehiscence, is not disrupted (Mitsuda et al., 2005; Mizuno et al., 2007; Yang et al., 2007b). However, the residual middle layer in mutants was highly lignified, which may contribute to nondehiscence (Supplemental Fig. S5).
Similar to tga9 tga10, mutations in roxy1 roxy2 differentially affect adaxial and abaxial lobe development. ROXY1 and ROXY2 encode two CC-type glutaredoxins that are expressed in flowers and that interact with several TGA bZIP transcription factors in yeast (Li et al., 2009). Primary parietal and sporogenous cells fail to differentiate in roxy1 roxy2 adaxial anther lobes, whereas in abaxial anther lobes, microspore development terminates prematurely. Pollen mother cells enter meiosis but eventually aggregate and degrade, giving rise to empty locules (Xing and Zachgo, 2008). Although the tga9 tga10 block in pollen development is less severe than in roxy1 roxy2, the differential effect of these mutations on anther lobe development suggests that ROXY-dependent promotion of anther development may occur in part via the regulation of TGA9 and TGA10, likely at the posttranscriptional level.
TGA9 and TGA10 Expression Overlaps with ROXY in Developing Anthers
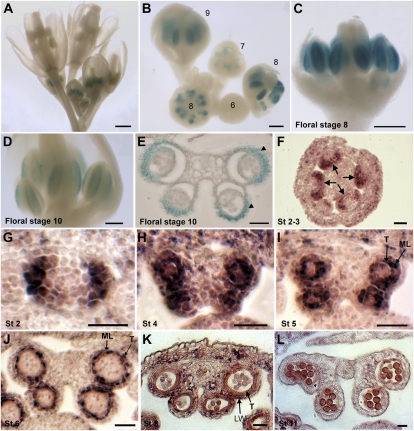
ROXY1 and ROXY2 are expressed in all four anther lobes beginning at stage 3 and in both meiocytes and the tapetum at stage 6 (Li et al., 2009). To examine TGA9 expression in developing anthers, ecotype Columbia-0 (Col-0) plants expressing a GUS reporter gene fused to a 3.9-kb TGA9 promoter sequence were generated. Whole-mount staining of inflorescences showed activation of GUS expression in stage 7 flowers (anther stage 4). Expression peaked in stage 8 flowers (anther stages 5–6) and declined in stage 9 to 11 flowers (anther stages 7–11; Fig. 5, A–E). However, this same promoter driving expression of the TGA9 coding sequence failed to restore fertility in tga9 tga10 mutants, suggesting that additional regulatory motifs present in the introns or 3′ untranslated region are needed for full expression. Therefore, in situ hybridization was used to monitor TGA9 transcript accumulation in anther cross-sections. This analysis showed activation of TGA9 in stage 2 anther primordia during archesporial cell specification. During stages 2 to 3 of anther development prior to the emergence of distinct locules, expression was in a horseshoe pattern associated with the lateral and adaxial portion of primordia (Fig. 5, F and G). During stages 4 and 5, expression was throughout sporogenic tissue and surrounding cells layers in adaxial and adaxial locules (Fig. 5, H and I). At stage 6, expression localized to the tapetum and middle layers, gradually fading postmeiosis with degeneration of these cell layers (Fig. 5, J–L). TGA10 was expressed in a similar pattern, albeit at lower levels (Supplemental Fig. S6). These data show that ROXY1/2 and TGA9/10 are coexpressed during stages 3 to 6 of anther development, consistent with functional overlap.
Figure 5.
Expression of TGA9 in wild-type inflorescences and flowers. Expression was monitored using a TGA9::GUS reporter gene (A–E) or by in situ hybridization (F–L). A to E, GUS activity is first detected in stage 7 flowers, peaks at stage 8, and declines during stages 9 to 10. E, Stage 10 flower cross section; expression persists in the locule wall (arrowheads). TGA9 in situ probe hybridized to anther cross-sections. F, Stage 2 to 3 anthers. Expression laterally and along the adaxial face of primordia (arrows) is shown. G, Stage 2. Anther primordia. H, Stage 4. Expression in all cell layers. I and J, Stages 5 to 6. Expression is restricted to the tapetum and middle layer. K and L, Stages 8 to 11. Expression declines. Bars = 100 μm except in A (0.5 mm) and F to L (25 μm). LW, locule wall; ML, middle layer; T, tapetum. [See online article for color version of this figure.]
