Abstract
Binding of hepatitis C virus (HCV) RNA to core, the capsid protein, results in the formation of the nucleocapsid, the first step in the assembly of the viral particle. A novel assay was developed to discover small molecule inhibitors of core dimerization. This assay is based on time-resolved fluorescence resonance energy transfer (TR-FRET) between anti-tag antibodies labeled with either europium cryptate (Eu) or allophycocyanin (XL-665). The N-terminal 106-residue portion of core protein (core106) was tagged with either glutathione-S-transferase (GST) or a Flag peptide. Tag-free core106 was selected as the reference inhibitor. The assay was used to screen the library of pharmacologically active compounds (LOPAC) consisting of 1,280 compounds and a 2,240-compound library from the Center for Chemical Methodology and Library Development at Boston University (CMLD-BU). Ten of the 28 hits from the primary TR-FRET run were confirmed in a secondary amplified luminescent proximity homogeneous assay (ALPHA screen). One hit was further characterized by dose–response analysis yielding an IC50 of 9.3 μM. This 513 Da compound was shown to inhibit HCV production in cultured hepatoma cells.
Introduction
Hepatitis C virus (HCV) infects over 170 million people worldwide,1,2 being the main cause of chronic liver disease in humans. The HCV pathogenic agent is a lipid-encapsulated RNA flavivirus,3 which despite its small 9.5 kb genome, is extremely effective at evading the immune system of its human host: after initial contact, 70% of individuals become persistently infected.2 This major infectious disease has recently worsened because the increasingly observed co-infection with HIV considerably affects prognosis of the patients.4 Despite intense efforts, there is still no anti-HCV vaccine and the only available treatment, based on the use of pegylated interferon α combined with ribavirin, cures less than half of the patients.5 The viral RNA encodes 10 proteins. Two of these, the NS3/NS4A protease and the NS5B polymerase enzymes, are the main focus of the development of specific inhibitors.6,7 HCV, however, undergoes multiple mutations during its life cycle, and escape mutants arise frequently during treatment with enzyme inhibitors.6,8 Alternate or complementary targets are therefore needed in order to implement treatments combining drugs acting on different proteins and pathways.
Core, HCV's most conserved (capsid) protein, is an attractive novel therapeutic target since its oligomerization plays an essential role in forming the viral particle.9 Inhibition of the dimerization might thus be a useful approach to find new anti-HCV drugs. We previously described an amplified luminescent proximity homogeneous assay (ALPHA screen)-based method to identify core-derived peptides that inhibit core dimerization and reduce production of HCV by hepatoma cells.10 We now report a new time-resolved fluorescence–resonance energy transfer (TR-FRET) based assay for high-throughput screening (HTS) for small molecule inhibitors of core dimerization.
Materials and Methods
Compound Libraries
LOPAC
The Library of Pharmacologically Active Compounds (LOPAC) contains 1,280 compounds representing all major target classes, was purchased from Sigma-Aldrich (St. Louis, MO). The molecular weights of all the compounds are <500 Da. The compounds are pre-solubilized and normalized in ready-to-use 100% DMSO stocks. The compounds were screened at 12.5 μM final concentration.
CMLD-BU Library
The Center for Chemical Methodology and Library Development at Boston University (CMLD-BU) library contains 2,240 compounds. The molecular weights of the compounds range from 200 to 700 Da. The average molecular weight is 500 Da. This library contains arrays of chemically diverse molecules derived from synthetic methods developed at the CMLD-BU.11–14 The compounds were solubilized in 100% DMSO and the library was formatted at the HTS facility at The Scripps Research Institute–Scripps Florida. The compounds were screened at 15 μM final concentration.
Cloning, Expression, and Purification of Proteins
The cloning, expression, and affinity purification of N-terminal 106-residue portion of core protein (core106), Flag-core106, and GST-core106 were done as per previously published protocols.10 Core106 was produced by expression of the gene in Escherichia coli, extraction by the appropriate buffers, and affinity purification using Nickel agarose. Several batches of protein have been produced on a regular basis, which have remained stable over several years.
ELISA for Confirming Core106 Dimerization
Dimerization between GST-core106 and Flag-core106 was initially confirmed by using a sandwich ELISA on a glutathione (GSH)-coated 96-well plate, as previously reported.10 Mouse anti-Flag antibody (Sigma Aldrich, St. Louis, MO), anti-mouse IgG (Jackson Immuno Research Laboratories, West Grove, PA), and Ultra TMB®-horseradish peroxidase (HRP) substrate (Thermo Fisher Scientific, Rockford, IL) were used for the detection of the complex.
