Summary
A series of papers in the last year reported major advances in our understanding of ABA signaling: the identification of soluble ABA receptors, the elucidation of a core ABA signaling pathway and structural insights into the mechanism of ABA perception and signaling. Here we summarize these advances, which have shown in atomic resolution that the ABA receptors PYR1, PYL1 and PYL2 function as allosteric switches that inhibit type 2C protein phosphatases (PP2Cs) in response to ABA. These receptors function at the apex of a core signaling pathway that regulates ABA responses by controlling SnRK2 kinase activity and the phosphorylation of downstream target proteins such as ABFs, which control nuclear responses, and the ion channel SLAC1, which mediates electrophysiological responses to ABA.
Introduction
Plants synthesize a diverse array of diffusible hormonal signals that work in concert to integrate growth, development and cellular physiology to environmental cues [1]. A key abiotic stress signal is the carotenoid derived molecule abscisic acid (ABA). Originally discovered in the 1960s, physiological, biochemical and genetic analyses have uncovered roles for ABA in numerous stress and developmental processes. Several reviews of ABA biosynthesis and signaling have been published recently[2–5]. Since May 2009, an unprecedented number of advances have occurred, including the discovery of a soluble ABA receptor family and the assembly of numerous pieces of the ABA signaling puzzle into a cohesive “core” pathway. We note that the last year also witnessed important advances in identifying ABA transporters [6**,7**], and the demonstration that ABA catabolism is coupled to high-humidity stress [8*], neither of which can be covered due to space limitations.
New ABA Receptors That Regulate PP2C Activity
Several ABA binding proteins have been described and implicated in ABA signaling including the plasma membrane localized GPCR-type G protein (GTGs) and the chloroplast localized Magnesium Cheletase subunit ChlH (For review see [3)]); given the flurry of activity on soluble receptors, we do not cover these proteins here. The PYR/PYL/RCAR family of ABA receptors was identified by 4 separate research groups [9**, 10**, 11**, 12**], and is comprised of a 14-member gene family in Arabidopsis [1–4]; of which at least 13 function in ABA perception [13**]. Three groups independently identified different members of this new receptor family by virtue of their physical interactions with clade A PP2Cs in yeast two hybrid [10**,11**] or immunoprecipitation experiments [12**]. The Arabidopsis genome encodes 76 PP2Cs. One subfamily of 9 “clade A” PP2Cs, which includes ABA INSENSITIVE 1 (ABI1), ABI2 and HOMOLOG OF ABI1 (HAB1) [14], are well characterized negative regulators of ABA signaling (reviewed in [3]). Taking a different approach, we identified pyrabactin, a selective ABA agonist and determined by genetic analysis that PYRABACTIN RESISTANCE 1 (PYR1) is necessary for pyrabactin action in vivo [9**]. A quadruple pyr1/pyl1/pyl2/pyl4 mutant shows defects in several ABA responses, including ABA-induced gene expression, ABA-mediated SnRK2 kinase activation [9**] and ABA-promoted guard cell closure [12**]. Transgenic plants overexpressing REGULATORY COMPONENT OF ABA RECEPTOR 1 (RCAR1/PYL9) are hypersensitive to ABA-promoted guard cell closure [10**] and overexpression of PYR1-LIKE 5 (PYL5) confers drought tolerance on transgenic Arabidopsis plants [11], which validates the new receptor family as a target for manipulating abiotic stress tolerance. PYR/PYL proteins bind ABA directly, and interestingly, their affinity for ABA is stimulated ~10-fold by the presence of PP2Cs [10**,11**,15**], a point we return to later. The single PYR/PYL mutants characterized to date do not possess ABA phenotypes [9**]; this redundancy likely explains why the gene family evaded detection by earlier genetic screens (reviewed in [16]). The selectivity of pyrabactin, a non-natural agonist, for the receptor PYR1 enabled the genetic redundancy observed for ABA to be bypassed, which illustrates the power of synthetic ligands for dissecting plant signaling networks [9**,17,18*].
The physical interactions of PYR1 and its 4 closest relatives (PYL1 – PYL4), with PP2Cs (ABI1, ABI2 and HAB1) are regulated by ABA, as measured using yeast two hybrid assays [9]. The interactions between PYLs 5 –12 and PP2Cs occur in the absence of exogenously added ABA in yeast two hybrid assays [9**, 10**, 11**], a point that requires further investigation. Upon binding ABA, PYR/PYL proteins inhibit the phosphatase activity of multiple clade A PP2Cs, with IC50 values measured in the range of 18 – 390 nM (+)-ABA, depending on the PYR/PYL-PP2C pair examined [9**, 10**, 11**,15*]. Since 9 clade A PP2Cs and 14 PYR/PYL proteins are encoded by the Arabidopsis genome, 126 PYR/PYL-PP2C combinations could potentially form. While the in vivo significance of this remains to be demonstrated, reports of differences in ABA sensitivity between different combinations are suggestive that the combinatorial interactions between receptors and PP2Cs enable a tunable response to stress signaling [10**,11**,15*].