TGA9 and TGA10 Function Genetically Downstream of SPOROCYTELESS/NOZZLE and Promote Its Expression in Adaxial Anther Lobes
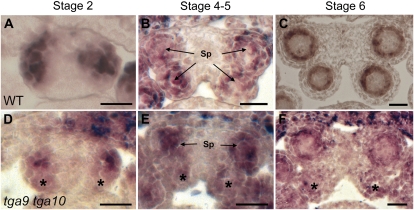
Initiation of sporogenesis requires SPOROCYTELESS/NOZZLE (SPL/NZZ), a MADS box-like transcription factor regulated by the floral homeotic factor AGAMOUS (Ito et al., 2004). SPL/NZZ is expressed in archesporial and sporogenous cells and later in pollen mother cells and the tapetum (Schiefthaler et al., 1999; Yang et al., 1999). In situ hybridization showed an accumulation of SPL/NZZ transcript in the stage 2 anthers of both the wild type and tga9 tga10 mutants, when archesporial cells are specified (Fig. 6, A and D; Sanders et al., 1999). SPL/NZZ transcript was uniform throughout all four anther lobes in the wild type, but in tga9 tga10 mutants, expression was often less intense in one or both adaxial lobes, consistent with variable specification of sporogenous cells in these lobes (Fig. 6, D–F; stages 2, 4–5, and 6). A similar but more pronounced reduction of SPL/NZZ expression in adaxial anther lobes was reported for roxy1 roxy2 mutants, whose activity lies downstream of SPL/NZZ but upstream of DYSFUNCTIONAL TAPETUM1 (DYT1), a key regulator of tapetal differentiation (Zhang et al., 2006; Xing and Zachgo, 2008).
Figure 6.
In situ analysis of SPL/NZZ expression in wild-type (WT) and tga9 tga10 anthers. Anther stages are as indicated. A to C, The wild type. SPL is uniformly expressed in adaxial versus abaxial anther lobes. D to F, The tga9 tga10 mutant. SPL expression is reduced in adaxial anther lobes (asterisks). Sp, Sporogenous cells. Bars = 25 μm. [See online article for color version of this figure.]
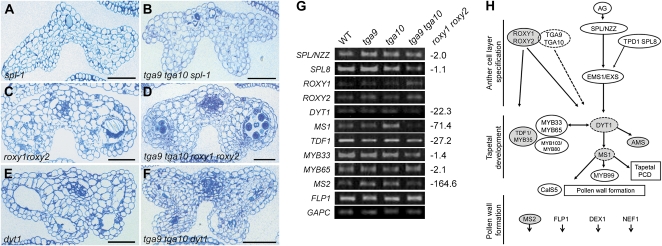
To further examine the position of TGA9 and TGA10 in the genetic hierarchy that controls anther development, we crossed tga9 tga10 mutants with spl/nzz, roxy1 roxy2, and dyt1 mutants and analyzed the resulting anther phenotypes. Anthers in the tga9 tga10 sp1-1 triple mutant were similar to those of the spl-1 parent, which lacks archesporial cells (Fig. 7, A and B; Yang et al., 1999). In tga9 tga10 roxy1 roxy2 mutants, there was a strong block in adaxial lobe development, typical of the roxy1 roxy2 parent (Fig. 7, C and D). Interestingly, a few aberrant pollen grains formed in the abaxial lobes of these quadruple mutants. This partial suppression of the roxy1 roxy2 phenotype by tga9 tga10 could indicate that TGA9/10 are repressors in their inactive form, as shown for TGA2 (Boyle et al., 2009). Alternatively, the roxy1 roxy2 phenotype may be milder in Col-0 versus the original Nossen-0 genetic background. These data indicate that TGA9 and TGA10 operate genetically downstream of SPL/NZZ and ROXY1/2 but are unlikely to be transcriptional targets of ROXY1/2, since their transcript levels are not significantly altered in roxy1 roxy2 inflorescence apices (Supplemental Fig. S7). In tga9 tga10 dyt1 triple mutants, microspores failed to develop in either adaxial or abaxial anther lobes, an earlier block than for either parent (Fig. 7, E and F). Collectively, these data indicate that TGA9 and TGA10 function downstream of SPL/NZZ and ROXY1/2 and upstream or in parallel with DYT1 in the genetic hierarchy that controls anther development.
Figure 7.
Double mutant analysis and RT-PCR analysis of anther transcripts in the wild type and tga9 tga10 mutants. A to F, Cross-sections of stage 11 anthers stained with toluidine blue for the indicated genotypes. Bars = 50 μm. G, RT-PCR analysis of anther transcripts (35 cycles) in wild-type (WT) and mutant inflorescence apices. Numerical values for transcript down-regulation in roxy1 roxy2 mutants (Xing and Zachgo, 2008) are shown. H, Genetic framework for control of anther development (modified from Wilson and Zhang, 2009) showing overlap for tapetal genes down-regulated in roxy1 roxy2 and tga9 tga10 mutants. Shaded ovals, Down-regulated in roxy1 roxy2; dashed perimeter, down-regulated in tga9 tga10; shaded ovals with dashed perimeter; down-regulated in both. Genes not discussed in the text are as follows: TAPETAL DETERMINANT1 (TPD1; Yang et al., 2003), EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS; Canales et al., 2002; Zhao et al., 2002), MYB103/MYB80 (Higginson et al., 2003; Zhang et al., 2007), MYB99 (Ito et al., 2007), ABORTED MICROSPORES (AMS; Sorensen et al., 2003), DEFECTIVE IN EXINE FORMATION1 (DEX1; Paxson-Sowders et al., 2001), NO EXINE FORMATION1 (NEF1; Ariizumi et al., 2004), and CALLOSE SYNTHASE5 (CalS5; Dong et al., 2005). [See online article for color version of this figure.]