TR-FRET for Monitoring Core106 Dimerization and Screening for Inhibitors
TR-FRET was used for the design of the core106 dimerization assay. This assay is based on fluorescence–resonance energy transfer between anti-tag antibodies alternatively labeled with europium cryptate (Eu) and allophycocyanin (XL-665) (Cisbio US, Bedford, MA). When 2 tagged proteins interact, a transfer of energy occurs between the Eu (donor) and the XL-665 (acceptor).15,16 Core106, the 106-residue core domain, was tagged with either GST or Flag peptide in addition to a 6-His tag for purification purposes. We also prepared the same core domain with a 6-His tag as a reference inhibitor/competitor in the core–core interaction assay. Free core106 was used as a control inhibitor at 1 μM. The proteins were diluted to working concentrations in “protein buffer” (100 mM HEPES pH 7.5, 1 mM EDTA, 5 mM DTT, 0.1% CHAPS, 10% glycerol). The antibody-tagged fluorophores were diluted to working concentrations in “TR-FRET buffer” (PBS pH 7.5, 100 mM HEPES, 0.4 M KF, 0.1% BSA). The tagged protein domains were mixed together (27 nM of GST-core106 and 34 nM of Flag-core106). Then Eu-conjugated anti-GST antibody [(part# 61GSTKLB) Cisbio US, Bedford, MA] was added at a concentration of 1.8 ng/well (0.09 ng/μL). Finally, a XL-665-conjugated anti-Flag antibody [(part# 61FG2XLB) Cisbio US, Bedford, MA] was added to the plate at a concentration of 20 ng/well (1 ng/μL). Both antibodies were added in relative concentrations recommended by the manufacturer (www.htrf-assays.com). The interacting proteins, together with the fluorophore-coupled antibodies, were incubated for 4 h at room temperature. Interaction between the 2 protein domains and illumination with 330 nm light resulted in transfer of fluorescence energy and emission at 665 nm. The assays were executed in small volume black 384-well polystyrene plates (20 μL) [(part# 784076) Greiner Bio-One International A.G. Kremsmuenster, Austria] and measured on a ViewLux Instrument (Perkin Elmer, Waltham, MA). To screen for inhibitors, the assay components were distributed with an automated liquid dispenser (Aurora FRD, Beckman Coulter, Fullerton, CA) and compounds to be screened were added using the Pin-Tool (Genomics Institute of the Novartis Research Foundation (GNF), San Diego, CA) diluted to the appropriate concentrations as DMSO solutions. The control response curve (CRC) plate was set up using another automated liquid dispenser (Biomek FX, Beckman Coulter, Fullerton, CA).
A screen was performed against the LOPAC collection using the core106 TR-FRET assay in four 384-well plates; interlaced with these plates were 5 DMSO control plates to monitor Z-factor, signal-to-background ratio, and compound carryover. Two CRC plates were also prepared for a screen that involved 11 plates total. Plates were analyzed sequentially and the data were examined for artifacts and systematic signal degradation/changes over time.
Additionally, the core106 TR-FRET assay was used in a screen against the CMLD-BU compound collection. The CMLD-BU collection consists of 6 plates in a 384-well format. Two DMSO control plates were added at the beginning and end of the run.
TR-FRET Data Calculations
The ViewLux instrument used for reading the results of the TR-FRET assay (Perkin Elmer, Waltham, MA) includes a filter set with 617 and 671 nm, which are within the range of emission wavelength for Eu. The assay components were measured at 620 nm for the specific Eu signal (background) and 665 nm for the fluorescence transfer signal between the interacting fluorophores-tagged proteins. The TR-FRET signal was calculated as (665/620) × 10,000.15,16
Amplified Luminescent Proximity Homogeneous Assay (ALPHA screen)
ALPHA screen is based on the use of photoactive donor and acceptor beads that recognize specific tags on interacting proteins.17 GSH-coated donor beads and anti-Flag antibody-coated acceptor beads (AlphaLISA® beads) for the assay were obtained from Perkin Elmer Lifesciences. Core106 dimerization was confirmed using ALPHA screen technology in which a core106 was tagged with either GST tag or a Flag peptide tag. Tag-free core106 was used as a reference competitor in the assay. The assays were designed as per a previously published protocol.10 This assay was used as a secondary confirmatory screen for the hit compounds identified in the primary TR-FRET screen.
HCV 2a J6/JFH-1 Culture in Huh-7.5 Cells
Infectious HCV 2a strain J6/JFH-1 was produced using a protocol previously published.18–20 The addition of SL201 (CMLD-BU library) was done as per previously published protocols10 for an initial 72 h period [early stage (T1)] and an additional 72 h [late stage (T2)]. The compound was dosed from 0.001 to 100 μM with 1:10 serial dilutions.
Real-Time RT-PCR Analysis of HCV 2a Infected Huh-7.5 Cell Lysate Treated With SL201
Real-time RT-PCR protocols, primers, and probes were used as previously published.21–22 RNA was purified from HCV 2a infected Huh-7.5 cell lysates after treatment with varying concentrations of SL201. Real-time RT-PCR was carried out using a Roche LightCycler RNA amplification kit (cat#12015145001, Roche Diagnostics, Indianapolis, IN).
