SUMMARY
Lung cancer is the leading cause of cancer-related deaths in the world, and non-small cell lung cancer (NSCLC) accounts for 80% of cases. MicroRNA-21 (miR-21) expression is increased and predicts poor survival in NSCLC. Although miR-21 function has been studied in vitro using cancer cell lines, the role of miR-21 in tumor development in vivo is unknown. We utilize transgenic mice with loss-of-function and gain-of-function miR-21 alleles combined with a model of NSCLC to determine the role of miR-21 in lung cancer. We show that over-expression of miR-21 enhances tumorigenesis and genetic deletion of miR-21 partially protects against tumor formation. MiR-21 drives tumorigenesis through inhibition of negative regulators of the Ras/MEK/ERK pathway and inhibition of apoptosis.
INTRODUCTION
MicroRNAs are evolutionarily conserved, endogenous, non-protein coding, ~20–23 nucleotide single-stranded RNAs that negatively regulate gene expression in a sequence-specific manner (Cho, 2007; Esquela-Kerscher and Slack, 2006; Voorhoeve and Agami, 2007). The human genome is predicted to encode as many as 1000 miRNAs, or about 3% of the total number of human genes (Bartel, 2004; Esquela-Kerscher and Slack, 2006). The 5′ portion of miRNA sequence containing bases two to eight, termed the “seed” region, is important in target mRNA recognition. miRNAs negatively regulate target gene expression through complementarity between the miRNA seed sequence and the target mRNA 3′ UTR. miRNAs that bind with perfect complementarity to the protein encoding messenger RNA (mRNA) target the mRNA for destruction, whereas miRNAs with imperfect complementarity to the 3′ untranslated region (UTR) of the mRNA target repress mRNA translation. Expression of approximately 30% of human proteins appears to be regulated by miRNAs (Lewis et al., 2005). Through interactions with 3′UTRs, miRNAs can modulate the expression of many genes simultaneously, often regulating individual signaling pathways at multiple levels (Baek et al., 2008; Selbach et al., 2008).
An integral role of miRNAs in cancer pathogenesis has begun to emerge. MiRNA expression profiling reveals characteristic signatures for many tumor types including NSCLC (Volinia et al., 2006) and are predictive of tumor classification, prognosis and response to therapy (Calin and Croce, 2006). MiRNA expression patterns are remarkably reliable markers of cancers; in some cases they have even proven more reliable than conventional histology (Subramanian et al., 2007). MicroRNAs are capable of functioning as classical tumor suppressors or oncogenes, thus actively participating in human cancer pathogenesis (Ventura and Jacks, 2009). Recently, gain and loss-of-function studies in mice demonstrate critical roles for miR-26a and miR-9 in hepatocellular carcinoma and breast cancer metastasis, respectively (Kota et al., 2009; Ma et al., 2010). These data suggest that the pattern of miRNA expression contributes to fundamental aspects of tumor biology.
A large scale survey to determine the miRNA signature of 540 tumor samples including lung, breast, stomach, prostate, colon, and pancreatic tumors and their respective normal adjacent tissue revealed miR-21 was the only miRNA up-regulated in all these tumors (Volinia et al., 2006). Further miRNA profiling in tumor samples and cancer cell lines show increased miR-21 expression in glioblastoma (Chan et al., 2005; Ciafre et al., 2005), head and neck carcinomas (Tran et al., 2007), ovarian cancer (Iorio et al., 2007), B-cell lymphoma (Lawrie et al., 2007), hepatocellular (Meng et al., 2007), and cervical carcinoma (Lui et al., 2007). These studies clearly illustrate miR-21 dysregulation in tumors; however, the studies do not prove a causal role for miR-21 in cancer pathogenesis.
Functional studies in cancer cell lines suggest that miR-21 has oncogenic activity. Knockdown of miR-21 in cultured glioblastoma cells activates caspases leading to apoptotic cell death, suggesting miR-21 is an anti-apoptotic factor (Chan et al., 2005). In breast cancer MCF-7 cells, miR-21 knock-down results in suppression of cell growth in vitro and tumor growth in xenografts (Si et al., 2007). Knock-down of miR-21 in the metastatic breast cancer MD-MBA-231 cells reduced invasion and metastasis (Zhu et al., 2008). Targeted deletion of miR-21 in RKO and DLD1 colon cancer cells revealed miR-21 contributes to tumorigenesis through compromising cell cycle progression and DNA damage-induced checkpoint function through the Cdc25a target gene (Wang et al., 2009). These studies indicate knock-down of miR-21 expression in cancer cell lines results in phenotypes important for tumor biology. However, the potential role of miR-21 in tumorigenesis in vivo has not yet been explored.