TGA9/10 and ROXY1/2 Regulate an Overlapping Set of Genes with Functions in Early and Middle Tapetal Development
Both ROXY1/2 and TGA9/10 are coexpressed in the tapetum during stages 4 to 7 of anther development, and their genetic interactions are consistent with parallel activities downstream of SPL/NZZ in anther development. Microarray analysis has identified a set of genes functioning primarily in tapetal development and pollen wall formation whose expression is severely down-regulated in roxy1 roxy2 inflorescence apices (Xing and Zachgo, 2008). To examine the possibility that some of these genes might also be targets of TGA9/10 activity, we used RT-PCR to monitor the transcripts of representative genes acting during the early, middle, and late stages of tapetal development in wild-type and tga9 tga10 inflorescence apices (Fig. 7, G and H). The expression of genes at the top of the hierarchy required for early archesporial cell formation and specification of anther cell layers, represented by SPL/NZZ and SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE8 (SPL8; Unte et al., 2003), were not markedly altered in tga9 tga10 inflorescence apices.
In contrast, a marked reduction was observed in the transcripts of DYT1 and MALE-STERILITY1 (MS1), tapetal genes operating before and during the meiosis of pollen mother cells (Wilson et al., 2001; Zhang et al., 2006; Ito et al., 2007; Yang et al., 2007a). Similar to tga9 tga10 mutants, ms1 mutants show an abnormally vacuolated tapetum and defects in microspore development, resulting in pollen with an altered exine pattern (Ito et al., 2007; Yang et al., 2007a). No obvious changes were observed in the expression of the MYB transcription factors encoded by MYB35/DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1), or MYB33 and MYB65, of which double mutants show tapetum hypertrophy leading to premeiotic abortion of pollen development (Millar and Gubler, 2005; Zhu et al., 2008). Tapetal genes with post-meiotic functions in pollen coat formation, represented by MS2 and FACELESS POLLEN (FLP), were also expressed at wild-type levels in tga9 tga10 mutant apices (Aarts et al., 1997; Ariizumi et al., 2003). Quantitative RT-PCR in a separate experiment using roxy1 roxy2 mutant apices as a control confirmed these trends, showing a significant reduction in DYT1 and MS1 transcript in tga9 tga10 mutants (Supplemental Fig. S8; Supplemental Materials and Methods S1).
Therefore, we concluded that ROXY1/2 and TGA9/10 function at similar levels in the genetic hierarchy that controls anther development and are likely to coregulate a set of genes expressed in the tapetum before and during meiosis of pollen mother cells, represented by DYT1 and MS1. This result is compatible with the overlapping expression patterns of ROXY1/2 and TGA9/10 in the tapetum during stages 4 to 7 of anther development.
TGA9/10 and ROXY1/2 Interact in Yeast and in Planta
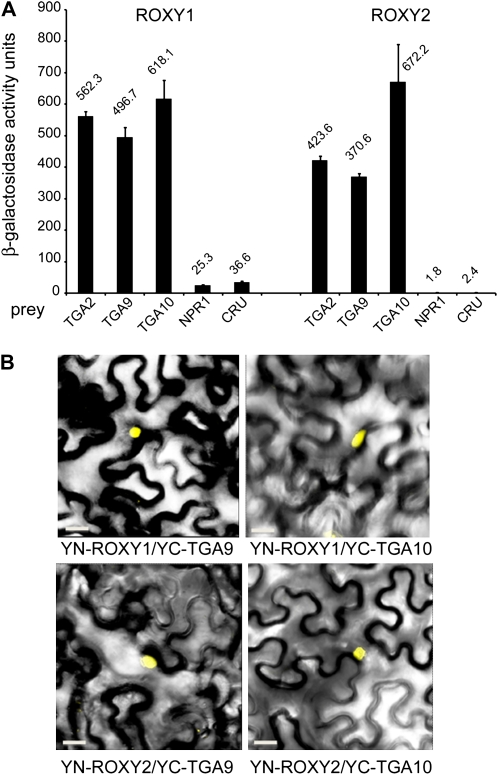
Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays indicate that ROXY1 interacts with several TGA transcription factors in yeast and in planta, including PAN, TGA2, TGA3, and TGA7 (Li et al., 2009). We first used a two-hybrid assay to test for a direct interaction between ROXY1/2 and TGA9/10, obtaining a positive result (Fig. 8A). As a further test, we used BiFC to examine the interaction of these proteins in planta. This assay showed reconstitution of yellow fluorescent protein (YFP) fluorescence in the nuclei of transformed tobacco leaf cells, indicating that ROXY proteins directly interact with TGA9 and TGA10 in plant cells (Fig. 8B). Conversely, ROXY1/2 (fused to the N terminus of YFP) or TGA9/10 (fused to the C terminus of YFP) coexpressed with the C or N terminus of YFP alone, respectively, failed to reconstitute YFP fluorescence (data not shown; Li et al., 2009). Previously, BiFC was used to confirm a nuclear interaction between ROXY1 and PAN, which is important for exerting their function (Li et al., 2009). Our prediction is that ROXY1/2 modification of TGA9/10 in the nucleus promotes their transcription factor activity, since loss-of-function mutations in either set of genes leads to a similar deficiency in adaxial anther lobe development and altered transcriptional profile. Collectively, these results provide new evidence that redox signaling plays a critical role in anther specification and dehiscence and that TGA factors mediate these signals.