Results
To accurately measure core dimerization and its inhibition, we developed several quantitative assays of increasing sensitivity: ELISA, TR-FRET, and ALPHA screen. Each of the 3 assays provides different advantages: the ELISA is simplest and least expensive, demonstrating that the dimerization occurs but requires a washing step; TR-FRET is most amenable to HTS format; and ALPHA screen is most sensitive (signal-to-noise ratio over 50-fold), allowing study of compounds with an IC50 lower than 20 μM. However, the cost-per-assay point was higher for ALPHA screen than for TR-FRET.
Purification of core106 Fragments and Verification of Dimerization
Each of the different HCV core106 fragments tagged with an N-terminal GST or Flag and a C-terminal sequence of 6 or 8 His residues was expressed in E. coli and purified to homogeneity by affinity chromatography on Ni-NTA agarose columns. The composition and mass of core106 were confirmed by mass spectrometry. The size of the tagged core proteins was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and was confirmed by immunoblot analysis using rabbit anti-core antibody generated against core106.10
Heterodimerization of Core106 Shown by ELISA
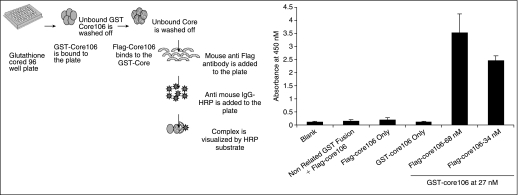
A sandwich ELISA was used for the original confirmation of the GST-core106/Flag-core106 heterodimerization. GST-core106 was adsorbed on a microtiter plate coated with GSH. Flag-core106 was added and mouse anti-Flag antibody, anti-mouse IgG-HRP, and an HRP substrate were used to visualize core106 heterodimerization. As shown in Figure 1, heterodimerization of GST-core106 and Flag-core106 was shown as ∼30-fold signal over the background. Buffer only, GST-core106 alone, and Flag-core106 alone were included as the background controls in the assay. An unrelated GST-tagged protein (1 μg) was adsorbed on a GSH-coated plate and when mixed with Flag-core106 gave a signal similar to the background in the assay, confirming that the GST-core106/Flag-core106 interaction was specific.
Fig. 1.
ELISA scheme: glutathione-coated 96-well plates were coated with GST-core106 and incubated overnight at 4°C. The unbound protein–antibody was washed off at every coating step. Different concentrations of Flag-core106 were added to the plate and incubated for 2 h at 37°C. Then, mouse anti-Flag antibody was added and incubated for 2 h at room temperature. Anti-mouse IgG was added and incubated for 1 h at room temperature. The complex was visualized by the addition of Ultra TMB-HRP substrate. Figure 1 data: confirming GST-core106/Flag-core106 dimerization: GST-core106 was kept constant at 27 nM/well and Flag-core106 was dosed at 68 and 34 nM/well. Buffer only, GST-core106 alone, Flag-core106 alone were included in the assay as background controls. An unrelated GST-tagged protein (1 μg) was adsorbed on a glutathione-coated plate and when mixed with Flag-core106 gave a signal similar to the background in the assay. Error bars represent standard deviations (SDs) of n = 3 values in 3 assays.
