Abstract
Anthocyanin accumulation is regulated negatively by ethylene signaling and positively by sugar and light signaling. However, the antagonistic interactions underlying these signalings remain to be elucidated fully. We show that ethylene inhibits anthocyanin accumulation induced by sucrose (Suc) and light by suppressing the expression of transcription factors that positively regulate anthocyanin biosynthesis, including GLABRA3, TRANSPARENT TESTA8, and PRODUCTION OF ANTHOCYANIN PIGMENT1, while stimulating the concomitant expression of the negative R3-MYB regulator MYBL2. Genetic analyses show that the ethylene-mediated suppression of anthocyanin accumulation is dependent upon ethylene signaling components responsible for the triple response. Furthermore, these positive and negative signaling pathways appear to be under photosynthetic control. Suc and light induction of anthocyanin accumulation was almost fully inhibited in wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia and ethylene (ethylene response1 [etr1-1]) and light (long hypocotyl1 [hy1], cryptochrome1/2, and hy5) signaling mutants treated with the photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea. The transcript level of the sugar transporter gene SUC1 was enhanced in ecotype Columbia treated with the ethylene-binding inhibitor silver and in etr1-1, ethylene insensitive2 (ein2-1), and ein3 ein3-like1 mutants. In contrast, 3-(3,4-dichlorophenyl)-1,1-dimethylurea treatment reduced SUC1 expression, which indicates strongly that SUC1 represents an integrator for signals provided by sugar, light, and ethylene. SUC1 mutations lowered accumulations of anthocyanin pigment, soluble sugar content, and ethylene production in response to Suc and light signals. These data demonstrate that the suppression of SUC1 expression by ethylene inhibits Suc-induced anthocyanin accumulation in the presence of light and, hence, fine-tunes anthocyanin homeostasis.
Anthocyanins play key roles in many plant physiological processes; for instance, they form photoprotective screens in vegetative tissue, act as visual attractors to aid pollination and seed dispersal, and function as antimicrobial agents and feeding deterrents in the defense response (Winkel-Shirley, 2001; Steyn et al., 2002). The anthocyanin biosynthetic pathway is well described in plants. In Arabidopsis (Arabidopsis thaliana) and other plants, including Antirrhinum majus (snapdragon) and Petunia hybrida (petunia), early biosynthesis genes (EBGs) such as chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3′-hydroxylase (F3′H), which are common to different flavonoid subpathways, are induced prior to late biosynthesis genes (LBGs) such as dihydroflavonol 4-reductase (DFR), leucoanthocyanidin oxygenase (LDOX), anthocyanidin reductase (ANR), and UDP-glucose:flavonoid 3-O-glucosyltransferase (UF3GT; Pelletier et al., 1997). EBGs and LBGs are transcriptionally activated by R2R3-MYBs, such as PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2. R2R3-MYBs form transcription complexes with TRANSPARENT TESTA GLABRA1 (TTG1; a WD40 protein) and several basic helix-loop-helix (bHLH) proteins such as TRANSPARENT TESTA8 (TT8; bHLH042), GLABRA3 (GL3; bHLH001), and ENHANCER OF GLABRA3 (EGL3; bHLH002), which have overlapping roles in protein-protein interactions and anthocyanin production in seedlings, stems, and leaves of Arabidopsis (for review, see Quattrocchio et al., 2006). MYB/bHLH/TTG1 (MBW) transcription complexes regulate anthocyanin biosynthesis in a spatiotemporal manner. Recently, TTG1-independent MYBs, including MYB11, MYB12, and MYB111 (Mehrtens et al., 2005; Stracke et al., 2007), and PAP1 homologs such as MYB113 and MYB114 (Gonzalez et al., 2008) were shown to regulate EBGs and LBGs, respectively.
In contrast to the positive transcription factors (TFs) mentioned above, the R3-MYB protein MYBL2 is a negative regulator of anthocyanin biosynthesis. In addition to being regulated developmentally, MYBL2 expression also depends upon environmental cues such as high light levels and, presumably, nitrogen deficiency (Dubos et al., 2008; Matsui et al., 2008). MYBL2 is thought to inhibit anthocyanin biosynthesis by interacting with TT8 to form a transcriptional inhibitory complex, MYBL2/bHLH/TTG1 (L2BW). Hence, the anthocyanin content in a specific cell type is proposed to be regulated by a balance between MBW and L2BW complexes (Dubos et al., 2008).
Anthocyanin biosynthesis genes are regulated tightly by both the quantity and quality of light (Solfanelli et al., 2006; Cominelli et al., 2008). In Arabidopsis, light signaling is perceived and mediated by photoreceptors such as the UV-B receptor, cryptochrome 1 (CRY1), and phytochrome A (PHYA; Neff and Chory, 1998) and PHYB (Ahmad and Cashmore, 1997; Wade et al., 2001). LONG HYPOCOTYL5 (HY5), a Leu-zipper TF, serves as a point of convergence for PHY and CRY signaling (Gyula et al., 2003), functioning as a positive component in anthocyanin biosynthesis (Chattopadhyay et al., 1998). In addition to HY5, PHYTOCHROME-INTERACTING TRANSCRIPTION FACTOR3 (PIF3), a bHLH protein, interacts directly with PHYs, and PIF3 positively regulates anthocyanin biosynthesis (Kim et al., 2003; Shin et al., 2007). HY5 and PIF3 bind directly to the promoters of anthocyanin structural genes such as CHS, CHI, F3H, F3′H, DFR, and LDOX (Lee et al., 2007; Shin et al., 2007). In addition to photoreceptor-mediated anthocyanin regulation, photosynthesis also contributes to anthocyanin production in turnip (Brassica rapa) seedlings (Schneider and Stimson, 1971) and nonchlorophyllous corn (Zea mays) leaves (Kim et al., 2006). Treatment with the photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a diuron, significantly inhibits light-dependent anthocyanin accumulation. In Arabidopsis, residual anthocyanin production is still detected in the photoreceptor mutants hy5 and PIF3 (Shin et al., 2007). However, the regulatory pathways involved in this process are largely unknown, and many questions remain to be answered. For example, does photosynthesis affect anthocyanin synthesis in a HY5-independent manner?
Sugar is a common regulator for the expression of genes encoding metabolic enzymes and proteins involved in photosynthesis, carbohydrate metabolism, pathogenesis (Rolland et al., 2006), and anthocyanin biosynthesis (Mita et al., 1997; Baier et al., 2004). Suc induces a key TF for anthocyanin biosynthesis, PAP1 (Lloyd and Zakhleniuk, 2004; Teng et al., 2005). Further support for the role played by Suc can be found in the positive correlation between increases in PAP1 expression and several anthocyanin structural genes in a phosphoglucomutase-deficient Arabidopsis mutant that accumulates high levels of Suc (Solfanelli et al., 2006). Based on observations of Suc- and maltose (Mal)-dependent anthocyanin accumulation, Solfanelli et al. (2006) indicated that Suc-induced anthocyanin accumulation is sensed either by Suc transporters (SUCs) or proteins closely associated with SUCs rather than by a membrane-bound hexose transporter or the activity of HXK1, an internal Glc sensor. This view has been supported further by a recent report showing that suc1-defective mutants grown in 3%, but not in 5%, Suc-containing growth medium had diminished anthocyanin accumulation (Sivitz et al., 2008). However, SUC1 expression and anthocyanin accumulation are separated spatially; SUC1 is expressed preferentially in the roots (Sivitz et al., 2008), while anthocyanin accumulates predominantly in the subepidermal cell layers of leaves (Kubo et al., 1999). Therefore, how SUC1 expression in roots is involved in anthocyanin accumulation in shoots needs to be answered.
There are nine putative SUCs in Arabidopsis. SUC1 is expressed in roots, pollen, and trichomes (Sivitz et al., 2008). SUC2 and SUC4 are thought to be involved in Suc loading in companion cells (Gottwald et al., 2000) and minor veins (Meyer et al., 2000; Weise et al., 2000), respectively. SUC3 (SUT2) has been characterized as a weak low-affinity transporter (Barker et al., 2000). Expression of SUC5 (Baud et al., 2005) and SUC8/9 (Sauer et al., 2004) is restricted to the endosperm during early seed development and to floral tissues. Both SUC6 and SUC7 encode aberrant proteins in various Arabidopsis ecotypes (Sauer et al., 2004). In addition to developmental cues, other factors such as sugars, light, and hormones also influence the expression of SUCs. Sugars can positively or negatively modulate expression levels of SUCs. For example, SUCs in the clade 2 symporter family (Lalonde et al., 2004) are up-regulated by Suc (Aoki et al., 1999; Barker et al., 2000; Zhou et al., 2009) or Glc (Matsukura et al., 2000), while those in the SUT1 clade respond negatively to increasing Suc (Chiou and Bush, 1998; Vaughn et al., 2002) or Glc (Li et al., 2003; Zhou et al., 2009) concentrations. Light is responsible for the expression of LeSUT1 and StSUT1, and expression levels are enhanced by light treatment (Kühn et al., 1997). Furthermore, the expression of SUCs may be regulated by the plant hormone abscisic acid (Saftner and Wyse, 1984). In Arabidopsis, SUC1 expression appears to increase in response to Suc treatment (Sivitz et al., 2008). However, no direct connection has yet been identified between Suc, light, hormone signaling, and SUC regulation in Arabidopsis.
Plant hormones such as auxin and abscisic acid (Jeong et al., 2004; Hoth et al., 2010), gibberellins (Weiss et al., 1995), cytokinin (Deikman and Hammer, 1995), and ethylene (Morgan and Drew, 1997) differentially regulate anthocyanin biosynthesis in whole plants as well as in cell suspensions (Ozeki and Komamine, 1986). Ethylene markedly suppresses anthocyanin accumulation (Craker and Wetherbee, 1973; Kang and Burg, 1973), while the Co2+-mediated inhibition of ethylene biosynthesis and the prevention of ethylene activity by silver increases the anthocyanin content of corn seedlings (Rengel and Kordan, 1987). Likewise, the petals of transgenic tobacco (Nicotiana tabacum) plants expressing a mutated melon (Cucumis melo) ethylene receptor gene, ethylene response1 (ETR1H69A), accumulate higher levels of anthocyanins than control plants (Takada et al., 2005). In contrast, the constitutive triple response1 (ctr1) mutant, which exhibits a constitutive response to ethylene (Kieber et al., 1993), contains similar concentrations of anthocyanin to wild-type ecotype Columbia (Col0) in the presence of high concentrations of Suc (Gibson et al., 2001). Thus, certain positive and negative regulatory factors in ethylene signaling, such as ETR1, CTR1, ETHYLENE INSENSITIVE2 (EIN2), EIN3, or EIN3-like1 (EIL1; for recent reviews, see Kendrick and Chang, 2008; Yoo et al., 2009), may play roles in the regulation of anthocyanin biosynthesis.
Ethylene modulates Suc and Glc sensitivity during Arabidopsis seedling development and controls anthocyanin biosynthesis (Gibson et al., 2001). However, the molecular and cellular mechanisms underlying antagonistic interactions between Suc/light and ethylene signaling remain to be elucidated for anthocyanin regulation. In this paper, we report the presence of an anthocyanin induction pathway that is independent of HY5 but dependent upon photosynthetic electron transport in acyanic mesophyll cells. Furthermore, we also present experimental evidence showing that ethylene is, at least in part, responsible for the suppression of sugar-inducible anthocyanin synthesis in Arabidopsis plants grown under light and that this control is mediated via down-regulation of SUC1 expression in roots.