Figure 8.
TGA9 and TGA10 directly interact with ROXY1/2. A, Yeast two-hybrid assay. ROXY proteins served as bait. NPR1 and CRUCIFERIN were negative controls. Error bars indicate se (three replicates). B, ROXY proteins interact with TGA9/10 in the nuclei of transiently transformed tobacco leaves. Images show reconstitution of YFP fluorescence 3 to 5 d after coexpression of protein pairs. The N terminus of YFP (YN) was fused in-frame upstream of ROXY1 and ROXY2. The C terminus of YFP (YC) was fused in-frame upstream of TGA9 and TGA10. As a negative control, coexpression of YN-ROXY1 and YC alone failed to reconstitute a fluorescent YFP chromophore (data not shown). Bars = 50 μm. [See online article for color version of this figure.]
DISCUSSION
We show here that two previously uncharacterized Arabidopsis TGA bZIP family members, TGA9 and TGA10, have overlapping functions in anther development. Loss-of-function tga9 tga10 mutations differentially disrupt the development of adaxial anther lobes and are required for microspore maturation, pollen viability, degeneration of the middle layer, and dehiscence. A similar differential block in adaxial anther lobe development is seen in roxy1 roxy2 double mutants, suggesting that TGA9 and TGA10 may be substrates of the CC-type floral glutaredoxins, ROXY1 and ROXY2. In support of this model, we show that ROXY1/2 and TGA9/10 have an overlapping pattern of expression in developing anthers and function downstream of SPL/NZZ, where they positively regulate the expression of a set of genes with tapetal functions. Finally, we show that ROXY1/2 and TGA9/10 directly interact in the nuclei of plant cells.
Differential Requirement for TGA9 and TGA10 in Abaxial and Adaxial Anther Lobes
In contrast to most early-acting gene mutants (e.g. spl/nzz, tpd1, and spl8), where development is equally dysfunctional in all four anther lobes, mutations in ROXY1/2 and TGA9/10 differentially impair SPL/NZZ expression and the specification and/or proliferation of sporogenous cells in adaxial anther lobes (Schiefthaler et al., 1999; Yang et al., 1999; Unte et al., 2003; Xing and Zachgo, 2008). Compatible with this, TGA9 is expressed during stages 2 to 3 in a horseshoe pattern, laterally (overlapping with SPL) and on the adaxial face of the anther primordium (Fig. 5F). In wild-type anthers, cell development in anther lobes is not synchronous until stage 5, with the adaxial lobes smaller than their abaxial counterparts (Sanders et al., 1999). Specification and proliferation of sporogenous cells in adaxial lobes, therefore, may depend in part on interactions with region-specific factors. In Arabidopsis and rice, abaxial and adaxial polarity determinants, similar to those in leaves, are differentially expressed in the anther and are associated with morphological asymmetry (Sessions et al., 1997; Dinneny et al., 2006; Toriba et al., 2010). At stages 2 to 3, prior to outgrowth of the four anther lobes, abaxial markers ETTIN/AUXIN REPONSE FACTOR3 (ETT/ARF3) and the YABBY gene FILAMENTOUS FLOWER are expressed on the abaxial side of anther primordia in a pattern complementary to adaxial markers such as PHABULOSA (PHB), encoding a class III homeodomain Leu-zipper transcription factor and the zinc-finger transcription factors encoded by JAGGED and NUBBIN (Sessions et al., 1997; Dinneny et al., 2006). PHB expression in Arabidopsis and rice rearranges to the notch region at stage 4, presaging the site of stomium development (Dinneny et al., 2006; Toriba et al., 2010). Mutations in ETT, whose activity is auxin responsive, and the auxin transport mutants pin formed1 and pinoid also perturb anther structure biased toward the loss of adaxial features (Bennett et al., 1995; Sessions et al., 1997), indicating that adaxial-abaxial patterning relies in part on a gradient of auxin, as in leaves (Pekker et al., 2005; Chitwood et al., 2007). The existence of region-specific factors in the anther, therefore, may lessen the requirement for ROXY-TGA9/10 activity in abaxial anther lobes.