Dimerization of Core106 Quantified by TR-FRET
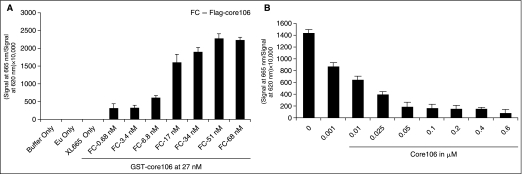
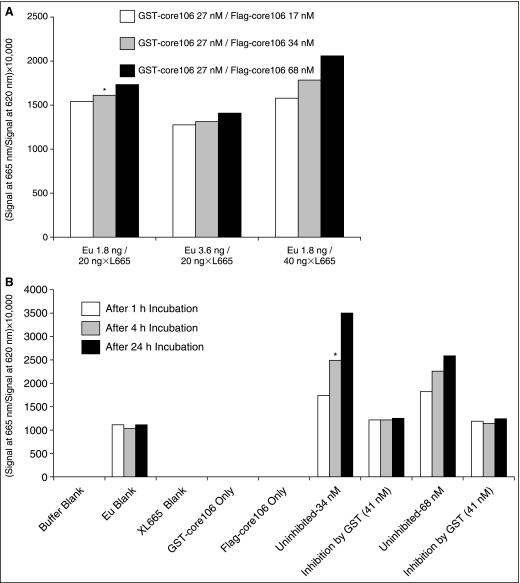
A TR-FRET assay was designed to validate core106 dimerization in vitro. Eu-tagged anti-GST and XL-665-tagged anti-Flag antibodies were used to measure the core–core interaction. GST-core106 was used at a concentration of 27 nM and Flag-core106 at 34 nM. This optimal ratio of 0.8 was based on the dose–response studies (Fig. 2). Use of higher ratios did not significantly improve the signal-to-background window. The concentrations of Eu-tagged anti-GST and XL-665-tagged anti-Flag antibodies (Fig. 3A) were optimized to identify the ratio of concentrations that produced the highest TR-FRET signal-to-background value. Using 1.8 ng/well of Eu-tagged anti-GST and 20 ng/well of XL-665-tagged anti-Flag antibodies produced a robust TR-FRET signal and was chosen as the optimal concentrations for the CMLD-BU run. Using 40 ng/well of XL-665-tagged anti-Flag antibodies produced a somewhat better signal than 20 ng/well but a lower concentration was selected to keep the cost-per-well down. A 24 h incubation time–course study suggested that incubation times longer than 4 h marginally improved the signal-to-background window (Fig. 3B). Nevertheless, the 4 h incubation was preferred in the interest of the gain of time. The final 384-well assay protocol is summarized Table 1 As shown in Figure 2A, dimerization of the 2 fusion proteins could be quantified over a wide range of concentrations. Dose–response analysis showed that core106 completely inhibited Flag-core106 binding to GST-core106, with an IC50 of 89 nM (Fig. 2B).
Fig. 2.
(A) Time-resolved fluorescence–resonance energy transfer (TR-FRET) assays confirming core dimerization. Concentration of GST-core106 was kept constant at 27 nM. Flag-core106 concentrations ranged from 0.068 to 680 nM. The transfer of fluorescence was measured as ratio of (signal at 665 nm/signal at 620 nm) × 10,000. (B) TR-FRET assay showing the reference inhibition/competition of core dimerization by N-terminal 106-residue portion of core protein (core106). Core106 concentrations ranged from 0.001 to 0.6 μM. The graph was plotted after europium cryptate (Eu) background correction. This TR-FRET assay is representative of 5 independent experiments. Error bars represent standard deviation (SD) of n = 7 values in 1 assay.
Fig. 3.
(A) Optimization of europium cryptate-tagged anti-GST-and XL 665-tagged anti-Flag- in 384-well format. Eu-anti-GST antibody was analyzed at 2 different concentrations: 1.8 ng/well and 3.6 ng/well. Allophycocyanin (XL-665)-anti-Flag antibody was analyzed at 20 ng/well and 40 ng/well. The asterisk indicates the condition used for the medium-throughput Center for Chemical Methodology and Library Development at Boston University (CMLD-BU) run. The reaction conditions are discussed in the results section. (B) Optimization of incubation times for N-terminal 106-residue portion of core protein (core106) time-resolved fluorescence–resonance energy transfer (TR-FRET) assay in 384-well format. GST-core106 and Flag-core106 were kept constant at 27 and 34 nM, respectively. The assay was analyzed at 1, 4, and 24 h. Free GST at 41 nM was included as a control inhibitor. The asterisk indicates the incubation time selected for the CMLD-BU run.
Table 1.
Summary of 384-Well Format Assay Protocols for TR-FRET Assay
| |
|
|
|
Final Concentration or Incubation Time |
|---|---|---|---|---|
| Step # | Description | Reagents | Total Volume Added, μL | TR-FRET |
| 1 | Protein addition | GST-core106 | 2.5 | 0.027 μM |
| 2 | Control addition | corel06 | 5 | 1 μM |
| 3 | Test compound addition | NA | 0.01 | 15 μM |
| 4 | Protein addition | Flag-core106 | 2.5 | 0.034 μM |
| 5 | HTRF reagents addition | Eu-Ab and Flag-XL665 | 10 | 1.8 and 20 ng/well |
| 6 | Incubation | NA | NA | 4 h |
Screening of the LOPAC and CMLD-BU Libraries for Identification of core106 Dimerization Inhibitors
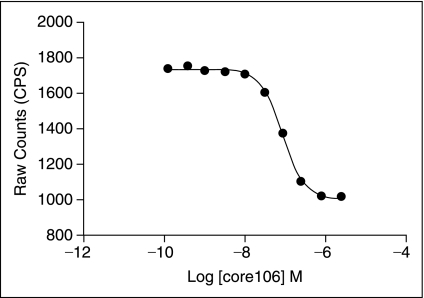
Screening of the LOPAC library was done with a hundred percent effect (HPE) inhibitor control using 1 μM core106 as a reference inhibitor to 27 nM GST-core106 and 34 nM Flag-core106. The zero percent effect (ZPE) uninhibited control was 27 nM GST-core106 and 34 nM Flag-core106 only. The LOPAC run was performed twice in triplicate in the presence of 0.5% DMSO. The compounds were screened at 12.5 μM final concentration. The average Z′ was 0.73 ± 0.07. The average signal-to-background in the run was 2.28 ± 0.32. A CRC titration was performed twice in the first (CRC-1) and the very last (CRC-2) plate in the 11 plate series. The reproducibility of the inhibition response was 98% with an estimated IC50 of ∼89 nM for untagged core inhibition of core dimerization (Fig. 4). No biochemical degradation or systematic changes were evident over the 6 h run. No false hits could be identified on any DMSO plate within 3 standard deviations (SDs) of the 100% inhibition control. Thus, the DMSO controls provided a gauge for identifying hits on the LOPAC collection. Additionally, no compound carryover was evident in the run. Detergent (0.1% CHAPS) was present in the assay buffer to reduce to a certain extent, the likelihood of false-positive signal due to aggregation of the proteins by compounds acting non-specifically.