Structural Insights into Receptor Function
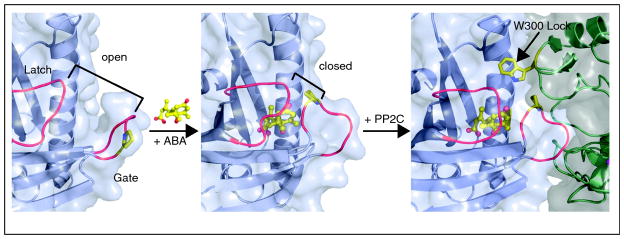
The structures of PYR1, PYL1 and PYL2 in apo, ABA bound and ABI1 or HAB1 complexed forms were reported in late 2009 [19**, 20**, 21**, 22**, 23**]; the conclusions from 5 studies are in general agreement and here we present a consensus view of these studies. The PYR/PYL receptors are members of the START-domain superfamily of lipophilic ligand binding proteins [24–26], which exhibit a “helix-grip” structure, in which N-terminal and C-terminal α helices enclose a seven-stranded β sheet to create a ligand-binding pocket [24]. In the published PYR/PYL structures, two highly conserved loops flank the ligand binding pocket: the SGLPA “gate” loop (also called the “cap”, or CL2 loop) between the β3 and β4 strands and the HRL “latch” loop (also called the lock or CL3 loop) between the β5 and β6 strands. The gate and latch loops undergo significant conformational rearrangement upon ABA binding, which is the primary allosteric mechanism that underlies information transfer (Figure 1). Two mutations in PYR1’s gate (P88S) and latch (H115A) abolish ABA-mediated PP2C inhibition without disrupting ABA binding (as measured using NMR methods) [9**,19**], showing that ABA binding and PP2C inhibition can be uncoupled.
Figure 1. Abscisic acid mediated formation of thePYR/PYL-ABA-PP2C ternary complex.
PYR/PYL proteins contain a central hydrophobic pocket that is flanked by two mobile loops called the “gate” and “latch”, shown in red. ABA binding triggers closure of the gate, this in turn creates an interaction surface for binding to the PP2Cs, which dock onto the closed form of PYR/PYL proteins. The site of docking is adjacent to the magnesium ion containing active site of PP2Cs (shown in magenta). A conserved tryptophan in the PP2Cs, called the “lock” inserts between the gate and latch and makes a water mediated contact to ABA. Docking of the PYR/PYL proteins into the PP2C active site inhibits PP2C activity by occluding access of target proteins. This figure was made using the coordinates for apo-PYL2, ABA-bound PYL2 and the PYL2-ABA-HAB1 ternary complex (3KAZ, 3KBO 3KB3), described in (**19).
The binding of ABA to PYR/PYL proteins is mediated by a combination of hydrogen bonds and hydrophobic interactions, including direct contacts between ABA and residues in the gate and latch, which stabilizes their closure. A conserved lysine (corresponding to K59 in PYR1, K86 in PYL1, K64 in PYL2) forms a charge interaction with the acidic head group of ABA, which explains the critical necessity of a COOH noted in ABA structure activity relationships (for review see [3,27]). Many of the residues that make contacts to ABA are highly conserved between receptor proteins; however subtle sequence variation in ABA contacting residues does exist. Due to space limitations, we cannot cover recently published structural data for PYR/PYL-pyrabactin complexes [28*, 29**, 30**, 31**], but we note that sequence variation in pocket-lining residues contributes to differences in ligand sensitivity between receptor family members [29**, 30**].
Molecular modeling predicted[19**] and ternary structures confirmed [19**, 22**, 23**] that the altered protein surface created by the motion of gate and latch residues in response to ABA facilitates PP2C docking on to PYR/PYL receptors. ABA-bound PYR/PYL receptors are able to bind several clade A PP2Cs and inhibit their phosphatase activity in vitro [9**, 10**, 11**]. The gate loop of ABA-bound receptors is positioned with its centrally located SGLPA serine inserted into the PP2C catalytic site and apparently acts as a high affinity product mimic to inhibit the enzyme, by blocking normal substrate access (Figure 2); direct measurements have shown that PYL2 acts as a competitive inhibitor of HAB1’s phosphatase activity[19**]. Thus, ABA ultimately inhibits PP2C activity by inducing a conformational change in PYR/PYL proteins that converts them into PP2C inhibitors. Interestingly, clade A PP2Cs interact with PYR/PYL proteins via a small recognition loop that contains a conserved tryptophan (W300 in ABI1) that has been called the “lock” [19**] (Figure 2). This tryptophan inserts between the gate and latch loops [19**,22**,23**] and its indole NH makes a water-mediated hydrogen bond to ABA’s ketone group. Mutation of the tryptophan lock residue abolishes ABA-PYR/PYL-mediated PP2C inhibition [19**,23**]. With the exception of AHG1, the tryptophan lock residue is present in all clade A PP2Cs (Figure 3) and missing in other plant PP2Cs, suggesting that PYR/PYL regulation is restricted to clade A PP2Cs.
Figure 2. The SGLPA gate docks into the PP2C active site.