RESULTS
Anthocyanin Accumulation in Wild-Type Col0 Plants and in Ethylene Signaling Pathway Mutants Involved in the Triple Response
Arabidopsis Col0 plants were grown under white light (140 μmol m−2 s−1) on half-strength Murashige and Skoog (MS) medium supplemented with 60 mm (2.16%) Suc. The anthocyanin content started to increase 6 d after germination, became saturated by 9 d after germination, and then remained unchanged for another 3 d (data not shown). In contrast, no apparent anthocyanin accumulation was observed in Arabidopsis plants grown on the same medium under dark conditions. Furthermore, very little anthocyanin accumulation was detected in plants grown under light but on medium lacking Suc. These findings indicate that both light and Suc are required for anthocyanin induction.
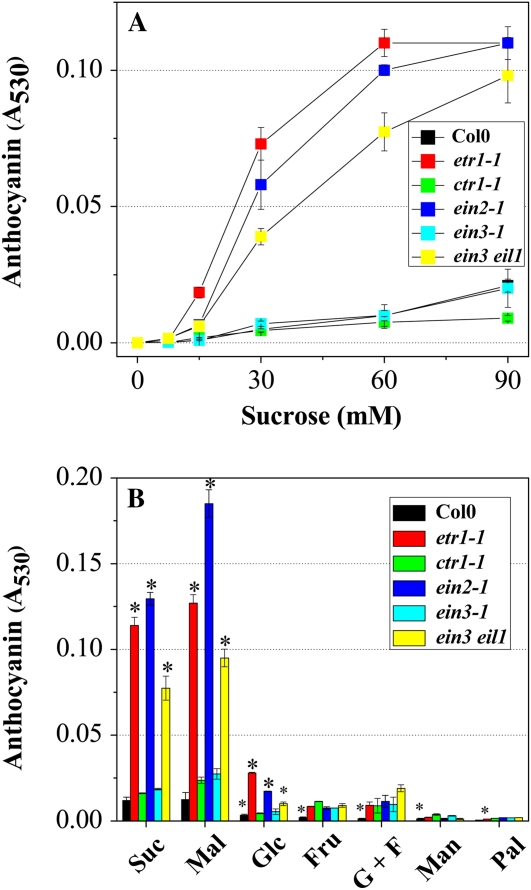
In comparison with the Col0 seedlings, ethylene signaling mutants, including etr1-1, ein2-1, and ein3 eil1, showed an apparent increase in anthocyanin content under the same conditions (Suc with light) as described above (Fig. 1A). Anthocyanin accumulation was observed mainly at the abaxial surface of the cotyledons and rosette leaves during the early seedling stage but was found on both the abaxial and adaxial sides at later stages. On a transverse section of ein2-1 leaf tissue, it can be seen that anthocyanin accumulated predominantly in the epidermis of the abaxial side (Fig. 1B). Although treatment with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) did not significantly decrease the anthocyanin content of Col0 plants, treatments with ethylene-binding (silver) and ethylene synthesis (aminoethoxyvinylglycine [AVG]) inhibitors did result in 4.2- and 1.6-fold increases in anthocyanin content, respectively (Fig. 1C). However, a stimulatory effect of silver on anthocyanin level was not observed in plants when they were grown on Suc-lacking medium under light (data not shown).
Figure 1.
Anthocyanin contents in Col0 and ethylene receptor and signaling mutants. A, Images of representative seedlings of Col0 and ethylene signaling mutants (etr1-1, ctr1-1, ein2-1, ein3-1, and ein3 eil1). B, Images of the abaxial side (left) and of the transverse (right, top) and longitudinal (right, bottom) sections of a rosette leaf from a representative ein2-1 plant. Bars = 500 μm (left), 200 μm (right, top), and 100 μm (right, bottom). In A and B, plants were grown for 12 d on half-strength MS medium containing 60 mm Suc under illumination (140 μmol m−2 s−1). C, Anthocyanin contents in Col0, ethylene receptor family mutants (etr1-1, ers1-2, etr2-1, ers2-1, and ein4), ETR1 mutant alleles (etr1-1, etr1-2, etr1-3, etr1-4, and etr1-7), and signaling mutant plants. Plants were grown on half-strength MS medium containing 60 mm Suc supplemented with 10 μm ACC (ACC), 10 μm AVG (AVG), 1 mm AgNO3 (Ag+), or without (CO for Col0 and ethylene mutants) the inhibitors indicated and incubated for 12 d under illumination (140 μmol m−2 s−1). Error bars represent sd values for the means of four or five independent replicates. Asterisks over bars indicate differences between control (CO) and treatment or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
These findings prompted further investigation into the roles played by ethylene signaling in anthocyanin biosynthesis. The Arabidopsis ethylene receptors ETR1, ETHYLENE RESPONSE SENSOR1 (ERS1), ETR2, ERS2, and EIN4 can directly bind and transfer ethylene signals (Kendrick and Chang, 2008; Yoo et al., 2009). Upon ethylene binding, they may act as negative regulators of anthocyanin accumulation. Indeed, seedlings with ethylene insensitivity resulting from mutations in the four ethylene receptors ETR1, ERS1, ETR2, and ERS2 exhibited greater than 3-fold higher anthocyanin levels than Col0 plants (Fig. 1C). Among the ethylene receptor mutants, etr1-1 conferred the highest level of anthocyanin accumulation (Fig. 1C). To investigate the specific domain function of ETR1 further, anthocyanin content was estimated in etr1-1 (Cys65Tyr; dominant ethylene strong insensitive), etr1-2 (Ala102Thr; dominant ethylene weak insensitive), etr1-3 (Ala31Val; dominant ethylene weak insensitive), etr1-4 (Ile62Phe; dominant ethylene weak insensitive), and etr1-7 (Trp74*; recessive ethylene hypersensitive) mutants. As shown in Figure 1C, the ethylene insensitive etr1-1 and three other alleles (etr1-2, etr1-3, and etr1-4) showed 5- to 10-fold increases in anthocyanin content when compared with Col0 plants. These findings imply that ethylene binding and its coupling to signal output may exert negative control on anthocyanin accumulation. Lower levels of anthocyanin were observed consistently in the ethylene-hypersensitive mutant etr1-7. EIN2, which functions downstream of the ETR1-CTR1 complex, acts via EIN3 and EIL1, which are primary transcription activators in ethylene signaling. Expression of EIN3 or EIL1 induces the up-regulation of secondary TFs, including ETHYLENE RESPONSE FACTOR1 (Kendrick and Chang, 2008; Yoo et al., 2009). As shown in Figure 1C, increased anthocyanin levels were observed in the ethylene-insensitive mutants etr1-1, ein2-1, and ein3 eil1. In contrast, the constitutive ethylene response mutant, ctr1-1, a partial ethylene-insensitive mutant, ein3-1, and a weak ethylene-insensitive mutant, eil1-3, showed more or less comparable anthocyanin levels to that of Col0 plants. A CTR1-independent ethylene signaling pathway, which may be mediated via ARABIDOPSIS RESPONSE REGULATOR2 (Hass et al., 2004), has also been reported in Arabidopsis (Larsen and Chang, 2001). However, since silver treatment did not induce as strong an anthocyanin production in ctr1-1 as in Col0 (Supplemental Fig. S1), it is unlikely that the alternative pathway contributes to ethylene repression. Notably, the ein3 eil1 double mutant exhibited a greater than 7-fold increase in anthocyanin levels, whereas increased production was not observed in ein3-1 and eil1-3 single mutants, which indicates that EIN3 and EIL1 are functionally redundant with respect to the regulation of anthocyanin accumulation.
Ethylene Repression of Sugar- and Light-Dependent Anthocyanin Accumulation
To investigate whether ethylene-mediated suppression of anthocyanin accumulation under light is influenced by changes in exogenously supplied Suc content, Col0 and ethylene signaling mutants were grown in the presence of various concentrations of Suc. Anthocyanin did not accumulate in Col0 and ethylene signaling mutants grown in Suc-free medium, but anthocyanin was observed as the Suc concentration of the medium increased (Fig. 2A). Below 15 mm Suc, the Col0 and ethylene signaling mutants investigated in this study showed almost no anthocyanin accumulation. However, above 30 mm Suc, the Col0, ctr1-1, and ein3-1 mutants showed proportional increases in anthocyanin content with respect to Suc concentration, whereas etr1-1, ein2-1, and ein3 eil1 mutants showed S-shaped curves.
Figure 2.
Induction of anthocyanin in Col0 and ethylene signaling mutants (etr1-1, ctr1-1, ein2-1, ein3-1, and ein3 eil1). A, Anthocyanin accumulates in plants as a function of Suc concentration. B, Effects of various sugars on anthocyanin content. Plants were grown on half-strength MS medium supplemented with various concentrations of Suc (0, 7.5, 15, 30, 60, and 90 mm; A) or with metabolic sugars such as Suc (60 mm), Mal (60 mm), Glc (60 mm), or Fru (60 mm), a 1:1 mixture of Glc:Fru (G+F; 30 mm Glc and 30 mm Fru), a sugar alcohol, Man (60 mm), or a nonmetabolic sugar, Pal (60 mm; B) for 12 d under light conditions of 140 μmol m−2 s−1. In A, the black square for Col0 is hardly visible because it overlaps with symbols for ein3-1 or ein3 eil1 mutants. Error bars represent sd values for the means of three or four independent replicates. Asterisks over bars indicate differences between control (60 mm Suc) and treatment (other sugars) or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
Col0 and ethylene signaling mutants were grown in the presence of various sugars to determine whether ethylene repression of Suc-induced anthocyanin accumulation is specific to Suc signaling. Palatinose (Pal) and mannitol (Man) were included in the experiment as osmotic controls. As shown in Figure 2B, and consistent with previous reports (Solfanelli et al., 2006), the highest levels of anthocyanin were produced by plants treated with Suc or Mal. Only a slight response was observed with Glc, Fru, or a combination of Glc + Fru, and no effect on anthocyanin accumulation was detected in seedlings treated with Man or Pal. Among the ethylene signaling mutants, etr1-1, ein2-1, and ein3 eil1 exhibited Suc- and Mal-specific anthocyanin accumulation, whereas ctr1-1 and ein3-1 were rather insensitive to these sugars, further supporting the notion that ethylene represses the Suc- and Mal-specific signaling pathway via ETR1, CTR1, EIN2, and EIN3 EIL1.
Transcript Levels of Structural and Regulatory Genes Involved in Anthocyanin Biosynthesis in Col0 and in Ethylene Signaling Mutants
Total RNAs were extracted from 12-d-old seedlings grown in the presence of 3% (83.3 mm) Suc, where full induction of anthocyanin was observed in etr1-1 mutants (Fig. 2A). Whole genome RNA expression analyses were performed using an Affymetrix DNA microarray, and the expression of anthocyanin biosynthesis genes was compared between etr1-1 and Col0. CHS, DFR, LDOX, and UF3GT transcript levels were more than 2-fold higher in etr1-1 mutants than in Col0, although the specific expression levels of each gene family member varied (Supplemental Table S1). With the exception of an EBG, CHS, the expression levels of three LBGs were at least 8-fold higher in etr1-1, which indicates that LBGs are regulated preferentially by ethylene signaling. Among the TFs involved in anthocyanin biosynthesis, the expression of EGL3, TT8, and PAP1 increased significantly in etr1-1 (P < 0.05), whereas MYBL2 expression decreased (P < 0.01).