Fig. 4.
Dose–response analysis of N-terminal 106-residue portion of core protein (core106) inhibition on GST-core106 and Flag-core106 dimerization. The x-axis shows the molar concentration of core106. The y-axis shows the time-resolved fluorescence–resonance energy transfer (TR-FRET) ratio. The inhibitory concentration at 50% (IC50) for the free core inhibition of the core dimerization was calculated to be 89 nM. The standard deviation (SD) was not significant hence not represented by error bars.
Five hits were selected based on 3 SDs from the ZPE control in the first LOPAC run. In the second LOPAC run, only the dequalinium analog with a C-linker, one of the 5 original hit compounds was confirmed. The C8 or C10 analogs of this compound, when analyzed in a dose–response study, did not show any significant inhibitory activity on GST-core106 and Flag-core106 oligomerization. The original compound probably acted non-specifically by virtue of its long aliphatic chain, flanked by 2 hydrophobic ends.
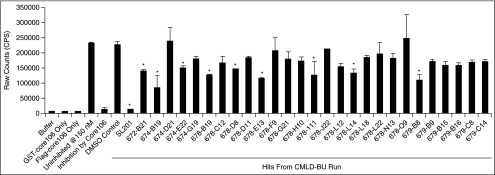
Screening of the CMLD-BU library (2,240 compounds) was done with the same HPE and ZPE controls as in the LOPAC assays. The CMLD-BU library was screened in duplicate in the presence of 0.5% DMSO at a final concentration of 15 μM. The average Z was 0.61 ± 0.04. The average signal-to-background ratio in the run was 1.8 ± 0.06 (Table 2). No false-positives were identified in any of the DMSO plates within 3 SDs of the 100% inhibition control. The consistency in the data from the CMLD-BU run confirmed that there was no biochemical degradation or systematic change evident over time and no compound carryover between plates. Detergent (0.1% CHAPS) was again present in the assay buffer. Twenty-eight hits were identified in the run using a cut-off point of 6 SDs from the ZPE control.
Table 2.
Plate Statistics for CMLD-BU Run
| Plate # | Z Value | S/B Value | # of Hits per Plate |
|---|---|---|---|
| 1 | 0.589 | 1.77 | 1 |
| 2 | 0.59 | 1.77 | 1 |
| 3 | 0.573 | 1.93 | 0 |
| 4 | 0.609 | 1.86 | 4 |
| 5 | 0.692 | 1.82 | 16 |
| 6 | 0.591 | 1.76 | 6 |
The data from the CMLD-BU run are shown as a table. Column 1 shows the plate number, column 2 shows the Z value of the plate, column 3 shows the signal-to-basal value, and the last column shows the number of hits per plate in the run.
A core106 ALPHA screen assay was used as a secondary confirmation for the 28 hit compounds identified from the primary TR-FRET screen of the CMLD-BU library (Fig. 5). The final 384-well assay protocol is summarized in Table 3 The GST-core106 and Flag-core106 were kept constant at 150 nM each. GSH-coated donor beads and anti-Flag antibody-coated acceptor beads were used for the detection of GST-core106 and Flag-core106 dimerization. The controls in the confirmatory screen were: buffer only, GST-core106 only, Flag-core106 only, ZPE: GST-core106 and Flag-core106, HPE: GST-core106 and Flag-core106 with 1 μM of core106 as inhibitor. The 384-well assay protocol is summarized in Table 3.
Fig. 5.