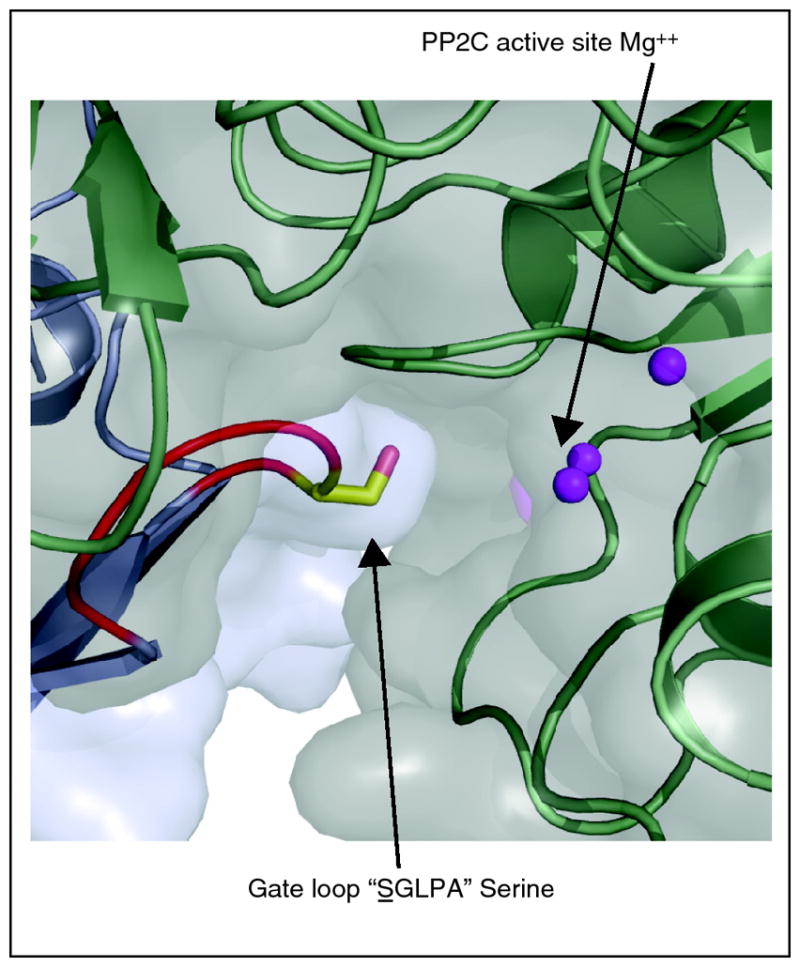
Serine of the SGLPA gate loop inserts adjacent to the PP2C active site, acting as a high affinity product-mimic.
Figure 3. The tryptophan lock is part of a plant specific recognition loop in clade A PP2Cs.
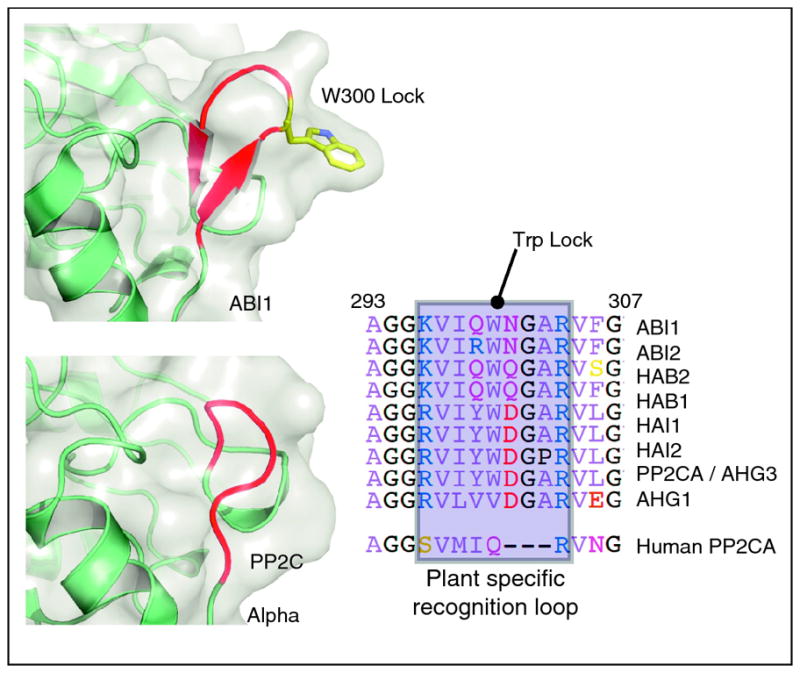
ABI1 and HAB1 dock onto PYR/PYL proteins and insert their conserved tryptophan lock residues between the gate and latch. This recognition module is absent from human PP2C structures and therefore a plant specific modulation of PP2C. Eight of the 9 clade A PP2Cs contain the tryptophan lock residue and it is absent from other plant PP2Cs.