Lung cancer is the most common form of cancer in the world, accounting for approximately 12.3% of all cancers with an estimated 1.2 million new cases each year (Parkin et al., 2001). Lung cancer is also the leading cause of cancer-related deaths in the world, wiht non-small cell lung cancer (NSCLC) accounting for 80% of all cases (Ramalingam et al., 1998). Despite novel therapies and advances in early detection, NSCLC is often diagnosed at an advanced stage and has a poor prognosis, with a median survival of 8 to 11 months and a five year survival rate in patients with NSCLC of only 13% using conventional cytotoxic chemotherapy (Soon et al., 2009). Recently, miR-21 expression levels have proven useful prognostic markers in non-small cell lung cancer (Markou et al., 2008; Yanaihara et al., 2006). The level of miR-21 expression in sputum distinguishes NSCLC patients from cancer-free controls with a greater sensitivity than conventional cytology (Xie et al., 2009). Currently, more experiments are needed to determine if miR-21 participates in driving the malignant phenotype or simply reflects the cellular stress.
In the current study, we used transgenic mice with both loss-of-function and gain-of-function miR-21 alleles in combination with the K-rasLA2 mouse model of non-small cell lung cancer to elucidate the role of miR-21 in NSCLC pathogenesis.
RESULTS
miR-21 over-expression promotes tumor development In vivo
Increased expression of miR-21 in human cancer could reflect malignant physiology or could determine important aspects of cancer biology. To address this question, we generated transgenic mice that over-express the miR-21 gene and mice with deletion of the miR-21 gene, and tested the consequences in a mouse model of NSCLC. We designed a strategy to conditionally over-express miR-21 using the Cre-lox system and the CAG-Z-EGFP vector (Figure S1A and Supplementary experimental procedures) (Fukuda et al., 2005). Global miR-21 over-expression was achieved by breeding the CAG-Z-miR21-EGFP mouse to transgenic mice expressing Cre recombinase from the ubiquitous CAG promoter resulting in the CAG-miR-21 mouse, which globally over-expressed miR-21 4–6 fold over normal levels of expression (Figure S1B–D). CAG-miR-21 transgenic mice were viable, fertile and born in expected Mendelian ratios. MiR-21 expression increases in mouse models of cardiac hypertrophy and following myocardial infarction; however, adult male CAG-miR-21 mice showed no signs of heart failure compared to littermate controls (Figure S1E)(van Rooij et al., 2006; van Rooij et al., 2008).
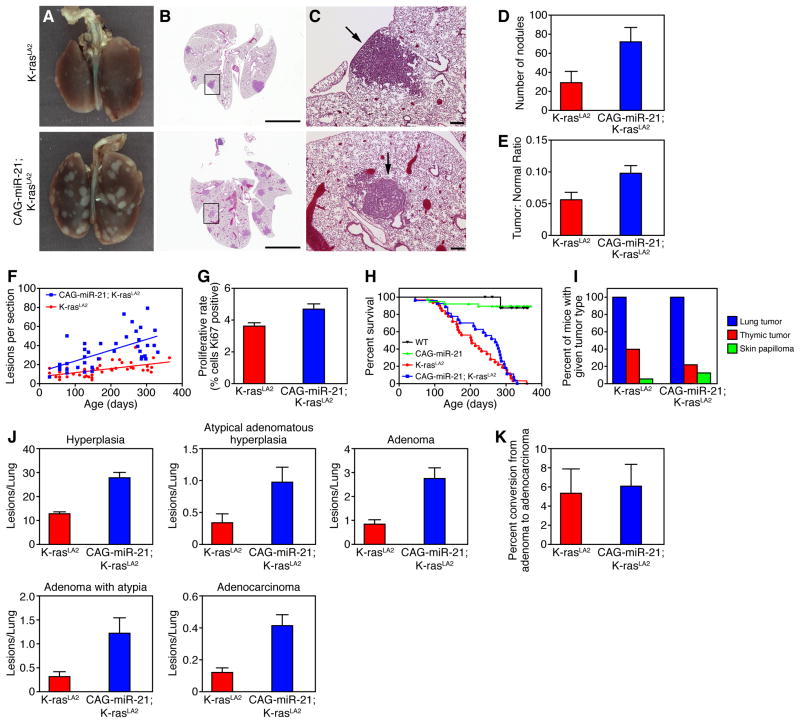
To explore the role of miR-21 in the pathogenesis of NSCLC, we utilized the K-rasLA2 murine lung cancer model, which harbors a targeted, latent K-ras G12D allele that is activated by two distinct recombination events (Johnson et al., 2001). The first occurs in ES cells, and the second occurs in vivo, resulting in the random activation of the mutant K-ras G12D allele in somatic cells. K-rasLA2 mice develop multi-focal lung tumors with 100% penetrance, and less frequently develop thymic lymphomas and skin papillomas (Johnson et al., 2001). CAG-miR-21;K-rasLA2 compound mutant mice were generated by breeding to compare the number, rate of formation and histology of tumors relative to littermate control K-rasLA2 mice. Remarkably, over-expression of miR-21 enhanced the number of tumors in this model of lung cancer (Figure 1A–C). Compared to K-rasLA2 littermates, CAG-miR-21;K-rasLA2 mice displayed significantly more lung tumors visible grossly at 18 weeks (Figure 1D) and microscopically on H&E cross-section over all time points (Figure 1F). Total tumor area as a proportion of total lung area was significantly increased with miR-21 over-expression (Figure 1E).