Quantitative (q)PCR analysis was conducted on Col0 and ethylene signaling mutants to verify the microarray data and identify anthocyanin biosynthesis genes regulated by ethylene signaling. During a preliminary experiment, we noticed that seedling growth of wild-type and etr1-1 plants started to decrease at concentrations of Suc above 30 mm (data not shown). Thus, in order to minimize potential side effects resulting from high concentrations of Suc on seedling growth while at the same time obtaining obvious effects of Suc on anthocyanin pigmentation, as shown in Figure 2A, we used plants grown at 60 mm Suc instead 83.3 mm Suc (equivalent to 3% Suc) for further analysis. qPCR was also used to analyze regulatory genes such as GL3 and PAP2, which were omitted from the microarray data analysis. Since we intended to characterize the TTG1-dependent regulation of EBG and LBGs during vegetative growth, the early embryo-specific R2R3-Myb, TT2 (Nesi et al., 2001), TTG1-independent MYBs such as MYB11, MYB12, and MYB111 (Mehrtens et al., 2005; Stracke et al., 2007), and two PAP1 homologs, MYB113 and MYB114 (Gonzalez et al., 2008), were not included in the analysis.
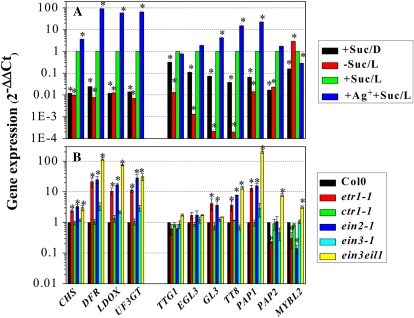
Induction of anthocyanin pigmentation requires disaccharides such as Suc and Mal as well as light signals (Fig. 2). Compared with Col0 grown on 60 mm Suc-containing medium in the light, plants grown with Suc in the dark, or in the light without Suc, showed negligible expression of either structural or positive regulatory genes. These findings indicate that both light and Suc are required for transcript accumulation. In contrast, expression of the negative regulatory gene MYBL2 was down-regulated by Suc under both light and dark conditions, whereas it was highly up-regulated by illumination alone (2.9-fold; Fig. 3A). Thus, it appears that sugars and light induce anthocyanin accumulation by enhancing the expression of both structural and positive regulatory genes while reducing the expression of the negative regulator MYBL2.
Figure 3.
Transcript levels of structural (CHS, DFR, LDOX, and UF3GT) and regulatory (TTG1, EGL3, GL3, TT8, PAP1, PAP2, and MYBL2) genes involved in anthocyanin biosynthesis in Col0 and ethylene signaling mutants (etr1-1, ctr1-1, ein2-1, ein3-1, and ein3 eil1). A, Col0 plants were grown on half-strength MS medium supplemented with (+Suc) or without (−Suc) 60 mm Suc in the presence of 1 mm AgNO3 (+Ag+). Seedlings were incubated for 12 d in continuous darkness (D) or under light (L; 140 μmol m−2 s−1) conditions. B, Plants were grown on half-strength MS medium supplemented with 60 mm Suc for 12 d under light. Transcript levels were quantified by reverse transcription (RT)-qPCR (for details, see “Materials and Methods”). Error bars represent sd values for the means of three or four independent replicates. Asterisks over bars indicate differences between control (+Suc/L) and treatment (A) or between Col0 and mutants (B), with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
In Col0 plants, significant increases in transcription of EBG and LBGs (P < 0.05) were observed when ethylene signaling was inhibited by silver treatment (Fig. 3A). These increases might be due to a greater than 2-fold accumulation of positive TF transcripts, including GL3, TT8 and PAP1, at the same time as a 3.3-fold decrease of the negative TF MYBL2. Consistently, the transcript levels of structural genes such as LBGs were significantly higher (10- to 118-fold) in the etr1-1, ein2-1, and ein3 eil1 mutants than in Col0, while their expression levels remained almost unchanged in ctr1-1 and ein3-1 (Fig. 3B). Among the TFs investigated, the expression levels of TTG1 and EGL3 remained almost unchanged, being insensitive to both silver treatment and mutations in the ethylene signaling components tested. However, the other TF genes were sensitive to ethylene signaling. For example, transcripts of the positive regulatory genes GL3, TT8, and PAP1 increased more than 2-fold when compared with Col0 seedlings grown under control conditions; the expression pattern of PAP2 was less clear than that observed with PAP1 (Fig. 3B). In sharp contrast to the induction of positive regulators, expression of the negative regulator MYBL2 decreased by more than 70% in etr1-1 and ein2-1 mutants but remained unchanged in ctr1-1 and ein3-1. Therefore, it appears that ethylene represses anthocyanin biosynthesis by down-regulating the expression of positive TFs such as GL3, TT8, and PAP1 while up-regulating expression of the negative TF MYBL2.
Effects of Photosynthetic Activity on Anthocyanin Induction in Light Signaling Pathway Mutants and Col0
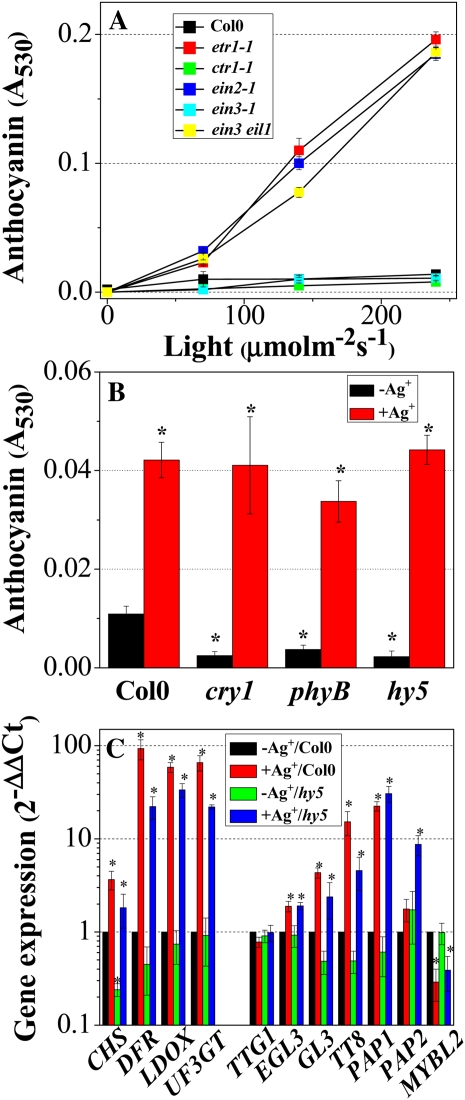
To determine whether the ability of ethylene to suppress anthocyanin biosynthesis is dependent upon light conditions, anthocyanin content was measured in ethylene signaling mutants grown under various light intensities on medium containing 60 mm Suc (Fig. 4A). As reported previously (Solfanelli et al., 2006; Cominelli et al., 2008), light-inducible anthocyanin accumulation is dependent upon light intensity in Col0, becoming saturated at 140 μmol m−2 s−1. In contrast, the etr1-1, ein2-1, and ein3 eil1 mutants showed proportional increases in anthocyanin levels with light intensity, while ctr1-1 and ein3-1 mutants showed comparable anthocyanin accumulation to Col0 (Fig. 4A). These findings imply that the magnitude of ethylene repression is not constant but varies according to light intensity. Moreover, when Col0 and ethylene signaling mutants were grown in Suc-free medium, there was negligible accumulation of anthocyanin, even under high light intensity (240 μmol m−2 s−1; data not shown). Thus, it appears that in the absence of sugar, light is not sufficient for anthocyanin induction.
Figure 4.
Anthocyanin contents and transcript levels of anthocyanin biosynthesis-related genes in Col0, ethylene signaling mutants (etr1-1, ctr1-1, ein2-1, ein3-1, and ein3 eil1), and anthocyanin biosynthesis-related light signaling mutants (cry1, phyB, and hy5). A, Anthocyanin contents of Col0 and ethylene signaling mutants grown at various light intensities. B, Anthocyanin contents in Col0 and light signaling mutants. C, Transcript levels of structural (CHS, DFR, LDOX, and UF3GT) and regulatory (TTG1, EGL3, GL3, TT8, PAP1, PAP2, and MYBL2) genes in Col0 and the hy5 mutant. Transcript levels were quantified by RT-qPCR. Plants were grown for 12 d on half-strength MS medium containing 60 mm Suc under various light intensities (0, 70, 140, and 240 μmol m−2 s−1; A) or complemented with (+Ag+) or without (−Ag+) 1 mm AgNO3 under growth light conditions (140 μmol m−2 s−1; B and C). Error bars represent sd values for the means of three or four independent replicates. Asterisks over bars indicate differences between control (−Ag+/Col0) and treatment or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
To establish whether or not the ethylene suppression observed in Figures 1 to 3 has an impact on the light signaling cascade (Shin et al., 2007), anthocyanin content was measured in several light signaling mutants, including cry1, phyB, and hy5, grown on 60 mm Suc medium with or without silver. cry1, phyB, and hy5 mutants exhibited significant (78%–96%) inhibition of anthocyanin accumulation in the presence of light (P < 0.01; Fig. 4B). This result supports earlier findings that CRY1 and PHYB are the major photoreceptors involved in anthocyanin accumulation and that HY5 is a downstream component of these photoreceptors (Shin et al., 2007). However, it is interesting that when cry1, phyB, and hy5 mutants were grown in the presence of silver, they could accumulate anthocyanin levels comparable to Col0 (Fig. 4B). This finding implies that HY5-independent anthocyanin biosynthesis operates in Arabidopsis and that it is under ethylene repression. Silver treatment significantly affected the transcript levels for ethylene-responsive structural genes (CHS, DFR, LDOX, and UF3GT) and TFs (GL3, TT8, PAP1, and MYBL2; P < 0.01; Fig. 4C). Therefore, it appears that ethylene suppresses the transcription of sugar-regulated anthocyanin biosynthesis genes via a HY5-independent light signaling cascade.