The tryptophan lock’s interaction with ABA’s ketone and enhancement of ABA binding affinity by PP2Cs has led to some discussion as to whether PYR/PYL proteins are best described as co-receptors rather than receptors [2,4,19**]. Since over 20 residues in the PYR/PYL proteins make direct or water-mediated contacts with ABA, and ABA is buried within the ligand-binding cavity of PYR/PYL proteins [20**, 21**, 22**, 23**], the structures imply that the primary site of ABA recognition is by the PYR/PYL receptors. Based on current structural data, the primary basis for the enhancements in ABA binding stimulated by PP2Cs occurs by PP2C-mediated stabilization of the closed form of the receptor [11**,19**,22**,23**], which is expected to lower Kd by lowering the Koff for ABA. Thus, the structures provide a rationalization for the ~10 fold increases in ABA affinity provided by PP2Cs and suggest that PYR/PYL proteins participate as the primary sites for ABA and pyrabactin recognition.
PYR1, PYL1, and PYL2 form homodimers in their ligand-free states, but PYL1 and PYL2 bind PP2Cs at 1:1 stoichiometry in the presence of ABA [19**,22**,23**]. Additionally, most of the residues that make contact in the PYL2 homodimer are also involved in binding to the PP2C in the ternary ABA-bound complex [22**], implying that a receptor dimer dissociation step occurs prior to PP2C binding [22**]. Importantly, PYR1 has been shown to form a dimer in vivo [21**], suggesting that dimer breaking in response to ABA is biologically relevant, at least for PYR1. Both Melcher et al. [19**] and Yin et al. [22**] have noted that ABA binding reduces the buried surface area between PYL protomers, which is also supported by small-angle X-ray scattering measurements made in solution [21**]. Furthermore, Yin et al. have suggested dimer breaking may be initiated by tryptophan lock of the PP2C inserting between the gate and latch loops of ABA bound PYR/PYL proteins [22**]. Elucidating the mechanism and functional relevance of dimer breaking will likely be an important line of future investigation.
Since PP2Cs are conserved throughout eukaryotes and have no known direct protein regulators (besides PYR/PYL proteins), it is interesting to ask if the structural and functional insights from plants illuminate mechanisms of PP2C regulation outside of plants? Human PP2CA aligns very closely with ABI1 (Figure 3) and inspection of the aligned structures reveals that the human PP2CA does not contain the recognition loop that ABI1 and HAB1 use to dock onto ABA-bound PYL proteins (Figure 3). Thus, the recognition loop and conserved tryptophan lock appear to be a plant specific innovation exploited in selective regulation of clade A PP2Cs. However, it will be interesting to determine if PP2C inhibition by product-mimics unrelated to PYR/PYL proteins is exploited in the regulation of other PP2Cs.
Elucidation of The Core ABA Response Pathway
The structural studies have revealed how ABA binding to PYR/PYL receptor proteins leads to PP2C inhibition, but how is this event then conveyed to other outputs? A key clue came from the observation that SnRK2 kinases are not properly activated by ABA in the pyr1/pyl1/pyl2/pyl4 quadruple mutant [9**] and this suggested a model for regulation of SnRK2 kinase activity by the PYR/PYL-PP2Cs [9**], which has been validated and greatly extended by two seminal studies described below [13**,31**].
Members of the plant SnRK2 family were originally identified in wheat as an ABA induced kinase transcript [32], and in Vicia faba as a rapid ABA-activated kinase activity [33]. The participation of SnRK2s in ABA signaling has since been established in many species [33–35] and loss-of-function alleles of OPEN STOMATA 1 (OST1; also known as SnRK2.6 or SRK2E) demonstrated the key role of the SnRK2s in vivo [36]. The Arabidopsis genome encodes 10 SnRK2s, of which OST1 and its two closest relatives (SnRK2.2 and 2.3) participate in ABA signaling. Triple mutants lacking these three kinases are deficient in almost all ABA responses, demonstrating the centrality of these kinases to ABA signaling [37*, 38*, 39*]. Active, phosphorylated OST1 immunoprecipitated from plants can be dephoshphorylated by recombinant ABI1, which in turn reduces OST1’s kinase activity [31**]. The mechanism of SnRK2 activation in vivo may be mediated by autophosphorylation [40], but this point is currently unresolved. In vitro, OST1 can autophosphorylate on at least 5 sites [40], but it appears that a single residue, Ser-175, is critical for OST1 kinase activity. Serine 175 is located within the SnRK2 activation loop, proximal to the kinase catalytic site, and the mutation S175A disrupts OST1 autoactivation and cannot complement an ost1-1 mutant in planta [40]. LC-MS studies suggest that the activation loop (and Ser 175 inside this loop) is both phosphorylated in vivo by ABA stimulation and directly dephosphorylated by ABI1 [31]. Coupled to experiments documenting dephosphorylation and concomitant deactivation of OST1 kinase activity by HAB1 in vitro [41*], it is now well established that clade A PP2Cs directly inhibit SnRK2 kinase activity by dephosphorylating them. Thus, ABA pathway activation leads to the accumulation of active and phosphorylated SnRK2s via PP2C inhibition. Once activated, the SnRK2s are poised to directly phosphorylate numerous target proteins involved in ABA responses, including transcription factors that bind to abscisic acid-responsive promoter elements (ABREs) called ABFs (for ABRE-binding factors) [42,43]. These bZIP-class transcription factors are direct substrates of SnRK2 kinases, as documented in several plant species [44–46].