TGA9 and TGA10 Contribute to Tapetal Development
The tapetum is essential for normal microspore and pollen development, as it provides nutrients and secretes enzymes and structural components for the pollen coat. Indeed, mutations in tapetally expressed genes often cause defects in microspore development leading to male sterility (Scott et al., 2004; Ma, 2005). Consistent with this, transcript analysis in tga9 tga10 mutants identified a significant reduction in DTY1 and MS1, representative genes operating in the tapetum before and during meiosis (Wilson et al., 2001; Zhang et al., 2006; Ito et al., 2007). During this time, TGA9/10 are strongly expressed in the middle layer and tapetum (Fig. 5, stages 5–6). Unexpectedly, tapetal genes with postmeiotic roles in pollen coat development represented by MS2 and FLP1 (Aarts et al., 1997; Ariizumi et al., 2003) were not markedly affected in this assay. MS2 is down-regulated 164.6-fold in roxy1 roxy2 mutants, and we expected to see a similar result for tga9 tga10 but did not. Microspores degenerate completely in roxy1 roxy2 but continue to develop in tga9 tga10 mutants, which may account for this difference. Tapetal programmed cell death is initiated at about the time that microspores are released from tetrads (Sanders et al., 1999), during which time TGA9 and TGA10 expression in the tapetum declines.
TGA9 and TGA10 Are Required for Anther Dehiscence
Dehiscence is a multistep program that begins with degeneration of the middle layer (stages 7–8) and the tapetum (stages 8–11) and is accompanied by the expansion and reinforcement of the endothecium (stage 11). Degradation of the septum between anther compartments and stomium differentiation are complete by the end of stage 12. At stage 13, breakage of the stomium releases the mature pollen (Sanders et al., 1999, 2005). Our results show that multiple steps of the dehiscence program require TGA9/10. First, dissolution of the middle layer is incomplete in the double mutant. Second, thickening and lignification of the endothecium, essential for dehiscence (Steiner-Lange et al., 2003; Mitsuda et al., 2005; Yang et al., 2007b), is similar to the wild type at stage 11, but the middle layer remnant becomes ectopically lignified, which may interfere with dehiscence (Supplemental Fig. S5). Septums in the mutant are disorganized and fail to degenerate, even when immature pollen grains form in adaxial lobes (Supplemental Fig. S4), and finally, stomium-like cells form at the notch between locules but are nonfunctional (Supplemental Fig. S4).
Relatively few nondehiscent mutants have been described to date (Ma, 2005; Wilson and Zhang, 2009). Mutations that impair jasmonic acid (JA) biosynthesis or signal transduction tend to delay rather than block anther dehiscence (Xie et al., 1998; Sanders et al., 2000; Ishiguro et al., 2001; Mandaokar and Browse, 2009). Jasmonate treatment fails to rescue dehiscence in tga9 tga10 mutants (data not shown), suggesting that TGA9/10 function independently or downstream of a jasmonate signal. Auxin similarly regulates the timing of dehiscence, by at least two mechanisms: ARF6 and ARF8 promote dehiscence via JA biosynthesis, and premature lignification of the endothecium occurs in auxin receptor mutants (Nagpal et al., 2005; Cecchetti et al., 2008; Tabata et al., 2010). Mutation of the tapetally expressed RECEPTOR-LIKE PROTEIN KINASE2 (RPK2) gene blocks dehiscence, but its mode of action is unknown (Mizuno et al., 2007). In rpk2 mutants, the middle layer fails to differentiate from the tapetum, which grows hypertrophically during meiosis and does not fully degenerate. Reinforcement of the endothecium and degeneration of the septum and stomium are also blocked in this mutant, suggesting important functions for the middle layer in dehiscence.
Degeneration of specific cell types during dehiscence relies on programmed cell death (Wu and Cheung, 2000; Sanders et al., 2005), which in senescence and plant defense are dependent on reactive oxygen species as signaling intermediates (Apel and Hirt, 2004; Gechev et al., 2006). Hence, it is unsurprising that the dehiscence program relies on TGA9/10, whose activity is likely to be redox regulated. Jasmonates, produced by plants in response to biotic and abiotic stresses, are known to promote the production of reactive oxygen species such as hydrogen peroxide (H2O2), which can be stress indicators but also important development signals (Buchanan and Balmer, 2005; Gapper and Dolan, 2006; Huffaker and Ryan, 2007; Wasternack, 2007). Indeed, the promoter elements to which TGA factors bind respond to various signals, including auxin, JA, and H2O2 (Xiang et al., 1996, 1997; Chen and Singh, 1999). Microarray studies show that TGA factors contribute in large part to the plant’s transcriptional response to stress, as mimicked by treatment with oxylipins such as 12-oxo-phytodienoic acid and JA (Mueller et al., 2008). In plant defense, the SA-inducible CC-type glutaredoxin ROXY19/GRX480 interacts with TGA factors and plays a role in SA/JA cross talk by repressing JA-responsive PDF1.2 gene transcription (Ndamukong et al., 2007). Thus, TGA9/10 may respond to redox changes induced by the localized production of auxin, JA, or H2O2. Indeed, significant changes in the redox status of protein thiols are seen by two-dimensional gel electrophoresis following treatment of Arabidopsis tissues with JA (Alvarez et al., 2009). These reversible modifications, often in the form of disulfide bridges, can function as redox switches to control protein activity, as shown for TGA1 and TGA4 (Després et al., 2003).