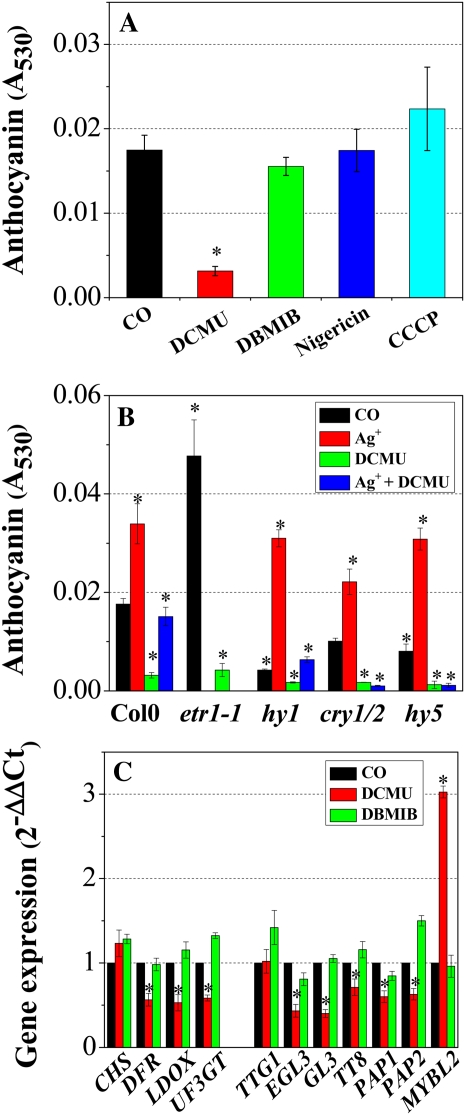
Next, the feasibility of using photosynthetic electron transport-generated signaling (Pfannschmidt et al., 2009) as an alternative, HY5-independent light signaling pathway was examined. Col0 seedlings grown for 9 d in Suc-free growth medium were transferred to filter paper soaked with liquid growth medium containing 60 mm Suc alone, with photosynthetic electron transport inhibitors, or with H+ uncouplers. These seedlings were then subjected to illumination (140 μmol m−2 s−1) for 48 h. The photosynthetic electron transport inhibitors used were DCMU (an electron transport inhibitor that functions at the QB binding site of PSII) and 2,5-dibromo-3-methyl-6-isopropyl-pbenzoquinone (DBMIB; an inhibitor of PQH2 oxidation). The H+ uncouplers were nigericin and carbonyl cyanide m-chlorophenylhydrazone (CCCP; Trebst, 1980). As shown in Figure 5A, anthocyanin accumulation was inhibited strongly (approximately 80%) by DCMU treatment but not by treatment with DBMIB or uncouplers. Moreover, this DCMU effect was not detected in plants incubated in Suc-free medium (data not shown). These findings suggest the involvement of the redox state of the plastoquinone (PQ) pool in sugar and light signaling for anthocyanin accumulation, since DCMU keeps the PQ pool oxidized, whereas it is reduced by DBMIB. Furthermore, anthocyanin accumulation was almost completely inhibited by DCMU treatment in the hy1 (phytochrome chromophore defective mutant), cry1/2 (cry1 and cry2 double mutant), and hy5 mutants (Fig. 5B), which suggests that, with respect to anthocyanin accumulation, photosynthesis-dependent redox status functions as an alternative signal to photoreceptors. DCMU treatment also inhibited anthocyanin accumulation in silver-treated Col0 as well as in etr1 mutant seedlings (Fig. 5B), which provides a further indication that ethylene represses photosynthesis-mediated Suc and light signaling for anthocyanin induction.
Figure 5.
Effects of photosynthesis inhibitors on anthocyanin content and transcript levels of anthocyanin biosynthesis genes in Col0, an ethylene receptor mutant (etr1-1), and light signaling mutants (hy1, cry1/2, and hy5). A, Anthocyanin accumulation in Col0 in the presence of various photosynthesis inhibitors. B, Anthocyanin accumulation in Col0, ethylene receptor mutant, and light signaling mutants in the presence of silver or the photosynthesis inhibitor DCMU. C, Transcript levels of structural (CHS, DFR, LDOX, and UF3GT) and regulatory (TTG1, EGL3, GL3, TT8, PAP1, PAP2, and MYBL2) genes in Col0 in the presence of the photosynthesis inhibitors DCMU and DBMIB. Transcript levels were quantified by RT-qPCR. Nine-day-old seedlings grown on half-strength MS medium containing 7.5 mm Glc were transferred to filter papers soaked with liquid growth medium containing 60 mm Suc supplemented with 10 μm DCMU (DCMU), 5 μm DBMIB (DBMIB), 5 μm nigericin (Nigericin), 5 μm CCCP (CCCP), 1 mm AgNO3 (Ag+), or a mixture of 1 mm AgNO3 and 10 μm DCMU (Ag+ + DCMU) or without (CO) the inhibitors indicated and then incubated for 2 d under growth light conditions (140 μmol m−2 s−1). Anthocyanin content was not measured for silver-treated etr1-1 mutants, in which ethylene signaling is intrinsically blocked by ETR1 mutation. Error bars represent sd values for the means of three or four independent replicates. Asterisks over bars indicate differences between control (CO) and treatment (DCMU, DBMIB, nigericin, CCCP, silver) or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
Transcript levels of structural and regulatory genes involved in anthocyanin accumulation were also investigated to determine whether expression is responsive to photosynthetic electron transport. As shown in Figure 5C, DCMU treatment reduced the expression of three LBGs and regulatory genes, (E)GL3, TT8, and PAP1(2), by 30% to 60% (P < 0.05), while MYBL2 expression was stimulated by about 3-fold over control plants. Consistent with its negligible effect on anthocyanin content, DBMIB treatment did not significantly affect transcript levels of the structural or regulatory genes investigated in this study. Presumably, light and sugar signaling are mediated by the redox state of the photosynthetic electron transport PQ pool.
Transcript Levels of Suc Transporters in Col0 and Ethylene and Light Signaling Mutants
Recently, Sivitz et al. (2008) showed that a SUC1 mutation could lower Suc-inducible anthocyanin accumulation in Arabidopsis. SUC1 expression is highly dynamic at the transcriptional level (Vaughn et al., 2002; Li et al., 2003; Sivitz et al., 2008; Zhou et al., 2009); thus, regulation of SUC1 expression might represent a target for ethylene. Transcript levels were examined for four SUC genes, SUC2, -3, and -4 in addition to SUC1, which are expressed at the seedling stage, as well as in the adult leaves (Gottwald et al., 2000; Meyer et al., 2000; Weise et al., 2000), where anthocyanin accumulation is observed (Supplemental Fig. S2A). Endosperm-specific SUC5 (Baud et al., 2005), SUC8/9 (Sauer et al., 2004), and SUC6/7 coding for aberrant proteins (Sauer et al., 2004) were not examined further in this study. Indeed, DNA microarray analysis comparing the etr1-1 mutant with Col0 showed that ethylene negatively regulates SUC1 and SUC4, whereas it positively regulates SUC2 (Supplemental Table S1). In contrast, expression of SUC3 remained unaltered by the mutation in ETR1 (Supplemental Table S1), implying that SUC1, -2, and -4 might be responsible for the sugar response under light and ethylene signaling.
If SUCs are a common target for Suc, light, and ethylene signaling, the transcript levels of SUCs should be responsive to all three signals. Col0 seedlings were grown in the dark (60 mm, equivalent to 2.16%) or under light (140 μmol m−2 s−1) in the presence of various concentrations of Suc (7.5–90 mm, equivalent to 0.27%–3.24%). As shown in Figure 6A, all four SUCs showed both light- and Suc-dependent expression in Col0 plants. However, as expected, transcriptional accumulation did not occur in plants treated with light or Suc alone, with the exception of SUC1, which showed partial induction of expression with light alone. Under the illumination conditions used here (140 μmol m−2 s−1), the expression levels of SUCs became saturated by Suc concentrations as low as 7.5 mm (0.27%). These findings are also consistent with the ATH GeneChip data analysis, which showed that expression of SUCs requires light (i.e. statistically significant levels of these transcripts were not induced in Col0 grown in 90 mm Suc in the dark; Supplemental Fig. S2B).
Figure 6.
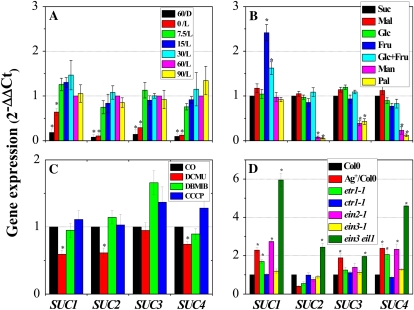
Transcript levels of SUC genes in Col0 and ethylene signaling mutants (etr1-1, ctr1-1, ein2-1, ein3-1, and ein3 eil1). A, Transcript levels of SUC genes in Col0 plants grown under various concentrations of Suc. B, Effects of various sugars on the transcript levels of SUC genes in Col0. C, Effects of photosynthesis inhibitors on the transcript levels of SUC genes in Col0. D, Transcript levels of SUC genes in Col0 and ethylene signaling mutants. For A, B, and D, plants were grown for 12 d on half-strength MS medium supplemented with various concentrations of Suc (0, 7.5, 15, 30, 60, and 90 mm) or various sugars (see Fig. 2B legend) in the dark (D) or under light (L; 140 μmol m−2 s−1) with (Ag+) or without 1 mm AgNO3. C, Growth and treatment conditions were as described in the legend to Figure 5. Transcript levels were quantified by RT-qPCR. Error bars represent sd values for the means of three to four independent replicates. Asterisks over bars indicate differences between control (60/L in A and Suc in B) and treatment or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
Next, we determined whether expression of SUCs could also be induced by Mal, but not by other sugars, as observed in anthocyanin accumulation (Fig. 2B). In contrast, Mal, Glc, Fru, and a combination of Glc + Fru had similar effects to Suc, inducing the expression of four SUCs (SUC1 to -4; Fig. 6B). Although Fru stimulation of SUC1 expression was approximately 2.5-fold higher than the other metabolic sugars, its induction of the other three genes was more or less comparable. It is also noteworthy that the expression of SUC1 differs from the other three genes in terms of Man and Pal specificity (i.e. these sugars induced similar levels of SUC1 expression to Suc while only exerting a marginal effect on expression of the other three SUCs). These findings indicate that there are distinct differences between the sugar signaling of SUC1 expression and SUC2, -3, and -4.
Photosynthetic electron transport mutants, inhibitors, and H+ uncouplers were used to determine whether the expression of SUCs is dependent upon photosynthesis-mediated light signaling, similar to anthocyanin biosynthetic genes. Indeed, the Suc- and light-dependent expression of the four SUCs was unaltered in the anthocyanin biosynthesis-related light signaling mutants cry1, phyB, and hy5 (Supplemental Fig. S3), a finding consistent with DNA microarray analyses for the expression of nine SUCs in cry1 and hy5 grown under low or high light intensity (Supplemental Fig. S2C). However, treatment with the photosynthetic electron transport inhibitor DCMU decreased the expression of SUC1, -2, and -4 by 25% to 50%, while SUC3 expression appeared almost insensitive to the photosynthesis-related signal (Fig. 6C). DCMU exerted greater inhibitory effects on the expression of SUC1 and SUC2 than on SUC4. However, none of the SUCs investigated here exhibited any sensitivity to DBMIB or CCCP treatment at the transcript level, a finding similar to that observed for anthocyanin biosynthetic genes.
Silver-treated Col0 and ethylene signaling mutants grown in the presence of 60 mm Suc were used to examine the ethylene dependency of expression in the four SUC genes. As shown in Figure 6D, the transcript levels of SUC1 and SUC4 were up-regulated more than 2-fold in response to silver treatment and by mutations in ETR1, EIN2, and EIN3 EIL1 compared with Col0, ctr1-1, and ein3-1. However, the expression levels of SUC2 and SUC3 were not affected significantly by the modulation of ethylene signaling. Therefore, it appears that ethylene signaling primarily controls the expression of SUC1 and SUC4.
Anthocyanin and Sugar Contents in suc1-2
Since the expression levels of SUC1 and SUC4 are dependent upon sugars, photosynthesis-mediated light, and ethylene signaling, it is possible that the regulation of transporter activities could represent a convergent target for all three signals. If this is the case, then anthocyanin induction should be influenced by mutations to SUC1 and SUC4. Anthocyanin contents were estimated in SUC1- and SUC4-defective mutants grown for 12 d in 60 mm Suc-containing medium with or without silver. The anthocyanin contents of suc1-2 plants treated with or without silver were 40% to 60% lower than Col0 (Fig. 7A). In contrast, mutation of SUC4 elicited no detectable differences in anthocyanin induction in response to Suc and silver under light (data not shown). Thus, it is likely that SUC1 is involved in anthocyanin regulation by Suc, as reported previously (Sivitz et al., 2008), as well as by ethylene. However, it appears that sugar transport systems other than SUC1 are also involved in regulation, since the SUC1 mutation could not fully interfere with photosynthesis-dependent anthocyanin accumulation (Fig. 7A).
Figure 7.