Two groups [13**,31**] have documented the sufficiency of the core PYR/PYL-PP2C-SnRK2 pathway for mediating an ABA response. In one set of experiments, recombinant PYR1, ABI1 and OST1 (immunoprecipitated from ABA stimulated plants) were sufficient for eliciting ABA-mediated OST1 kinase activation, as measured using phosphorylation of a histone substrate [31**], or in independent experiments by ABA-mediated phosphorylation of the ABRE-binding transcription factor ABF2 [13**]. Since the phosphorylation of ABFs by SnRK2s is critical for their ability to activate ABA-mediated gene transcription [45,47], the recent reconstitution experiments demonstrate that the core pathway provides a minimal set of proteins for linking ABA perception to a nuclear output (Figure 4).
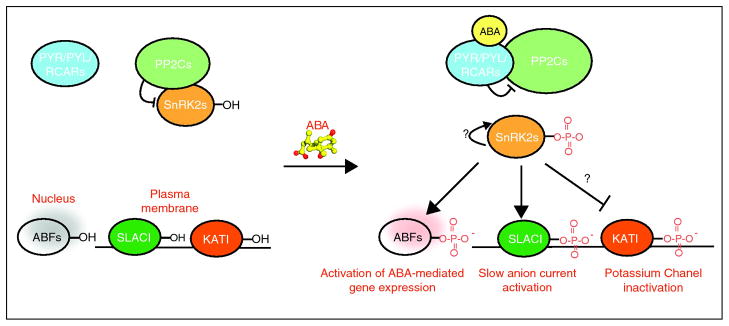
Figure 4.
The core PYR/PYL —| PP2C —| SnRK2 signaling pathway. In the absence of ABA, PP2C activity is high, and the PP2Cs prevent accumulation of phosphorylated SnRK2 kinases by directly dephosphorylating them. In the presence of ABA, PYR/PYL proteins bind to and inhibit PP2Cs, which leads to the accumulation of phosphorylated and active SnRK2s, possibly by auto-phosphorylation, however this is not currently clear. Once activated by phosphorylation, the SnRK2s can then directly phosphorylate downstream targets, such as the ABFs, SLAC, KAT1, and probably other proteins. Phosphorylation of ABFs is necessary for their ability to activate transcription. OST1 stimulates SLAC channel activity in Xenopus experiments and has been hypothesized to inhibit KAT1 channel activity.
What about the diverse non-transcriptional responses triggered by ABA? Key advances in the last year include the demonstration that OST1 phosphorylates and activates the anion channel SLAC1 expressed in Xenopus oocytes [48**, 49**, 50**] and phosphorylates and deactivates the potassium channel KAT1 when expressed in S. cerevisiae [51*]. These observations provide an appealing mechanism for ABA-mediated control of guard cell physiology by the core pathway, which is consistent with observations that the pyr1/pyl1/pyl2/pyl4 quadruple mutant posses defects in ABA promoted guard cell closure [12**]. Moreover, a recent report has demonstrated OST1-mediated phosphorylation of the Arabidopsis RESPIRATORY BURST OXIDASE SUBUNIT HOMOLOG F (RbohF) in in vitro assays [52*], which suggests a mechanism for ABA-mediated ROS production by the core pathway; however, further work will be needed to investigate the in vivo significance of these observations.
While the core pathway has impressive explanatory power, much work remains to be done towards understanding whether and how numerous well-characterized second messengers, such as Ca++ and NO are integrated with ABA signaling. Towards this, an important paper has shown that the calcium dependent kinase CPK23 can phosphorylate the N-terminus of the anion channel SLAC1 in vitro, and that this response is antagonized by ABI1 [53**]. Moreover, in Xenopus oocytes expressing SLAC1, the introduction of CPK23 (and other CPKs) enhances SLAC1 channel activity [53**]. The addition of ABA to in vitro reactions containing recombinant PYL9, ABI1 and CPK23 promotes phosphorylation of SLAC1’s N-terminus [53**]. Furthermore, ABI1 inhibits CPK23 kinase activity in vitro, which suggests that CPK23 kinase activity is regulated directly by the PYR/PYL-PP2C signaling system. These results suggest that the core pathway may bifurcate at the PP2Cs to regulate both SnRKs and CPKs. While exciting these conclusions were drawn largely from in vitro experiments using recombinant proteins and will benefit from further investigation in planta.