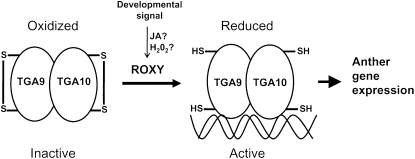
Posttranscriptional Control of TGA9 and TGA10 Activity by Glutaredoxin
In plants, there are now several examples of disulfide bridge formation or protein S-glutathionylation of Cys residues as mechanisms for regulating transcription factor activity (Dietz, 2008; Paulsen and Carroll, 2010). Our results support a model whereby modification of TGA9/10 by ROXY1/2 in response to a developmental signal such as JA biosynthesis promotes transcription factor activity (Fig. 9). Site-directed mutagenesis shows that Cys residues in the conserved C terminus of TGA1 (Cys-260 and Cys-266) and PAN (Cys-340) are crucial for function (Després et al., 2003; Li et al., 2009). Transcriptional activation by TGA1 requires an interaction with NPR1, a BTB-ankryin transcriptional coactivator (Rochon et al., 2006). This interaction relies on the reduction of Cys-266 and Cys-260 to disrupt an intramolecular disulfide bridge, modifications that are induced in SA-treated plants by an unknown glutaredoxin (Després et al., 2003). In contrast, PAN interacts constitutively with BTB-ankryin cofactors BOP1 and BOP2 in yeast and in the nuclei of Arabidopsis leaf mesophyll protoplasts (Hepworth et al., 2005; Xu et al., 2010). A single Cys residue in PAN (Cys-340, equivalent to Cys-266 in TGA1) is crucial for function, making it unclear if PAN is regulated by an intermolecular disulfide bridge or by S-glutathionylation. Genetic analysis shows that the pan phenotype (extra petals) is epistatic to the roxy1 roxy2 phenotype (fewer petals), suggesting that ROXY1 negatively regulates PAN activity (Li et al., 2009). In support of a monothiol mechanism for PAN regulation, complementation studies show that only one of three Cys residues (Cys-49) in the active site of ROXY1 is crucial for its in vivo activity (Xing et al., 2005). Alignments show that TGA10 alone has an equivalent to Cys-340/Cys-260 in PAN/TGA1, respectively, and that both TGA9 and TGA10 have a unique C-terminal Cys residue (Cys-429 and Cys-435, respectively; Supplemental Fig. S9). Our inability at present to complement the tga9 tga10 mutant phenotype with TGA9 or TGA10 cDNAs prevents us from testing if these residues are important for activity.
Figure 9.
Model for redox control of TGA9/10 activity by ROXY1/2. In their inactive form, one or more oxidized C-terminal Cys residues block TGA9/10 transcription factor activity (a dithiol bridge is shown) by an unknown mechanism. In response to redox changes triggered by a developmental signal, ROXY1 and ROXY2 reduce TGA9 and TGA10, converting them into a transcriptionally active form.
Functional Redundancy among TGA Family Members
Biochemical studies show that bZIP factors bind to DNA as dimers (Lamb and McKnight, 1991), supporting the notion that TGA9 and TGA10 may function redundantly as homodimers or heterodimers. Moreover, the tga9 tga10 phenotype is less severe than that of roxy1 roxy2 in that pollen formation is not completely blocked (Xing and Zachgo, 2008). A likely possibility is that other TGAs expressed in the anther work in conjunction with TGA9 and TGA10 as downstream effectors of ROXY1 and ROXY2. Candidates include TGA2, TGA6, and PAN, which interact with ROXY1 with differing affinities in yeast or in plants (Xiang et al., 1997; Li et al., 2009). The best candidate for redundancy is TGA2, but tight linkage to TGA10 hampers the genetic testing of this model. Given the broad expression pattern of TGA9, PAN, and TGA2 in tissues other than the flower (Fig. 1, D and E; Chuang et al., 1999; Li et al., 2009), it is likely that genetic redundancy continues to mask additional roles for this redox-regulated family of transcription factors in plant development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants were grown in growth chambers on agar plates and/or in soil at 21°C in 24 h of light (100 μmol m−2 s−1). The wild type was the Col-0 ecotype of Arabidopsis (Arabidopsis thaliana) unless otherwise stated. T-DNA insertion lines tga9-1 (SALK_057609), tga9-2 (SALK_091349), tga10-1 (SALK_124227), and tga10-2 (SALK_039672) were obtained from the Arabidopsis Biological Resource Center. The double mutant tga9-1 tga10-1 was characterized in this study. The position of T-DNA inserts was determined by sequencing, and homozygous mutants were identified by PCR genotyping (http://signal.salk.edu). Mutants roxy1 roxy2, spl-1, and dyt1 were described previously (Xing and Zachgo, 2008). All mutant combinations were constructed by crossing and confirmed by genotyping. Floral and anther development stages were determined according to Smyth et al. (1990) and Sanders et al. (1999), respectively. Supplemental Tables S1 and S2 list the sequences of most primers used in this study. All other primer sequences are available by request.