Anthocyanin and sugar contents in Col0 and the Suc transporter (suc1-2), the ethylene receptor (etr1-1), and the signaling (ein2-1) mutants. A, Anthocyanin contents in Col0 and suc1-2 mutants. B and C, Soluble sugar (Glc, Fru, and Suc) contents in Col0, suc1-2, etr1-1, and ein2-1 plants. For A and C, 9-d-old plants grown on half-strength MS medium containing 7.5 mm Glc were transferred to filter papers soaked with liquid growth medium containing either 0 (−Suc) or 60 mm Suc (CO) complemented with 1 mm AgNO3 (Ag+), 10 μm DCMU (DCMU), or a mixture of 1 mm AgNO3 and 10 μm DCMU (Ag+ + DCMU) or without the inhibitors indicated and incubated for 2 d under growth light conditions (140 μmol m−2 s−1). For B, plants were grown for 12 d on half-strength MS medium supplemented with 60 mm Suc under growth light conditions (140 μmol m−2 s−1). Error bars represent sd values for the means of four or five independent replicates. Asterisks over bars indicate differences between control (CO) and treatment or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
If the root-abundant SUC1 (Sivitz et al., 2008) is involved in the increase in shoot anthocyanin content, then the endogenous levels of soluble sugars such as Suc and its immediate cleavage products, Glc and Fru, might be lower in suc1-2 than in Col0. Furthermore, it might be expected that the soluble sugar contents of ethylene signaling mutants such as etr1-1 and ein2-1 would be higher than that of Col0, since SUC1 expression levels were enhanced when ethylene signaling was inhibited by silver treatment in Col0 seedlings and by mutations in ETR1 and EIN2 (Fig. 6D). Indeed, the soluble sugar (Glc, Fru, and Suc) contents of shoots were 44% lower in suc1-2 but 2.9- and 2.4-fold higher in etr1-1 and ein2-1, respectively (Fig. 7B). In Col0 and the mutants investigated in this study, Glc is the most abundant form of soluble sugar, representing 70% to 90% of total sugar, probably due to rapid Suc hydrolysis before or after entry into the cell (Chaudhuri et al., 2008).
To investigate whether DCMU-mediated down-regulation of SUC1 expression in Col0 and ethylene signaling mutants (Fig. 6C) correlates with soluble sugar contents, Col0 seedlings were grown for 9 d on a solid agar plate without Suc and then transferred to liquid medium containing 60 mm Suc with or without DCMU and silver for 2 d under light. As shown in Figure 7C, DCMU treatment reduced the soluble sugar content of Col0 treated with or without silver by 55% to 60% (Fig. 7C), indicating that photosynthetic electron transport is perhaps responsible for changes in sugar contents via the regulation of SUC1 expression. Alternatively, enhanced sugar contents upon exposure to light could be due to the stimulation of photosynthesis and carbon assimilation. Thus, we estimated sugar contents in Suc-free-grown plants, where change in sugar contents is almost attributable to photosynthesis. Soluble sugar contents from plants grown in Suc-free growth medium were about 20% of that from plants grown in 60 mm Suc-containing medium (data not shown). Thus, light-induced increases in sugar levels in the presence of exogenous Suc are mainly due to exogenous Suc uptake rather than photosynthesis-mediated sugar production.
Suc-Induced Production of Ethylene
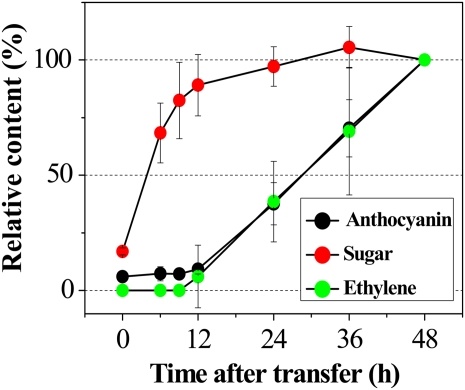
Exogenously supplied Suc enhances ethylene production in a dose-dependent manner in tobacco (Philosoph-Hadas et al., 1985) and rice (Oryza sativa; Kobayashi and Saka, 2000) leaves. Therefore, a time-course analysis was used to investigate the induction kinetics of sugar, anthocyanin, and ethylene production in response to exogenous Suc treatment. Col0 seedlings were cultured for 9 d on growth medium without Suc and then transferred to liquid medium containing 60 mm Suc and grown under light conditions. The sugar content of seedlings started to increase immediately after the onset of Suc treatment and showed almost complete saturation by 12 h after transfer (Fig. 8). Accumulation of anthocyanin pigments and ethylene production started to increase linearly 12 h after transfer. These findings indicate that Suc signaling results in concurrent induction of anthocyanin pigmentation and ethylene production.
Figure 8.
Relative changes in sugars, anthocyanin, and ethylene contents in Col0. Nine-day-old plants grown on half-strength MS medium supplemented with 7.5 mm Glc under illumination (140 μmol m−2 s−1) were transferred to filter papers soaked with liquid growth medium containing 60 mm Suc and incubated for a further 2 d. After 48 h of treatment, the anthocyanin, sugar, and ethylene contents were 0.019 ± 0.001, 7.81 ± 0.73, and 4.92 ± 0.90 nmol plant−1 h−1, respectively. Error bars represent sd values for the means of three or four independent replicates. [See online article for color version of this figure.]
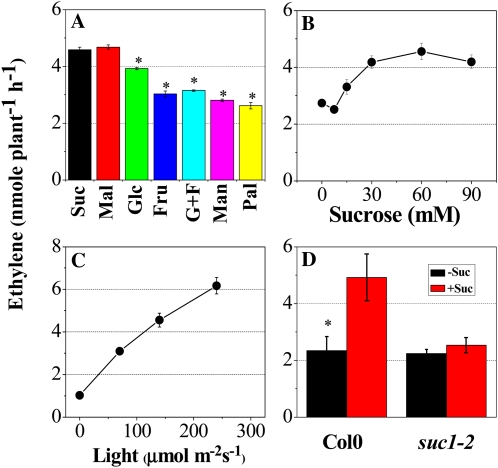
In order to determine whether ethylene is induced preferentially by Suc and Mal relative to other monosaccharides and disaccharides and Man, ethylene production was examined in Col0 seedlings treated with various sugars. Compared with Col0 seedlings treated with Fru, Glc + Fru, Man, and Pal, seedlings grown in Suc, Mal, and Glc produced approximately 1.3- to 1.8-fold more ethylene (Fig. 9A). Thus, as observed in anthocyanin accumulation, ethylene is induced preferentially by Suc and Mal. In Col0 seedlings treated with various concentrations of Suc (0–90 mm) at a given light intensity (140 μmol m−2 s−1), ethylene production increased rapidly at concentrations above 15 mm Suc and reached a near plateau at 30 mm Suc (Fig. 9B). Growth light intensity also influenced ethylene production, generating an almost linear increase in ethylene production with increasing light intensities (Fig. 9C). Finally, to identify whether SUC1 is involved in increased ethylene production, ethylene production was estimated in suc1-2 mutant seedlings. Although Col0 seedlings responded to Suc treatment by increasing ethylene production, this did not occur in the suc1-2 mutant, which suggests that SUC1 is primarily responsible for Suc-induced ethylene production (Fig. 9D).
Figure 9.
Sugar- and light-dependent ethylene production in Col0 and suc1-2 mutants. A, Ethylene produced from Col0 plants grown on half-strength MS medium supplemented with various metabolic and nonmetabolic sugars (see Fig. 2B legend). B, Ethylene produced from Col0 plants cultured on growth medium containing various concentrations of Suc (mm) for 12 d under illumination (140 μmol m−2 s−1). C, Ethylene produced from Col0 plants cultured on growth medium containing 60 mm Suc for 12 d under different light intensities (0, 70, 140, and 240 μmol m−2 s−1). D, Ethylene produced from Col0 and suc1-2 mutants cultured on growth medium containing 7.5 mm Glc (−Suc) or 60 mm Suc (+Suc) for 12 d under illumination (140 μmol m−2 s−1). Error bars represent sd values for the means of three or four independent replicates. Asterisks over bars indicate differences between control (Suc) and treatment or between Col0 and mutants, with statistical significance set at P < 0.05 (t test). [See online article for color version of this figure.]
DISCUSSION
The Ethylene Pathway That Suppresses Anthocyanin Biosynthesis Shares Signaling Components with a Pathway Involved in the Triple Response
Ethylene inhibits anthocyanin accumulation in Arabidopsis seedlings grown in the presence of Suc and light. Anthocyanin accumulation is promoted when ethylene binding is blocked by silver or when its synthesis is inhibited by AVG (Craker and Wetherbee, 1973; Kang and Burg, 1973; Rengel and Kordan, 1987). As in the ethylene triple response (Kendrick and Chang, 2008; Yoo et al., 2009), ethylene suppression of anthocyanin accumulation is probably initiated by the binding of ethylene to redundant receptors such as ETR1, ETR2, ERS1, and ERS2, with ETR1 possibly playing a dominant regulatory role, as suggested previously (Fig. 1C; Takada et al., 2005). Ethylene binds to the ETR1 transmembrane domain, and the hormone signal is transduced to downstream components via the C-terminal region of the receptor. This view is substantiated by the observation that several mutant alleles defective in ethylene binding (i.e. etr1-1, etr1-3, and etr1-4) or coupling to signal output (i.e. etr1-2) exhibit high levels of anthocyanin accumulation (Fig. 1C). In contrast, an ethylene-hypersensitive etr1 null mutant, etr1-7, exhibits constitutive repression of anthocyanin pigmentation. CTR1 is likely to represent a component immediately downstream of or within the ethylene receptor complex, since ctr1 loss-of-function mutants exhibit slightly reduced anthocyanin levels because of constitutive activation of ethylene repression (Fig. 1C). The Arabidopsis response regulator 2-mediated, CTR1-independent alternative ethylene signaling pathway (Hass et al., 2004) is unlikely to contribute to ethylene repression of anthocyanin accumulation. Instead, it is more likely that EIN2, which acts downstream of CTR1 and functions in a wide range of ethylene responses in plants (Cao et al., 2007), is involved in the ethylene signaling pathway that suppresses anthocyanin accumulation. This hypothesis is supported by the finding that pigment accumulation is elevated in the ein2-1 mutant, similar to the situations in Col0 treated with silver and etr1-1 plants (Fig. 1C). Both EIN3 and its homolog EIL1 appear to function redundantly in the ethylene pathway, since anthocyanin accumulation is only elevated significantly in the ein3 eil1 double mutant and not in the ein3-1 or eil1-3 single mutant. Taken together, these results indicate that ethylene represses sugar- and light-induced anthocyanin biosynthesis via a pathway involved in the triple response.
Ethylene Represses Sugar- and Light-Mediated Induction of Anthocyanin Accumulation by Regulating Sugar Transport
Ethylene may suppress anthocyanin biosynthesis by affecting Suc sensing and signaling pathways. The most likely potential target areas for regulating sugar signaling include transporters of monosaccharides and disaccharides or closely related SUC homologs as well as cytosolic hexokinase I, which phosphorylates the Suc metabolite Glc (Rolland et al., 2006). Sugar transport via SUC appears critical, since ethylene repression of anthocyanin accumulation is Suc and Mal specific, but is not observed with Pal, a nonmetabolizable and nontransportable Suc analog (Chandran et al., 2003), with Man, an osmoticum, or with its cleavage products, Glc and Fru (Fig. 2B). This view is further strengthened by the fact that endogenous sugar content, an indirect indicator of Suc transport activity and hence a proxy measure for Suc signal amplitude, correlates positively with anthocyanin content (Supplemental Fig. S4).