Conclusions
A burst of activity has led to enormous advances in our understanding of ABA signaling. Structural data show that ABA binds within the ligand binding pockets of PYR1, PYL1 and PYL2 and in so doing triggers a conformational change in the gate and latch loops of PYR/PYL receptors. The ABA bound receptors interact with clade A PP2Cs and inhibit their activity by docking within the PP2C active site. This in turn allows the accumulation of active, phosphorylated SnRK2s, and possibly CPKs, which can then phosphorylate and modulate the activity of downstream factors including ABFs, SLAC1, KAT1 and Rboh1. The core pathway provides a powerful starting point for developing an integrated picture of ABA action at the mechanistic level.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lumba S, Cutler S, McCourt P. Plant Nuclear Hormone Receptors: A Role for Small Molecules in Protein-Protein Interactions. Annual Review of Cell and Developmental Biology. 2010:26. doi: 10.1146/annurev-cellbio-100109-103956. [DOI] [PubMed] [Google Scholar]
- 2.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Cutler S, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic Acid: Emergence of a Core Signaling Network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 4.Klingler JP, Batelli G, Zhu JK. ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Bot. 2010 doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nambara E, Marion-Poll A. ABSCISIC ACID BIOSYNTHESIS AND CATABOLISM. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 6**.Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci U S A. 2010;107:2361–2366. doi: 10.1073/pnas.0912516107. This paper, along with (7) identifies two new ABC class transporters that can mediate ABA transport into and out of cell. In (6) the plasma membrane localized AtABCG25 that is proposed to mediate ABA efflux from cells. It was identified because loss-of-function mutations cause ABA hypersensitivity. The expression of AtABCG25 in vascular tissue is suggestive that it may mediate transport from sites of ABA biosynthesis, however whole plant measurements of ABA transport in mutant lineswere not reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci U S A. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. By screening for PDR mutant lines with ABA phenotypes, the plasma membrane localized AtABCB14 was identified. The gene is broadly expressed but also highly expressed n guard cells. Expression of AtABCB14 in BY2 cells leads to increase ABA uptake, suggestion that this protein mediates ABA uptake. Numerous defects in ABA responses suggest that AtABCB14 plays a role in ABA transport in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol. 2009;149:825–834. doi: 10.1104/pp.108.130823. The biosynthesis and catabolism of ABA are integrated with stress responses and therefore critically important to our understanding of stress physiology. This elegant paper demonstrates that ABA levels decrease under conditions of high humidity by a mechanism that involves catabolism of ABA by CYP707A3/A1, two cytochrome P450s involved in ABA degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. This paper, along with (10) made the first reports of the new soluble PYR/PYL/RCAR ABA receptor family and the demonstration that the receptors regulate PP2C and SnRK2 activities, which led to the formulation of the “core” signaling model for ABA action. In addition, the paper descried the isolation of pyrabactin, the first synthetic ABA agonist (that is not an ABA analog). The identification of pyrabactin demonstrates that the ABA pathway (and ABA receptors in particular) is “druggable”, which may have long-term importance in agrochemical development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. In addition to identifying the new PYR/PYL/RCAR ABA receptor family, providing quantitative measurements of ABA binding and demonstrating that these proteins regulate PP2C activity, this paper made the important observation that PP2Cs stimulate ABA binding and apparent Kds for ABA ~10 fold. [DOI] [PubMed] [Google Scholar]
- 11**.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park S-Y, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. The Plant Journal. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. This paper, along with (11) and (12) reports the independent identification of PYR/PYL proteins. In this paper, PYL5, PYL6 and PYL8 were identified in yeast two hybrid screens using HAB1 as a bait protein. Stimulation of ABA binding by PP2Cs is also observed, in agreement with (10). Importantly, Overexpression of PYL5 is shown to enhance drought tolerance of Arabidopsis plants, which validates the biotechnological significance of the new receptor family. [DOI] [PubMed] [Google Scholar]
- 12**.Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. In this paper, several PYR/PYL proteins were identified because they co-purify with ABI1 immunoprecipitated from plants. Importantly, immunoprecipitation of ABI1 was shown to lead to both PYR1 and SnRk2.3 co-immunoprecipitation, suggesting that a signaling complex could exist in vivo. Moreover, analysis of the pyr1/pyl1/[pyl2/pyl4 quadruple mutant generated in (9) revealed a defect in ABA-mediated guard cell closure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S-Y, Cutler SR, Sheen J, Rodriguez PL, Zhu J-K. In vitro Reconstitution of an ABA Signaling Pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. This paper, along with (31)are landmarks in ABA signaling. They demonstrate the sufficiency of the core pathway (originally proposed in reference (9)) to mediate information transfer from receptor to a nuclear output (in the case of (13)) or SnRK2 activation in the case of (31). In addition (13) developed a protoplast assay to probe the core pathway and used this to show that 13 of the 14 PYR/PYL genes are sufficient for initiating ABA responses in protoplasts. In apparent contrast to the signalosome model presented in (12), paper (13) suggests that ABA-bound PYR/PYL proteins/block the physical association of PP2Cs and SnRK2s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15*.Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2009;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. This paper investigates the biochemical properties of RCAR3/PYL8 and demonstrates that different receptors posses different sensitivities in their ability to inhibit PP2Cs in response to ABA, as measured in vitro. It is additionally shown that ABA sensitivity is influenced by the particular PP2C, as well. These observations lend support to the hypothesis that combinatorial interactions between different PYR/PYL/RCAR receptors and PP2Cs may have functional relevance, as originally proposed in ref.(10) [DOI] [PubMed] [Google Scholar]