Construction of TGA9::GUS Reporter Lines
A TGA9::GUS translational fusion gene was created using the strategy described by Hepworth et al. (2002). A sequence representing the putative promoter of TGA9 (nucleotides –3,895 to +5) was amplified by PCR using Arabidopsis bacterial artificial chromosome T27G7 DNA as the template. Wild-type plants were transformed with pTGA9::GUS by floral dipping (Clough and Bent, 1998) using the Agrobacterium tumefaciens strain C58C1 pGV101 pMP90 (Koncz and Schell, 1986). Basta-resistant transformants were selected on soil using the herbicide Finale (AgrEvo).
Pollen Germination and Viability Assays
For the in vitro assay of pollen germination, pollen grains from the anthers of late stage 12 flowers were excised into water and transferred to pollen germination medium presolidified onto a microscope slide (Fan et al., 2001). Slides were incubated at 22°C in a high-humidity chamber (Johnson-Brousseau and McCormick, 2004), and pollen tubes were observed after 24 h by light microscopy. The germination of more than 300 pollen grains was monitored for each genotype. Pollen viability was also assessed with Alexander’s stain (Alexander, 1969).
Histological Analysis of Developing Anthers
Floral buds were fixed overnight at 4°C in 2% (v/v) glutaraldehyde as described (Ogawa et al., 2009). Tissues were briefly rinsed in 25 mm NaPO4 buffer (pH 6.8) and dehydrated with ethanol prior to embedding in LR White resin (London Resin Company). Sections (1 μm) were heat fixed onto glass slides and stained with 0.05% (w/v) toluidine blue prior to imaging. To detect callose, dissected anthers were treated with 0.05% (w/v) aniline blue in 0.067 m phosphate buffer (pH 8.5) and imaged under UV illumination. To detect lignin, paraffin-embedded anther sections were dewaxed and dehydrated prior to successive incubations in 2% phloroglucinol (in 95% ethanol) and 6 n HCl for color development.
GUS Staining and in Situ Hydridization
Staining, sectioning, and embedding of GUS-stained tissues was performed essentially as described by Sieburth and Meyerowitz (1997). Sections (10 μm) were affixed to glass slides and dewaxed with tert-butanol prior to imaging. In situ hybridization and antisense probe synthesis was performed as described previously (Hepworth et al., 2005).
RT-PCR
Total RNA was isolated from inflorescence apices using the Trizol method (Invitrogen). Total cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen) using 1 μg of RNA as the template. PCR was subsequently performed using 2 μL of diluted cDNA as the template and Taq as the polymerase (Invitrogen). Primers bZIP21-F2 and bZIP21-R1, and bZIP21-F2 and bZIP21-R2, were used for amplification of TGA9 transcripts designated full and partial, respectively. Primers bZIP65cDNA-F1 and bZIP65cDNA-R2, and bZIP65cDNA-F1 and bZip65R1-3 were used for amplification TGA10 transcripts designated full and partial, respectively. GAPC encoding glyceraldehyde 3-phosphate dehydrogenase served as a control transcript (Hepworth et al., 2005).
Yeast Two-Hybrid Assay
We used the GAL4-based yeast two-hybrid system described by Kohalmi et al. (1998). To generate bait and prey constructs, the full coding sequences of TGA9, TGA10, ROXY1, and ROXY2 were amplified by PCR from cloned cDNA template. PCR products were subcloned into pCR-BluntII-TOPO (Invitrogen) and sequenced to ensure fidelity. Recognition sites for the restriction enzymes SalI and NotI were incorporated into the 5′ ends of primers to facilitate the subsequent directional cloning of ROXY1/2 and TGA9/10 cDNA inserts into the corresponding sites of bait (pBI-880) and prey (pBI-881) plasmids, respectively. Construction of TGA2 in pBI-880, yeast transformation, and β-galactosidase assays were described previously (Després et al., 2000).