Of the four Arabidopsis SUCs investigated here, SUC1 is the most likely target of ethylene signaling in anthocyanin regulation, since SUC1 expression was negatively regulated by ethylene signaling in the presence of light and sugars (Figs. 6 and 7). Consistently, we observed a somewhat linear correlation between the SUC1 transcript level, a proxy measure for SUC activity (Zhou et al., 2009), and anthocyanin content (data not shown). Thus, it would appear that SUC1-mediated transport of Suc is responsible for the induction of anthocyanin by sugar.
The high-affinity SUC2 transporter found in companion cells (Srivastava et al., 2008) is not necessary for ethylene repression of anthocyanin accumulation, since both Suc and ethylene can control pigment levels normally in the suc2-1 null mutant (data not shown). SUC3 is expressed specifically in sieve elements, where it funnels Suc from the mesophyll toward phloem or into pod storage tissues (Meyer et al., 2000). SUC3 is considered to be a putative Suc sensor with similar functions to the yeast sugar sensors SNF3 and RGT2 (Barker et al., 2000). However, its expression is dependent neither on photosynthetic electron transport nor on ethylene repression (Fig. 6). Thus, it is unlikely that SUC3 is involved in integrating light and ethylene signaling pathways for the regulation of anthocyanin biosynthesis at the transcriptional level. However, since SUC activity is also regulated at the translational level (Roblin et al., 1998), we cannot exclude the possibility that SUC3 functions as an integrator for anthocyanin regulatory signals until we have addressed this genetically using suc3 null mutants. SUC4 is localized primarily at the tonoplast of leaf mesophyll cells (Endler et al., 2006), and as with SUC1, SUC4 transcript levels are regulated by both light and ethylene. However, mutation of SUC4 had little if any effect on anthocyanin accumulation, which suggests that SUC4 is unlikely to function as a major factor in sugar and light induction of anthocyanin. Instead, it appears to control Suc transport between subcellular compartments (i.e. from cytosol to vacuole) for further storage or metabolism.
We propose that light and sugar signals perceived by chloroplasts in leaf mesophyll cells first induce SUC1 expression in the roots. Enhanced SUC1 transcript levels in root would be responsible, at least partially, for increased sugar levels in the shoot, which in turn would trigger anthocyanin accumulation in cyanic cells as well as ethylene production (Fig. 10). Currently, it is not known how elevated SUC1 expression in the root induces sugar accumulation in the shoot. If we assume that SUC1 is also regulated at the transcriptional level as for other SUCs (Zhou et al., 2009), the increase in SUC1 transcript levels induced by light and sugar would result in enhanced SUC1 activity. Thus, it is most likely that SUC1 plays a role in transporting exogenous Suc taken by proton-independent sugar transporters in the root tips (Chaudhuri et al., 2008) to the shoot. In accordance with this viewpoint, the sugar content in seedlings after root excision (Fig. 7) was more than 45% lower in the suc1 mutant than in the wild type. However, this view that Suc taken up by the roots would be transported to the leaves via the phloem in Arabidopsis seems implausible, considering that sugar transports from source (shoot-leaf) to sink (root) via the phloem. Rather than acting as a transporter, SUC1 might play a role as a sugar sensor, as purported previously for the yeast sugar sensor SNF3 (Barker et al., 2000). In this context, SUC1-generated sugar sensing in root would be signaled to the shoot. If this were the case, then seedlings without root would show diminished anthocyanin pigmentation in response to sugar treatment, since SUC1-mediated sugar sensing and signaling is expected not to be generated. However, Col0 seedlings with detached roots produced anthocyanin to the extent seen in intact seedlings (data not shown), ruling out a role for SUC1 as a sugar sensor in the root. Thus, at present, SUC1 is more likely responsible for the increased sugar content of the shoot than for sugar sensing/signaling; nonetheless, its specific role in increasing shoot sugar content remains elusive and requires further characterization. However, it is important to note that SUC1 alone does not fully account for the anthocyanin accumulation observed in this study. In particular, it is very likely that an ethylene-repressible, SUC1-independent Suc transport pathway exists, since mutation of SUC1 does not completely inhibit the effect of exogenous Suc on either anthocyanin accumulation or increases in sugar content (Fig. 7), findings consistent with a previous report (Sivitz et al., 2008). Indeed, SUC1-independent Suc uptake pathways in the root have been proposed previously (i.e. one mediated by endocytosis and another by pH-independent sugar transporters; Chaudhuri et al., 2008). However, we failed to inhibit anthocyanin induction using the endocytic inhibitor wortmannin (data not shown, Emans et al., 2002; Etxeberria et al., 2005) or the uncouplers CCCP and nigericin (Fig. 5; Chaudhuri et al., 2008). Several pH-independent sugar transporters are expressed in roots, including the sugar transporter family ERD6-like homologs (At1g08920 and At1g08930), two plastidic Glc transporters (At1g79820 and At5g16150), and four members of the aquaporin gene family (PIP1:2, PIP1:3, PIP2:8, and SIP1:1; Lalonde et al., 2004; Kaldenhoff et al., 2007; Chaudhuri et al., 2008). Although the microarray expression data did not indicate that any of the root-abundant, pH-independent sugar transporter genes were influenced significantly by a mutation in ETR1 (data not shown), further work, including molecular genetic studies, will be required to investigate whether they are involved in anthocyanin pigmentation.
Figure 10.
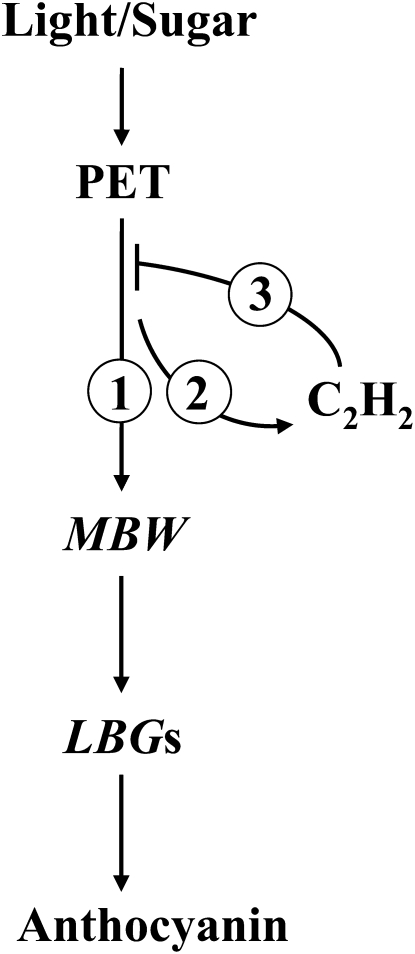
Schematic diagram of the relationship between light, Suc, ethylene signals, and anthocyanin biosynthesis. Step 1, Light and sugar signals generated from photosynthetic electron transport (PET) activates the MBW regulatory complex. It also down-regulates MybL2 expression, which in turn leads to the specific up-regulation of several LBGs, resulting in the accumulation of anthocyanin. Step 2, At the same time, sugar stimulates ethylene production in the presence of light. Step 3, This, in turn, triggers the repression of anthocyanin biosynthesis by interfering with the light-induced sugar signaling that is at least partially mediated by SUC1. The ethylene signaling pathway comprises the ethylene receptors CTR1, EIN2, EIN3, and EIL1.
Sugar Induction of SUC1 Expression Is Distinct from Anthocyanin Accumulation
Although SUC1 expression and anthocyanin accumulation are both regulated by sugars, these processes differ with respect to sugar specificity. Anthocyanin induction is preferentially induced by the metabolizable disaccharides Suc and Mal (Fig. 2). However, induction of SUC1 expression is not restricted to Suc and Mal but can be induced effectively or preferentially by monosaccharide sugars such as Glc and Fru (Fig. 6B). Thus, SUC1 expression is similar to CitSUT2 expression, in that its expression remains unaltered between various sugar treatments (Li et al., 2003). Furthermore, there are no apparent differences between sugars and the osmotica Man and Pal, which also induce SUC1 expression strongly in Col0 plants. These findings suggest that SUC1 induction requires the presence of an osmotic sensor rather than receptors localized at a membrane or intracellularly, such as the Glc sensor HXK1 (Rolland et al., 2006).
Although sugar induction of SUC1 expression requires the presence of light, light signaling components known to be involved in anthocyanin biosynthesis, such as CRY1, PHYB, and HY5, are not involved in SUC1 induction (Supplemental Fig. S3). Thus, it is likely that the light signaling pathway that leads to SUC1 expression differs from the well-characterized, HY5-dependent direct anthocyanin biosynthesis pathway (Shin et al., 2007). In Arabidopsis, photoreceptor-independent SUC1 expression could be induced by photosynthesis-related signaling, and especially the redox state of PQ pools. DCMU treatment causes the PQ pool to become oxidized, and since DCMU inhibited SUC1 expression, it is possible that anthocyanin accumulation might require the PQ pool to be in a reduced state. In contrast, DBMIB treatment, which renders the PQ pool more reduced by inhibiting PQH2 reoxidation by the cytochrome b6f complex, caused little if any inhibition of light-induced SUC1 expression or anthocyanin accumulation (Fig. 5, A and C). Thus, it is possible that the redox state of the PQ pool functions as a sensor for light and sugar signaling for sugar transport and anthocyanin induction (Fig. 10). This view is consistent with earlier reports that indicate that anthocyanin biosynthesis is inhibited by photosynthesis inhibitors in other plants such as turnip seedlings (Schneider and Stimson, 1971) and corn leaves (Kim et al., 2006).
DCMU-mediated inhibition of photosynthetic electron transport would prevent plants from forming chemical energy (ATP) and reducing power (NADPH), resulting in the inhibition of the carbon fixation process and, eventually, sugar biosynthesis. Thus, the effect of DCMU might be attributed to the diminished generation of sugar species such as Glc and Suc. If this were the case, then DBMIB-mediated inhibition of photosynthetic electron transport should also inhibit sugar production and cause an inhibition of anthocyanin accumulation. However, DBMIB treatment results in a negligible reduction of the Suc- and light-mediated induction of SUC1 or anthocyanin genes. Furthermore, light-induced anthocyanin accumulation was not significantly affected by treatment with uncouplers such as nigericin or CCCP, which also inhibit NADPH generation and, hence, prevent sugar production (Fig. 5A). Taken together, these results suggest that events downstream of photosynthetic electron transport such as carbohydrate metabolism are not involved in SUC1 expression, but the results do imply that the redox status of photosynthetic electron transport represents a signal for anthocyanin induction.
How do mesophyll cells sense enhanced levels of sugars such as Suc and Mal and not just their cleavage metabolites, Glc and Fru? The sugar-sensing mechanism is not yet known, but it is likely that enhanced sugar levels outside mesophyll cells induce feedback inhibition of photosynthetic electron transport (Oswald et al., 2001). Such inhibition could result in the PQ pool attaining a more reduced state and thereby acting as a plastid signal for anthocyanin induction. Whatever the detailed sensing mechanism, ethylene negatively regulates HY5-independent, photosynthesis-related anthocyanin accumulation, since DCMU treatment almost completely abolishes anthocyanin accumulation in silver-treated Col0 and etr1-1 plants (Fig. 5B).