- 16.Finkelstein R, Gampala S, Rock C. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutler S, McCourt P. Dude, where’s my phenotype? Dealing with redundancy in signaling networks. Plant Physiol. 2005;138:558–559. doi: 10.1104/pp.104.900152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.McCourt P, Desveaux D. Plant chemical genetics. New Phytol. 2010;185:15–26. doi: 10.1111/j.1469-8137.2009.03045.x. An excellent review of plant chemical genetics. The connectionmade between the chemical dissection of the Lac operon and modern chemical genetics is a gem. [DOI] [PubMed] [Google Scholar]
- 19**.Melcher K, Ng L-M, Zhou XE, Soon F-F, Xu Y, Suino-Powell KM, Park S-Y, Weiner JJ, Fujii H, Chinnusamy V, et al. A Gate-Latch-Lock Mechanism for Signal Transduction by Abscisic Acid Receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. This paper,along with papers 20 – 23,provides the first atomic level portraits of ABA perception. This paper captured the ABA receptor PYL2 in three relevant states-apo, ABA-bound and its complex with ABA and the PP2C HAB1 and coined the “gate”, “latch” and “lock” terminology adopted in this review. This paper and both (22) and (23) provide ternary complexes and reveal the importance of the “lock” tryptophan located on the PP2C for interactions with PYR/PYL receptors. This paper shows that the mechanism of PP2C inhibition is competitive and additionally that mutations in PYR1’s latch domain block receptor function in vitro and in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. This paper, along with (21) providesthe structure of dimeric PYR1 complexed with a single molecule of ABA. Interestingly, the dimer reveals two states of PYR1; the “open” conformation of the non-ABA bound protomer and the “closed” conformation of the ABA-bound protomer. [DOI] [PubMed] [Google Scholar]
- 21**.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. This paper, along with (20) provides the structure of dimeric PYR1 complexed with a single molecule of ABA, revealing conclusions similar to those reported in (20). Importantly, this paper shows that PYR1 exists as a dimer in planta and small angle X-ray scattering experiments on ABA-saturate PYR1, conducted ion solution, suggest that ABA converts the PYR1 dimer into a compact structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. Like (23) this paper reports the structure of apo and ABA-bound PYL2, and the complex of PYL1-ABA with ABI1. The conclusions of this paper are similar overall to (19) and (23) however, the authors uniquely note the importance of dimer breaking to ABA signaling and propose a model for PP2C triggered dimer disruption. [DOI] [PubMed] [Google Scholar]
- 23.Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. Like (22), this paper reports the structure of ABA-bound PYL1 and its complex with ABI1. The conclusions of this paper are similar overall to (19) and (23) [DOI] [PubMed] [Google Scholar]
- 24.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins: Structure, Function, and Genetics. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 25.Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- 26.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evolutionary Biology. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milborrow BV. The Chemistry and Physiology of Abscisic Acid. Annual Review of Plant Physiology. 1974;25:259–307. [Google Scholar]
- 28*.Hao Q, Yin P, Yan C, Yuan X, Li W, Zhang Z, Liu L, Wang J, Yan N. Functional mechanism of the ABA agonist pyrabactin. J Biol Chem. 2010;285:28946–28952. doi: 10.1074/jbc.M110.149005. This paper reports the structure of pyrabactin-bound to PYL1. One of the pyrabactin occupied subunits of the PYL1 dimer retains an open gate conformation while the other has a closed gate conformation. The data show that pyrabactin activates PYL1 in a manner analogous to ABA, that isby stabilizing gate closure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Melcher K, Xu Y, Ng L-M, Zhou XE, Soon F-F, Chinnusamy V, Suino-Powell KM, Kovach A, Tham FS, Cutler SR, et al. Identification and Mechanism of ABA Receptor Antagonism. Nature Stuctural and Molecular Biology. 2010;17:1102–1108. doi: 10.1038/nsmb.1887. This paper, along with (30), investigates the differences between PYR/PYL receptors in their response to pyrabactin. (29) reports the structure of pyrabactin bound to PYL1 and PYL2, a pyrabactin-PYL1-ABI1 ternary complex, as well as the structures of mutant proteins. While pyrabactin is an agonist of PYR1 and PYL1, it is shown in (30) to be a weak (and partial) agonist on PYL2. In (29) it is shown that pyrabactin antagonizes the ABA effects of PYL2 at high concentrations. This is exploited to investigate the structural basis for receptor antagonism, and it is shown that pyrabactin binds to PYL2 without eliciting gate closure. Results of docking screens show that new agonist can be identified using computational screens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson FC, Burgie ES, Park S-Y, Jensen DR, Weiner JJ, Bingman CA, Chang C-A, Cutler SR, George N. Phillips J, Volkman BF. Structural basis for selective activation of ABA receptors. Nature Stuctural and Molecular Biology. 2010;17:1109–1113. doi: 10.1038/nsmb.1898. The focus of this paper is on the underlying molecular basis for selective receptor activation by pyrabactin. Using PYR1 and PYL2 as models, genetics screens in yeast identified two residues (I110 and I62) critical for pyrabactin’s agonism of PYR1. These residues are pocket-lining residues that vary between family members.In PYL2 these residues are Valines, instead of Isoleucines. PYL2 can be converted to a pyrabactin-sensitivereceptor by mutatingits valine residues to isoleucines. These changes are shown to alter pyrabactin’s orientation within the pocket. Study (29) shows that these residues control pyrabactin orientation and activity as well. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. This paper, along with (13) are landmarks in ABA signaling. They demonstrate the sufficiency of the core pathway (originally proposed in reference (9)) to mediate information transfer from receptor to a nuclear output (in the case of (13) or SnRK2 activation in the case of (31). In addition, this study locates the critical site of phosphorylation on ABA stimulated OST1 to the activation loop by LC-MS/MS proteomics, conclusions which are very complementary to those reported in (41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci U S A. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- 34.Mikolajczyk M, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- 35.Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. This paper, along with (38) and (39) show the critical importance of the SnRK2 kinases to ABA signaling. A triple mutant lacking OST1, SnRK2.2 and SnRk2.3 is defective in almost all ABA response measured. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. This paper and (39) constructed the same SnRK2 triple mutant reported in (37). The results are in agreement. The SnrK2s rock ABA’s world! [DOI] [PubMed] [Google Scholar]
- 39*.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling are Essential for the Control of Seed Development and Dormancy. Plant and Cell Physiology. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. See comment for (38) [DOI] [PubMed] [Google Scholar]
- 40.Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. This paper demonstrates that the SnRK2 kinase OST1 is a direct substrate of the PP2C phosphatase HAB1. Serine 175 in OST1’sactivation loop is definedas a critical phosphorylation site. The experimental identification of a PP2C substrate consensus sequence was used to identify OST1 as a substrateusing bioinformatic searches for proteins containing the consensus sequence. Moreover, the prevalence of this sequence in many proteins suggests that there may be many PP2C substrates that are potentially regulated by the PYR/PYL core ABA signaling system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi H, Hong J, Ha J, Kang J, Kim S. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 43.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 46.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci U S A. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. This paper, along with (49) shows that the anion channel activity of SLAC1 is stimulated by the SnRK2 OST1, using heterologous experiments in Xenopus oocytes. As expected, ABI1 antagonizes this. Moreover, the N-terminus of SLAC1 can be phosphorylated invitro by OST1 and this is antagonized by ABI1’s inhibitory effect on OST1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci U S A. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. The results in (49) are in general agreement with (48) the core pathway regulates SLAC1 anion channel activity, however the direct reconstitution of ABA-mediated activation by incorporating PYR/PYL proteins remains to be demonstrated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Vahisalu T, Puzorjova I, Brosche M, Valk E, Lepiku M, Moldau H, Pechter P, Wang YS, Lindgren O, Salojarvi J, et al. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 2010;62:442–453. doi: 10.1111/j.1365-313X.2010.04159.x. As observed in (48) and (49), the N-terminus of SLAC is can be phoshorylated by OST1 in vitro. In in vitro experiments followed by LC-MS/MS analyses of phosphopeptides, multiple serines were phosphorylated by OST1, including S120. A tilling allele was isolated that contains an S120F point mutation in SLAC1, and showed a strong guard cell phenotype, suggesting that phosphorylation at this site is physiologically important. [DOI] [PubMed] [Google Scholar]
- 51*.Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J. 2009;424:439–448. doi: 10.1042/BJ20091221. The C-terminus of the potassium channel KAT1 is shown to be phosphorylated by the SnRK2 kinase OST1 invitro on residues T306 and T308. Several mutations at T306 impair channel activity when assayed in Xenopus. In functional complementation assays in yeast however, T306A, T306N and T306S mutants allow complementation of a K+ transport defective yeast strain, however T306D, T306E and T306Q do not complement the potassium uptake defect of the yeast strain. The authors propose that the non-complementing mutants are phospho-mimics that inactivate the channel and that OST1’s function is to negatively regulate channel activity. While the model is appealing and “fits”, discrepancies between the Xenopus and S. cerevisiae data, lack of in planta or other complementary data leave this critical point unresolved. [DOI] [PubMed] [Google Scholar]
- 52*.Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. OST1 is shown to phosphorylate S174 and S13 on the respiratory burst oxidase subunit homolog subunit F (AtrbohF) and it is speculated that OST1 phosphorylation may modulate rbohF activity. Because ABA has long been known to trigger ROS accumulation in guard cells, this experiment provides a tantalizing link between the core ABA signaling pathway and ROS generation. In vivo and functional experiments will be needed to fully appreciate the significance of these observations to ABA signaling and ROS production, however as a betting man, I’ll wager that they’re right. [DOI] [PubMed] [Google Scholar]
- 53**.Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci U S A. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. This paper is the first to link calcium dependent kinases into the core pathway and suggests that the PYR/PYL-PP2C signaling module controls activity of CPKs, in particular CPK23. CPK21 and CPK23 were identified as interacting partners with SLAC1 in split two hybrid experiments. SLAC1 channel activity was previously shown to be modulated by OST, and here it is shown that CPK23 can stimulate SLAC1 channel activity when coexpressed with SLAC1 in Xenopus oocytes, and this is antagonized by coexpressing ABI1or ABI2 (but interestingly not HAB1 or HAB2). In vitro phosphorylation assays show that the N-terminus of SLAC can be phosphorylated by CPK23 and that this is antagonized by ABI1, suggesting regulation of CPK activity by PP2Cs. Moreover, inclusion of RCAR1 leads to ABA modulated phosphorylation of SLAC1’s N-terminus. [DOI] [PMC free article] [PubMed] [Google Scholar]