BiFC Assay
BiFC assays were performed as described by Li et al. (2009). pE-SPYNE and pE-SPYCE (Walter et al., 2004) were used to generate expression vectors using a Gateway cloning strategy (Invitrogen). The N terminus of YFP was cloned in-frame upstream of ROXY1 and ROXY2 cDNAs. The C terminus of YFP was fused in-frame to the N termini of TGA9 and TGA10 cDNAs. Fusion genes were expressed under the control of a cauliflower mosaic virus 35S promoter. Infiltrated Nicotiana benthamiana leaves were examined for reconstitution of YFP fluorescence 3 to 5 d after inoculation using a LSM 510 Meta confocal microscope (Carl Zeiss).
Scanning Electron Microscopy
Samples were prepared for scanning electron microscopy as described previously (Hepworth et al., 2005) and imaged using a VegaII XMU VPSEM apparatus (Tescan).
Sequence data have been deposited in GenBank under accession numbers HQ132743 (TGA9 cDNA) and HQ132742 (TGA10 cDNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree for Arabidopsis TGA transcription factors.
Supplemental Figure S2. Analysis of pollen in wild-type and tga9 tga10 mutant plants.
Supplemental Figure S3. Callose deposition in wild-type and tga9 tga10 mutant plants.
Supplemental Figure S4. Comparison of wild-type and tga9 tga10 late-stage anther lobe development.
Supplemental Figure S5. Endothecium lignification of wild-type and tga9 tga10 anthers.
Supplemental Figure S6. In situ analysis of TGA10 expression in developing anthers.
Supplemental Figure S7. RT-PCR analysis of TGA9 and TGA10 transcript in wild-type (Nossen-0) and roxy1 roxy2 apices.
Supplemental Figure S8. Quantitative RT-PCR analysis of tapetum transcripts in wild-type, tga9 tga10, and roxy1 roxy2 apices.
Supplemental Figure S9. Alignment of selected TGA transcription factors.
Supplemental Table S1. List of primers.
Supplemental Table S2. List of primers for RT-PCR transcript analysis of anther development genes.
Supplementary Material
Acknowledgments
We thank Laurian S. Robert and Ann-Fook Yang at the Eastern Cereal and Oilseed Research Centre, Agriculture and Agri-Food Canada, Ottawa, for use of their ultramicrotome facility.
References
- Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alvarez S, Zhu M, Chen S. (2009) Proteomics of Arabidopsis redox proteins in response to methyl jasmonate. J Proteomics 73: 30–40 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR. (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8: 505–520 [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C. (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Singh KB. (1999) The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J 19: 667–677 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Guo M, Nogueira FTS, Timmermans MCP. (2007) Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 134: 813–823 [DOI] [PubMed] [Google Scholar]
- Chuang CF, Running MP, Williams RW, Meyerowitz EM. (1999) The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. (2008) Redox signal integration: from stimulus to networks and genes. Physiol Plant 133: 459–468 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. (2006) NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH. (2001) In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 52: 1603–1614 [PubMed] [Google Scholar]
- Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. (2005) BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW. (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35: 177–192 [DOI] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA. (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104: 10732–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K. (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F; bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S. (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X. (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohalmi SE, Reader LJW, Samach A, Nowak J, Haughn GW, Crosby WL. (1998) Identification and characterization of protein interactions using the yeast 2-hybrid system. Gelvin SB, Schilperoot RA, , Plant Molecular Biology Manual M1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–30 [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of T1-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lamb P, McKnight SL. (1991) Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci 16: 417–422 [DOI] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. (2009) Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Browse J. (2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F. (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50: 751–766 [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S. (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. (2010) Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol 5: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Després C. (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18: 3670–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Meyerowitz EM. (1996) Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122: 1261–1269 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Le BH, Goldberg RB. (2005) Differentiation and degeneration of cells that play a major role in tobacco anther dehiscence. Sex Plant Reprod 17: 219–241 [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB. (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiermeyer A, Thurow C, Gatz C. (2003) Tobacco bZIP factor TGA10 is a novel member of the TGA family of transcription factors. Plant Mol Biol 51: 817–829 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H. (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H. (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, Hirano HY. (2010) Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22: 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P. (2003) SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Freeling M. (1999) The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J 19: 489–495 [DOI] [PubMed] [Google Scholar]
- Walsh J, Waters CA, Freeling M. (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev 12: 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB. (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wu HM, Cheung AY. (2000) Programmed cell death in plant reproduction. Plant Mol Biol 44: 267–281 [DOI] [PubMed] [Google Scholar]
- Xiang C, Miao Z, Lam E. (1997) DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol Biol 34: 403–415 [DOI] [PubMed] [Google Scholar]
- Xiang C, Miao ZH, Lam E. (1996) Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol Biol 32: 415–426 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S. (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR. (2010) Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J 63: 974–989 [DOI] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. (2007a) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA. (2007b) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D. (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H. (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. (2008) Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.