Ethylene Repression of Sugar-Induced Anthocyanin Biosynthesis Is Mediated by Preferential Down-Regulation of PAP1 and Up-Regulation of MYBL2
In light-grown Arabidopsis, ethylene repression of anthocyanin induction is regulated at the transcriptional level. Anthocyanin levels correlate positively with transcript levels of LBGs such as DFR, LDOX, and UF3GT, which are mainly under the regulation of bHLH TFs (GL3 and TT8), an R2R3-MYB, PAP1, and an R3-MYB, MYBL2. PAP1 may act as a TF immediately upstream of LBGs, since the cis-element to which it binds (CCAC) is found upstream of the 5′ untranslated regions of DFR, LDOX, and UF3GT (Dare et al., 2008). Moreover, overexpression of PAP1 alone is sufficient to induce anthocyanin accumulation (Borevitz et al., 2000; Tohge et al., 2005). Since PAP1 expression is regulated directly by GL3 at the transcriptional level (Supplemental Fig. S5; Gonzalez et al., 2008), transcriptional down-regulation of GL3 is likely to be an early step in the transcriptional regulatory cascades for ethylene repression of anthocyanin accumulation.
A negative correlation was found between the transcription of genes for TFs that form an active MBW complex and MYBL2 transcript levels under the same conditions. The reciprocal correlation between positive TFs and a negative TF supports the hypothesis that the relative levels of activators and repressors determine whether the transcriptional complex is active, repressed, or inactive (Dubos et al., 2008; Matsui et al., 2008). Very little anthocyanin accumulation was observed in Col0 plants grown under illumination in the absence of Suc or in the presence of Suc supplemented with DCMU, because MYBL2 was expressed strongly (Figs. 3 and 5). Under these conditions, PAP1 expression was also reduced; hence, MYBL2 might form a transcriptional complex with TTG1 and TT8, resulting in an inactive L2BW complex. In contrast, PAP1 was induced significantly in the presence of Suc and light, while MYBL2 transcript levels were down-regulated. Under these conditions, an active MBW complex would be produced in addition to an inactive L2BW complex. Thus, two types of transcriptional regulatory complexes would work in concert to regulate anthocyanin accumulation (i.e. an active MBW complex that enables LBG induction and an inactive L2BW complex that suppresses this process). When silver treatment or mutation of ethylene signaling factors inhibited the ethylene promotion of MYBL2 expression, the inactive L2BW complex would not be formed, or alternatively, the inactive complex would be formed but competed out by the high concentration of the active complex. Under this condition, the active MBW complex would become dominant and hence result in high levels of anthocyanin accumulation.
The bHLHs (i.e. GL3, EGL3, and TT8) and MYBs (i.e. PAP1 and MYBL2) are transcriptionally regulated by Suc and ethylene. In contrast, ethylene signaling has little effect on the mRNA levels of TTG1 (Fig. 3B), which encodes an essential component for the induction of anthocyanin accumulation (Shirley et al., 1995). Taken together, these results suggest that ethylene suppresses the activity of the MBW complex through transcriptional regulation of bHLHs and MYBs. Unexpectedly, MYBL2 transcript levels were not reduced in the ein3 eil1 double mutant but actually stimulated 3-fold or less compared with Col0 plants. However, PAP1 was also significantly induced by approximately 200-fold in the ein3 eil1 double mutant compared with Col0 plants, which may account for the high level of anthocyanin accumulation despite elevated MYBL2 levels.
Suc-Induced Production of Ethylene Represses Anthocyanin Overaccumulation
In Arabidopsis, ethylene production is stimulated by Suc and light (Fig. 9) in a dose- and signal-dependent manner, respectively, similar to leaves in other plants, such as tobacco (Philosoph-Hadas et al., 1985) and rice (Kobayashi and Saka, 2000). Suc appears to regulate ethylene production as well as anthocyanin biosynthesis (Fig. 8), since a mutation in the SUC1 transporter resulted in a partial reduction in both processes (Fig. 9D). Indeed, increases in endogenous sugar levels occur about 9 h earlier than the accumulation of anthocyanin and ethylene (Fig. 8). However, at a molecular physiological level, it remains unclear how Suc induces the ethylene production that negatively regulates anthocyanin induction. Auxin might be involved in this signaling cascade, since exogenous application of Suc increases indole-3-acetic acid concentration by stimulating hydrolysis of the indole-3-acetic acid conjugate (Meir et al., 1989), which is known to induce ACC synthase expression. Alternatively, the reactive oxygen signal induced by elevated levels of Suc may lead to high levels of ethylene production (Chamnongpol et al., 1998). Indeed, hydrogen peroxide is known to induce the expression of some ethylene-responsive genes (Vandenabeele et al., 2003).
Sugar signaling may interfere with the ethylene suppression of anthocyanin accumulation, as observed with Glc repression of ethylene signaling, which mediates seed germination and seedling development (Zhou et al., 1998; Cho et al., 2010). However, this Glc-repressible branch of the ethylene pathway is uncoupled from the ethylene triple response, which shares common components with the anthocyanin repression pathway. Furthermore, if Suc suppresses the ethylene repression signal for anthocyanin induction, then it might be expected that the ethylene-insensitive mutants would show constant levels of anthocyanin, regardless of ethylene levels, since mutations in the ethylene signaling pathway would stop mutants from responding to varying ethylene levels. However, the anthocyanin content of ethylene-insensitive mutants exhibited rather linear and saturation relationships with light intensity (Fig. 4A) and Suc concentration (Fig. 2A), respectively, where ethylene production is not constant but variable (Fig. 9). Thus, it is unlikely that Suc inhibits ethylene repression, but it is possible that ethylene might repress the induction of anthocyanin pigmentation by Suc and Mal.
Although it remains unclear why sugars should induce anthocyanin and ethylene accumulation simultaneously, it is worth noting that the ability of ethylene to repress anthocyanin accumulation varies with respect to light intensity and sugar concentration. Higher levels of repression were observed with increasing Suc concentrations at a fixed light intensity or with higher light intensities at a given Suc concentration. Anthocyanin plays a vital role in protecting plants from the damaging effects of sunlight and, hence, is clearly beneficial to plants. However, the accumulation of anthocyanins above a certain level would be disadvantageous, since it would cause imbalances between primary and secondary metabolites. Assuming a strong selection pressure in favor of the economic usage of available resources and optimized usage of light, plants may accumulate only enough anthocyanin to provide photoprotection but avoid overproduction of the pigment, as excess anthocyanin may prevent light from reaching the underlying photosynthetic tissues, resulting in higher light saturation and compensation points (Albert et al., 2009).
In conclusion, we have demonstrated the presence of complex positive and negative networks that govern anthocyanin biosynthesis pathways in Arabidopsis. We also elucidated the presence of a mesophyll-derived intercellular signaling pathway, which is dependent upon the redox state of photosynthetic electron transport and induces anthocyanin biosynthesis mainly in the outer epidermal cells. Furthermore, we established that the ethylene signaling pathway, which shares some common components with the triple response, inhibits sugar- and light-mediated induction of anthocyanin accumulation via down-regulation of SUC1. In this study, we identified SUC1 as an integrating element of the signaling pathways initiated by light and ethylene. However, the SUC1-independent signaling cascades remain to be characterized.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The following Arabidopsis (Arabidopsis thaliana) lines were used: wild-type Col0 and Wassilewskija ecotypes; ethylene receptor mutants of ETR1 (etr1-1, etr1-2, etr1-3, etr1-4, and etr1-7), ERS1 (ers1-2) and ERS2 (ers2-1), and ETR2 (etr2-1); ethylene signaling mutants, including ctr1-1, ein2-1, ein3-1, eil1-3, and the ein3 eil1 double mutant; anthocyanin biosynthesis-related single (hy1, cry1, phyB, and hy5-221) and double (cry1/2) mutants; double mutants of TFs GL3 and EGL3 (egl3 gl3); and mutants of the Suc transporters 1, 2, and 4 (suc1-2, suc2-1, and suc2-4). Seeds were treated with 70% ethanol for 5 min and then sterilized with 15% bleach. After washing five times with sterilized water, seeds were plated onto solidified half-strength MS medium supplemented with various concentrations of Suc (0, 15, 30, 60, and 90 mm Suc), different metabolic sugars such as Mal (60 mm), Glc (60 mm), and Fru (60 mm), a 1:1 mixture of Glc:Fru (both at 30 mm), the sugar alcohol Man (60 mm), or the nonmetabolic sugar Pal (60 mm). In addition, these supplemented media contained ACC (10 μm), 1 mm AgNO3, or 10 μm AVG. Seedlings were grown at various light intensities (0, 70, 140, 200, and 240 μmol m−2 s−1) under an 18-h-light/6-h-dark photoperiod (22°C/20°C). When required, electron transport inhibitors (10 μm DCMU, 5 μm DBMIB, 10 μm CCCP, or 10 μm nigericin) were included in the growth media.
Affymetrix GeneChip Analysis
Analyses were performed on wild-type Col0 and the etr1 allele mutant etr1-1. Total RNAs were extracted from 12-d-old seedlings grown in the presence of 3% Suc. Hybridizations to the Arabidopsis ATH1 GeneChip arrays (Affymetrix) were performed by the Genomics Core at the Center for Reproductive Biology at Washington State University. Microarrays were scanned on a Hewlett-Packard Gene Array Scanner, and expression analyses were performed using Affymetrix microarray suite software (MAS version 5.0) with standard parameters. Three independent, replicated experiments were performed, and the output of Affymetrix MAS for each independent experiment was subjected to further analyses using Excel (Microsoft). Signal values (indicating the relative abundance of a particular transcript) and detection call values (indicating the probability that a particular transcript is present) were generated by Affymetrix MAS 5.0.
Real-Time Reverse Transcription-qPCR Analysis
Total RNA was extracted with TRI reagent (Molecular Research Center), followed by DNaseI (Takara) treatment. After a NucleoSpin RNA Cleanup (Macherey-Nagel), cDNA was synthesized from 1 μg of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using the CFX96 Real Time System (Bio-Rad), following the manufacturer’s instructions. All reactions were performed with the Dynamo HS SYBR Green qPCR Kit (Finnzymes), according to the procedure described by the manufacturer. Reactions were performed in triplicate using 5 μL of 2× Dynamo HS Master Mix, 0.5 μm of each primer (Supplemental Table S2), 2 μL of 20-fold-diluted cDNA, and nuclease-free water (Roche Diagnostics) to a final volume of 10 μL. A negative control using water was included in each run. Reactions were incubated at 95°C for 15 min, followed by 40 cycles of amplification at 94°C for 10 s and then 62°C for 30 s, after which a final extension step was performed at 72°C for 30 s. Fluorescence was measured at the end of each extension step. Amplification was followed by melting curve analysis with continual fluorescence data acquisition during the 65°C to 95°C melt. The raw data were analyzed with CFX Manager software (version 1.1), and expression was normalized to actin2 (At3g18780) to minimize variation in cDNA template levels. For each gene, a standard curve was generated using serial dilutions of cDNA, and the resultant PCR efficiency was in the 90% to 99.5% range. To ensure that transcripts of single genes were amplified, qPCR amplicons were sequenced. Relative expression levels were calculated using the comparative threshold (Ct; cycle value) method. Fold changes (2–ΔΔCt) were expressed relative to wild-type seedlings grown in Suc-containing medium in the light. Mean values were obtained from three to five biological replicates, each determined in triplicate.