Amplified luminescent proximity homogeneous assay (ALPHA screen) confirming hits from of primary time-resolved fluorescence–resonance energy transfer (TR-FRET)-based Center for Chemical Methodology and Library Development at Boston University (CMLD-BU) run. N-terminal 106-residue portion of core protein (Core106) ALPHA screen assay was used as a secondary confirmation to validate the 28 hits from the primary TR-FRET screen. GST-core106 (GC) and Flag-core106 (FC) were kept constant at 150 nM each. Core106 was added as a 100% inhibition control. DMSO was included as a control because the compounds were dissolved in DMSO. Ten of the 28 hits were confirmed as potential inhibitors of core106 dimerization, indicated with asterisks. Error bars represent standard deviation (SD) of n = 2 in 2 assays.
Table 3.
Summary of 384-Well Format Assay Protocol for ALPHA Screen Assay
| |
|
|
|
Final Concentration or Incubation Time |
|---|---|---|---|---|
| Step # | Descrption | Reagents | Total Volume Added, μL | ALPHA Screen |
| 1 | Protein addition | GST-core106 | 6 | 0.15 μM |
| 2 | Control addition | core106 | 1 | 1 μM |
| 3 | Test compound addition | NA | 0.015 | 15 μM |
| 4 | Protein addition | Flag-core106 | 6 | 0.15 μM |
| 5 | Incubation | NA | NA | 1 h |
| 6 | AS bead addition | GSH coated donor beads | 6 | 20 μg/mL |
| 7 | Incubation | NA | NA | 1 h |
| 8 | AS bead addition | Anti Flag ab coated acceptor beads | 6 | 20 μg/mL |
| 9 | Incubation | NA | NA | 1 h |
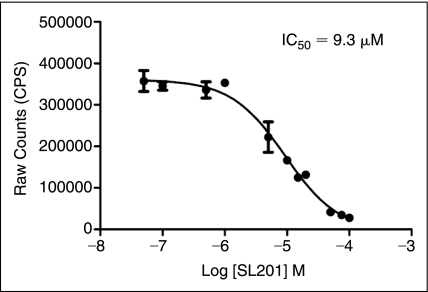
The 28 hit compounds were dissolved in DMSO. Hence, 1% DMSO was supplemented as a control to ZPE. Ten of the 28 TR-FRET confirmed hits were validated by the ALPHA screen as inhibitors of core106 dimerization based on 40% or higher inhibitory effect compared to the uninhibited control (Fig. 5). One of the hit compounds (SL201) was re-synthesized in sufficient quantities, and was analyzed in a dose–response study using the core106 ALPHA screen assay, yielding an IC50 = 9.3 μM (Fig. 6).
Fig. 6.
Dose–response analysis of compound SL201, using the N-terminal 106-residue portion of core protein (core106) amplified luminescent proximity homogeneous assay (ALPHA screen) assay. The compounds were dosed from 0.05 to 100 μM. The IC50 was calculated to be 9.3 μM. Error bars represent standard deviation (SD) of n = 2 in 2 assays.
Effect of SL201 on HCV 2a J6/JFH-1 Virus Production
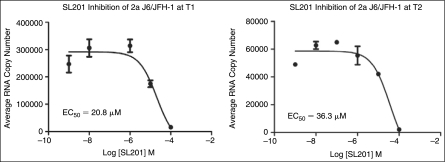
Compound SL201 was further analyzed in a biological screen to evaluate its inhibitory activity on the production of J6/JFH1 2a strain virus, as was done previously for core-derived peptides.10 In preparation for this secondary screening, the average toxicity (CC50) of SL201 for hepatoma Huh-7.5 cells was determined to be 320 μM. It was then tested in the same cells infected with HCV. Real-time RT-PCR was done on RNA purified from HCV 2a infected Huh-7.5 cell lysate treated with varying concentrations of SL201 (0.001–100 μM). The EC50 for SL201 was calculated to be 20.8 and 36.3 μM, respectively, at early stage (T1) and late stage (T2) of HCV-infected culture (Fig. 7).
Fig. 7.
Effect of SL201 on HCV 2a J6/JFH-1 virus. Inhibition assay of SL201 was performed on HCV 2a J6/JFH-1 virus by adding serially diluted SL201 and virus onto naïve Huh-7.5 cells and incubated for 3 days (early stage—T1). Culture supernatant (T1) was transferred onto naïve Huh-7.5 cells and incubated for an additional 3 days (late stage—T2). RNA was purified from lysed cells from both T1 and T2. Real-time RT-PCR was performed on purified RNA. EC50 for SL201 were calculated to be 20.8 and 36.3 μM, respectively, at T1 and T2.
Discussion
Inhibition of protein–protein interaction has long been considered as a desirable target for assay development and drug discovery. For this purpose, the use of peptides derived from one of the interacting partners has indeed been shown to be quite fruitful and a number of peptide inhibitors have been described for a variety of interacting protein pairs.23 In addition to providing proof-of-principle results, these peptides have subsequently often served as building blocks for synthesis of peptide mimetics acting as leads for drug development. However, drug-like small molecule inhibitors of protein–protein interactions have been considered more desirable, albeit more difficult to obtain.