Measurement of Anthocyanin and Soluble Sugar Contents
For anthocyanin extraction, 20 seedlings per sample were placed in 600 μL of 1% HCl in methanol (v/v) and then incubated overnight in the dark at 4°C with gentle shaking. After extraction, 400 μL of water and 400 μL of chloroform were added to the extract and mixed. After centrifugation at 12,000 rpm for 2 min, the absorbance of the supernatant was measured at 530 and 657 nm, and the concentration of anthocyanin was calculated using A530 − 0.25 A657 (Rabino and Mancinelli, 1986). To extract soluble sugars, 20 root-excised seedlings were ground to powder in liquid N2 and then extracted in 80% (v/v) ethanol at 80°C for 30 min. After centrifugation at 12,000 rpm for 2 min, the supernatant was decanted and stored on ice. This extraction procedure was performed on the pellet three more times, and the collected supernatants were combined. After depigmentation with chloroform (1:3, v/v, extract:chloroform), ketose sugars were degraded with 1.0 n NaOH (1:1, v/v) for 5 min. The sugar content was determined spectrophotometrically at 520 nm using the Resorcinol method, with Suc as the standard (Roe, 1934). Mean values were obtained from three or four independent replicates. Monosaccharide analysis was performed with high-performance anion-exchange chromatography using BioLC (DX 500 Chromatography System; Dionex) equipped with a pulsed amperometric detector (ED 50; Dionex) as described elsewhere (Park et al., 1999). Sugar extract (10 μL) was injected and fractionated on a CarboPac PA-1 column (4 × 250 mm; Dionex) preequilibrated with 200 mm NaOH. Fractions were eluted in isocratic mode at a rate of 1 mL min−1. d-Glc, d-Fru, and d-Suc (Sigma) were used as standard monosaccharides and disaccharides. All the reagents used for carbohydrate analysis were of American Chemical Society grade.
Measurement of Ethylene
For ethylene collection, petri dishes containing 11-d-old plants were opened to remove trapped air, and then the original lids were replaced by lids with silicone rubber seals. After a further 24 h of incubation under the same conditions, 1 mL of gas was withdrawn from each plate using a gas-tight syringe. The gas was injected into a gas chromatograph equipped with a flame ionization detector (GC-14B; Shimadzu). The carrier gas (N2) flow rate was 60 mL min−1. The detector response was standardized by injecting known amounts of ethylene prepared by serial dilution. Means and sd values were calculated from three experiments.
Bioinformatic Analyses
The microarray collection in Genevestigator (https://www.genevestigator.com) was searched in order to compare the different expression levels of SUC genes in the callus, flower, seed, adult leaf, root, and seedling. Furthermore, ATH GeneChip data were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) to compare gene expression levels in ethylene and light signaling-related mutants. The GSE7432, GSE12715, GSE18631, GSE3704, GSE3416, GSE5174, and GSE7743 series were used (16, 12, 4, 10, 18, 13, and 21 chips, respectively). The data generated using the Affymetrix ATH1 GeneChip were normalized using qspline and processed with cubic spline normalization using quantiles to adjust for signal variation between chips (Workman et al., 2002). Gene expression-level summarization was performed with Robust Multi-Chip Analysis using a median polish algorithm (Irizarry et al., 2003).
Data Analysis
The significance of differences between data sets was evaluated using Student’s t test. Calculations were carried out with OriginPro 8 (OriginLab).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Anthocyanin content in Col0 and the ctr1-1 mutant.
Supplemental Figure S2. Transcript levels of SUC in Col0, cry1, and hy5 seedlings.
Supplemental Figure S3. Transcript levels of SUC1, SUC2, SUC3, and SUC4 in Col0, hy1, cry1/2, and hy5 seedlings.
Supplemental Figure S4. Correlation between anthocyanin and soluble sugar contents.
Supplemental Figure S5. Anthocyanin content and transcript levels of anthocyanin biosynthesis-related genes in Col0 and egl3 gl3 seedlings.
Supplemental Table S1. Transcript levels of some genes involved in anthocyanin biosynthesis and Suc transport in Arabidopsis Col0 and etr1-1 seedlings.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. D. Pouchnik (Washington State University) for performing the GeneChip hybridizations and scanning. We are grateful to Drs. B. Binder (University of Tennessee), J. Sheen (Harvard Medical School), S.K. Song (Yonsei University), and J. Ward (University of Minnesota) for providing seeds of the ethylene signaling mutants, the gin2-1 mutant, the egl3 gl3 double mutant, and the suc1-2 mutants.
References
- Ahmad M, Cashmore AR. (1997) The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11: 421–427 [DOI] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Irving LJ, Jameson PE, Davies KM. (2009) Light-induced vegetative anthocyanin pigmentation in Petunia. J Exp Bot 60: 2191–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Takahashi S, Ono K, Ishimaru K, Ohsugi R. (1999) Molecular cloning and expression analysis of a gene for a sucrose transporter in maize (Zea mays L.). Plant Cell Physiol 40: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW. (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C. (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS. (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamnongpol S, Willekens H, Moeder W, Langebartels C, Sandermann H, Jr, Van Montagu M, Inzé D, Van Camp W. (1998) Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc Natl Acad Sci USA 95: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Reinders A, Ward JM. (2003) Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem 278: 44320–44325 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. (1998) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J 15: 69–77 [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Hörmann F, Lalonde S, Brady SM, Orlando DA, Benfey P, Frommer WB. (2008) Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J 56: 948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR. (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95: 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Sheen J, Yoo SD. (2010) Low glucose uncouples hexokinase1-dependent sugar signaling from stress and defense hormone abscisic acid and C2H4 responses in Arabidopsis. Plant Physiol 152: 1180–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C. (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165: 886–894 [DOI] [PubMed] [Google Scholar]
- Craker LE, Wetherbee PJ. (1973) Ethylene, light, and anthocyanin synthesis. Plant Physiol 51: 436–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare AP, Schaffer RJ, Lin-Wang K, Allan AC, Hellens RP. (2008) Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Hammer PE. (1995) Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol 108: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Emans N, Zimmermann S, Fischer R. (2002) Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria E, Baroja-Fernandez E, Muñoz FJ, Pozueta-Romero J. (2005) Sucrose-inducible endocytosis as a mechanism for nutrient uptake in heterotrophic plant cells. Plant Cell Physiol 46: 474–481 [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196–203 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P, Schäfer E, Nagy F. (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6: 446–452 [DOI] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al. (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Niedermeier M, Feuerstein A, Hornig J, Sauer N. (2010) An ABA-responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression. Planta 232: 911–923 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong ST, Goto-Yamamot N, Kobayashi S, Esaka M. (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci 167: 247–252 [Google Scholar]
- Kaldenhoff R, Bertl A, Otto B, Moshelion M, Uehlein N. (2007) Characterization of plant aquaporins. Methods Enzymol 428: 505–531 [DOI] [PubMed] [Google Scholar]
- Kang BG, Burg SP. (1973) Role of ethylene in phytochrome induced anthocyanin biosynthesis. Planta 110: 227–235 [DOI] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. (2008) Ethylene signaling: new levels of complexity and regulation. Curr Opin Plant Biol 11: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G. (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee BH, Kim SH, Ok KH, Cho KY. (2006) Response to environmental and chemical signals for anthocyanin biosynthesis in non-chlorophyllous corn (Zea mays L.) leaf. J Plant Biol 49: 16–25 [Google Scholar]
- Kobayashi H, Saka H. (2000) Relationship between ethylene evolution and sucrose content in excised leaf blades of rice. Plant Prod Sci 3: 398–403 [Google Scholar]
- Kubo H, Peeters AJM, Aarts MGM, Pereira A, Koornneef M. (1999) ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB. (1997) Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275: 1298–1300 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB. (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C. (2001) The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol 125: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Shi JX, Weiss D, Goldschmidt EE. (2003) Sugars regulate sucrose transporter gene expression in citrus. Biochem Biophys Res Commun 306: 402–407 [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV. (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Matsukura C, Saitoh T, Hirose T, Ohsugi R, Perata P, Yamaguchi J. (2000) Sugar uptake and transport in rice embryo: expression of companion cell-specific sucrose transporter (OsSUT1) induced by sugar and light. Plant Physiol 124: 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Riov J, Philosoph-Hadas S, Aharoni N. (1989) Carbohydrates stimulate ethylene production in tobacco leaf discs. III. Stimulation of enzymatic hydrolysis of indole-3-acetyl-l-alanine. Plant Physiol 90: 1246–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Melzer M, Truernit E, Hümmer C, Besenbeck R, Stadler R, Sauer N. (2000) AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J 24: 869–882 [DOI] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K. (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Morgan PW, Drew MC. (1997) Ethylene and plant responses to stress. Physiol Plant 100: 620–630 [Google Scholar]
- Neff MM, Chory J. (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. (2001) Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA 98: 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Komamine A. (1986) Effects of growth regulators on the induction of anthocyanin synthesis in carrot suspension cultures. Plant Cell Environ 27: 1361–1368 [Google Scholar]
- Park YI, Wood HA, Lee YC. (1999) Monosaccharide compositions of Danaus plexippus (monarch butterfly) and Trichoplusia ni (cabbage looper) egg glycoproteins. Glycoconj J 16: 629–638 [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Murrell JR, Shirley BW. (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis: further evidence for differential regulation of “early” and “late” genes. Plant Physiol 113: 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Bräutigam K, Wagner R, Dietzel L, Schröter Y, Steiner S, Nykytenko A. (2009) Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann Bot (Lond) 103: 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Meir S, Aharoni N. (1985) Carbohydrates stimulate ethylene production in tobacco leaf discs. II. Sites of stimulation in the ethylene biosynthesis pathway. Plant Physiol 78: 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Baudry A, Lepiniec L, Grotewold E. (2006) The regulation of flavonoid biosynthesis. Grotewold E, , The Science of Flavonoids. Springer, Berlin, pp 97–122 [Google Scholar]
- Rabino I, Mancinelli AL. (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengel Z, Kordan HA. (1987) Effects of growth regulators on light-dependent anthocyanin production in Zea mays seedlings. Physiol Plant 69: 511–519 [Google Scholar]
- Roblin G, Sakr S, Bonmort J, Delrot S. (1998) Regulation of a plant plasma membrane sucrose transporter by phosphorylation. FEBS Lett 424: 165–168 [DOI] [PubMed] [Google Scholar]
- Roe JH. (1934) A colorimetric method for the determination of fructose in blood and urine. J Biol Chem 107: 15–22 [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Saftner RA, Wyse RE. (1984) Effect of plant hormones on sucrose uptake by sugar beet root tissue discs. Plant Physiol 74: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N, Ludwig A, Knoblauch A, Rothe P, Gahrtz M, Klebl F. (2004) AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J 40: 120–130 [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Stimson WR. (1971) Contributions of photosynthesis and phytochrome to the formation of anthocyanin in turnip seedlings. Plant Physiol 48: 312–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. (2008) Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Ishimaru K, Minamisawa K, Kamada H, Ezura H. (2005) Expression of a mutated melon ethylene receptor gene Cm-ETR1/H69A affects stamen development in Nicotiana tabacum. Plant Sci 169: 935–942 [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Trebst A. (1980) Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation states. Methods Enzymol 69: 675–715 [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al. (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR. (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. (2001) Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J 25: 675–685 [DOI] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM. (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Van Der Luit A, Knegt E, Vermeer E, Mol JNM, Kooter JM. (1995) Identification of endogenous gibberellins in petunia flowers (induction of anthocyanin biosynthetic gene expression and the antagonistic effect of abscisic acid). Plant Physiol 107: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Saxild HH, Nielsen C, Brunak S, Knudsen S. (2002) A new non-linear normalization method to reduce variability in DNA microarray experiments. Genome Biol 3: research0048.1–research0048.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci 14: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Chan K, Wang TL, Hedley CL, Offler CE, Patrick JW. (2009) Intracellular sucrose communicates metabolic demand to sucrose transporters in developing pea cotyledons. J Exp Bot 60: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.