Virology is an area of great interest for inhibition of protein–protein interactions since it is often difficult to identify substances active against new emerging, difficult-to-grow viruses. There have been previous attempts to disrupt protein–protein interactions involving viral capsid proteins by identifying small molecules that interfere with the assembly of a virus. The group of Zlotnick has been most successful in this area by using a screening assay based on fluorescence quenching of dye-labeled Hepatitis B virus (HBV) capsid protein.24 In this work, we have used the capsid protein “core” of HCV, since core was known to self-associate, and since we had previously demonstrated its dimerization.10 In the TR-FRET core106 dimerization assay described here, we have shown that one can actually identify small molecules that block this protein–protein interaction with higher potency than core-derived 18- and 15-mer peptides that were previously identified in an ALPHA screen assay.10
The data presented here indicate that a robust and reproducible TR-FRET assay was developed to monitor dimerization of GST- or Flag-tagged core106, which contains the 106 N-terminal residues of the 191-residue long core protein. While the dynamic range of the assay is relatively modest, ranging from 1.7 to 3.0, the assay was confirmed to be robust based on the Z values from 0.56 to 0.72. Inclusion of detergent in all assays reduced to a certain extent, the likelihood of a false-positive signal due to protein aggregation. Core106 and its derivatives were easily produced in large amounts in E. coli: over the course of our studies we purified >500 mg and have shown that the core106 protein is stable over time, can be shipped without loss of activity, and is not altered by the presence of up to 10% DMSO.10 Anti-tag antibodies were readily obtained from various commercial sources. Direct coupling of the fluorophores to core106 proteins did not improve the signal-to-background ratios obtained using the commercially available fluorophore-labeled anti-GST or anti-Flag antibodies (unpublished results).
Tag-free core106 completely inhibited GST-core106/Flagcore106 dimerization with an IC50 of 89 nM. Inhibition of dimerization by compounds in the LOPAC library yielded a single hit; dequalinium was reproducibly found to inhibit dimerization but could not be further analyzed in a dose–response study due to lack of availability from the manufacturer. Available analogs of this compound, with a spacer of 8 or 10 methyl groups, compared to for the original compound, failed to show any inhibitory activity on core106 dimerization.
Results obtained with compounds from the CMLD-BU library confirmed 10 hits with a Z-factor of 0.52. One of these, SL201, was re-synthesized and was further characterized by dose–response analysis, yielding an IC50 = 9.3 μM. Furthermore, this compound was shown to inhibit HCV 2a production in culture after 24 h (EC50 = 20.8 μM) and after 72 h (EC50 = 36.3 μM). Interestingly, these effects were observed only after viral infection was established, suggesting that the compound did not affect viral penetration nor initial steps of infection, and most likely specifically affected assembly of the particle.
HTS compatibility of the TR-FRET assay was demonstrated in a HTS suite operational at our Institute. Since screening of even small libraries yielded several hits, it is expected that more inhibitors (compounds) will be discovered in a larger collection of products, possibly with even lower IC50s than the 9 μM concentration observed so far.
Abbreviations
- ALPHA screen
amplified luminescent proximity homogeneous assay
- CMLD-BU
Center for Chemical Methodology and Library Development at Boston University
- Core106
N-terminal 106-residue portion of core protein
- CRC
control response curve
- EC50
median effective concentration (required to induce a 50% effect)
- Eu
europium cryptate
- FC
flag-core106
- GC
GST-core106
- GSH
glutathione
- GST
glutathione-S-transferase
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HPE
hundred percent effect
- HRP
horseradish peroxidase
- HTS
high-throughput screening
- IC50
median inhibition concentration (concentration that reduces the effect by 50%)
- LOPAC
Library of Pharmacologically Active Compounds
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TMB
3,3′,5,5′–Tetramethylbenzidine
- TR-FRET
time-resolved fluorescence–resonance energy transfer
- XL-665
allophycocyanin
- ZPE
zero percent effect
Acknowledgments
We thank our various colleagues from the Scripps Research Institute—Scripps Florida (i) for helpful discussions: Prof. Weissmann and Dr. T. Tellinghuisen (Department of Infectology); (ii) for help with formatting the CMLD-BU library: Pierre Baillargeon and Lina Deluca (Department of Lead Identification); and (iii) for support at the HTS facility: Peter Chase (Department of Lead Identification); and the Factor Foundation (A.D.S.), NIGMS (P50 GM067041) (J.A.P.), and NIH (U54MH084512) (L.S., T.S., P.H.) for generous support.
AUTHOR DISCLOSURE STATEMENT
S.K., L.S., T.S., V.T., P.H., and A.D.S. are employees of the Scripps Research Institute. A.B.B., J.K.S., and J.A.P. are employees of Boston University.
References
- 1.WHO. Hepatitis C: global prevalence. Wkly Epidemiol Rec. 1997;72:341–344. [PubMed] [Google Scholar]
- 2.Giannini C. Brechot C. Hepatitis C virus biology. Cell Death Differ. 2003;10(Suppl. 1):S27–S38. doi: 10.1038/sj.cdd.4401121. [DOI] [PubMed] [Google Scholar]
- 3.Choo Q. Kuo G. Weiner A. Overby L. Bradley D. Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.McGovern BH. Hepatitis C in the HIV-infected patient. J Acquir Immune Defic Syndr. 2007;45(Suppl 2):S47–S56. doi: 10.1097/QAI.0b013e318068d190. [DOI] [PubMed] [Google Scholar]
- 5.Cristina J. Moreno-del Pilar M. Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Res. 2007;127(2):185–194. doi: 10.1016/j.virusres.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco R. Carfí A. Advances in the development of new therapeutic agents targeting the NS3-4A serine protease or the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Adv Drug Del Revs. 2007;59(12):1242–1262. doi: 10.1016/j.addr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Thibeault D. Bousquet C. Gingras R. Lagace L. Maurice R. White PW, et al. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J Virol. 2004;78(14):7352–7359. doi: 10.1128/JVI.78.14.7352-7359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courcambeck J. Bouzidi M. Perbost R. Jouirou B. Amrani N. Cacoub P, et al. Resistance of hepatitis C virus to NS3-4A protease inhibitors: mechanisms of drug resistance induced by R155Q, A156T, D168A and D168V mutations. Antivir Ther. 2006;11(7):847–855. [PubMed] [Google Scholar]
- 9.McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepat. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 10.Kota S. Coito C. Mousseau G. Lavergne J-P. Strosberg AD. Peptide inhibitors of Hepatitis C Virus core oligomerization and virus production. J Gen Virol. 2009;90:1319–1328. doi: 10.1099/vir.0.008565-0. [DOI] [PubMed] [Google Scholar]
- 11.Beeler AB. Acquilano DE. Su Q. Yan F. Roth BL. Panek JS, et al. Synthesis of a library of complex macrodiolides employing cyclodimerization of hydroxyesters. J Comb Chem. 2005;7:673–681. doi: 10.1021/cc050064b. [DOI] [PubMed] [Google Scholar]
- 12.Lei X. Zaarur N. Sherman MY. Porco JA., Jr. Stereocontrolled synthesis of a complex library via elaboration of angular epoxyquinol scaffolds. J Org Chem. 2005;70:6474–6483. doi: 10.1021/jo050956y. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y. Beeler AB. Cho S. Wang Y. Franzblau SG. Snyder JK. Library synthesis using 5,6,7,8-Tetrahydro-1,6-naphthyridines as Scaffolds. J Comb Chem. 2008;10:534–540. doi: 10.1021/cc800038r. [DOI] [PubMed] [Google Scholar]
- 14.Su S. Acquilano DE. Arumugasamy J. Beeler AB. Eastwood EL. Giguere JR, et al. Convergent synthesis of a complex oxime library using chemical domain Shuffling. Org Lett. 2005;7:2751–2754. doi: 10.1021/ol051023r. [DOI] [PubMed] [Google Scholar]
- 15.Preaudat M. Ouled-Diaf J. Alpha-Bazin B. Mathis G. Mitsugi T. Aono Y, et al. A homogeneous caspase-3 activity assay using HTRF technology. J Biomol Screen. 2002;7(3):267–274. doi: 10.1177/108705710200700310. [DOI] [PubMed] [Google Scholar]
- 16.Mathias G. HTRF technology. J Biomol Screen. 1999;4:309–313. doi: 10.1177/108705719900400605. [DOI] [PubMed] [Google Scholar]
- 17.Peppard J. Glickman F. He Y. Hu SI. Doughty J. Goldberg R. Development of a high-throughput screening assay for inhibitors of aggrecan cleavage using luminescent oxygen channeling (ALPHA screen) J Biomol Screen. 2003;8(2):149–156. doi: 10.1177/1087057103252308. [DOI] [PubMed] [Google Scholar]
- 18.Lindenbach BD. Evans MJ. Syder AJ. Wolk B. Tellinghuisen TL. Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 19.Wakita T. Pietschmann T. Kato T. Date T. Miyamoto M. Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong J. Gastaminza P. Cheng G. Kapadia S. Kato T. Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tellinghuisen TL. Foss KL. Treadaway J. Regulation of Hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4(3):e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CT. Murray CL. Eastman DK. Tassello J. Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells JA. McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 24.Stray SJ. Johnson JM. Kopek BG. Zlotnick A. An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nat Biotechnol. 2007;24:358–362. doi: 10.1038/nbt1187. [DOI] [PubMed] [Google Scholar]