Figure 1. miR-21 over-expression enhances tumor formation in the K-rasLA2 mouse model of NSCLC.
(A) Gross and (B, C) cross sectional H&E histology of lungs isolated from CAG-miR-21;K-rasLA2 mice and K-rasLA2 littermate controls at 18 weeks of age. (C) Magnification of boxed lesions in (B). Arrows in top and bottom panels indicate a hyperplastic lesion and adenoma, respectively. Scale bars, (B) 5 mm, (C) 200 μm.
(D) Nodules grossly visible were counted on the lung surface of CAG-miR-21;K-rasLA2 (n = 5) and K-rasLA2 (n = 4) mice sacrificed at 18 weeks of age. Results are mean ± SEM. P value 0.013 using two-tailed, unpaired Student’s t-test.
(E) Tumor burden measured as the ratio of total tumor area to total lung area of the lungs counted in (D). Results are mean ± SEM. P value 0.045 using two-tailed, unpaired Student’s t-test.
(F) Quantification of tumor lesions on lung H&E cross-section from CAG-miR-21;K-rasLA2 (n = 49) and K-rasLA2 (n = 40) mice over time. Slopes were determined by linear regression, P value 0.0027.
(G) Quantification of proliferating cells detected by Ki67 antibody immunostaining of lung tumors. Results are mean ± SEM (n = 12). P value 0.01 by two-tailed, unpaired Student’s t-test.
(H) Survival curve of CAG-miR-21;K-rasLA2 (n = 27) and K-rasLA2 (n = 32) mice (P = 0.367) with median survival of 270 and 203 days, respectively. WT (n = 10), CAG-miR-21 (n = 20).
(I) Tumor spectra in CAG-miR-21;K-rasLA2 (n = 27) and K-rasLA2 mice (n = 32).
(J) Lung H&E cross sections from mice in the survival cohort were analyzed and all lesions were classified for tumor grade. CAG-miR-21;K-rasLA2 (n = 27), K-rasLA2 (n = 32). Hyperplasia, P value < 0.0001. Atypical adenomatous hyperplasia, P value 0.017. Adenoma, P value < 0.0001. Adenoma with atypia, P value 0.0042. Adenocarcinoma, P value 0.033. All data represented as the mean ± SEM of the number of lesions per lung. (K) Rate of conversion of adenoma to adenocarcinoma, data represented as mean ± SEM, P value 0.83. See also Figure S1.
Tumors in CAG-miR-21;K-rasLA2 mice exhibited increased proliferation compared with K-rasLA2 control tumors (Figure 1G). Despite the increased tumor burden, the survival of CAG-miR-21;K-rasLA2 mutant mice did not differ from K-rasLA2 control mice (Figure 1H). This is may be explained by a decreased incidence of thymic tumors in CAG-miR-21;K-rasLA2 mutant mice compared to K-rasLA2 controls (Figure 1I), thus contributing to the enhanced survival. Unlike K-rasLA1;p53−/− compound mutant mice (Johnson et al., 2001), we did not observe an increased tumor spectrum or increased metastasis in the CAG-miR-21;K-rasLA2 mutant mice. MiR-21 over-expression increased the incidence of all tumor grades without increasing the rate of conversion to adenocarcinoma, suggesting that miR-21 participates in tumor promotion rather than progression and metastasis in this model (Figure 1J and 1K). MiR-21 over-expression did not increase the rate of recombination of the K-rasLA2 allele (Figure S1F). We followed CAG-miR-21 mice (n = 39) without K-rasLA2 for up to 555 days and performed full necropsy and histological analysis on H&E sections the following tissues: lung, heart, brain, spleen, liver, kidney, stomach, colon, skeletal muscle, and bone marrow. No tumors were observed in CAG-miR-21 mice without K-rasLA2, indicating that miR-21 over-expression alone is not sufficient for tumorigenesis. These data suggest that miR-21 over-expression in human tumors does not simply reflect cancer pathology, but instead enhances aspects of the pathology of NSCLC.
miR-21 deletion suppresses Ras-driven transformation In vitro
To address the necessity of miR-21 in the development of NSCLC, we generated miR-21 knock-out mice. Homozygous miR-21 knock-out mice are viable, fertile, and born in expected Mendelian ratios without any gross phenotypic differences compared to wild type or heterozygous littermates, and without a cardiac phenotype (D.P., E.V.R. and E.N.O., manuscript in preparation). To dissect the role of miR-21 in Ras-driven transformation, we isolated mouse embryonic fibroblasts (MEFs) from wild type and miR-21−/− mice. Northern blot and real-time PCR confirmed deletion of miR-21 (Figure S2A and S2B). These MEFs were immortalized and transformed with retroviruses expressing SV40 large T and small t antigens and H-rasG12V. There was no difference in proliferation between the miR-21−/− and wild type MEFs (Figure S2C). However, the miR-21−/− MEFs formed significantly less colonies in soft agar than wild type MEFs (Figure S2D) and developed smaller tumors in xenografts in nude mice (Figure S2E). These observations suggest that miR-21 actively participates in Ras-driven transformation.
miR-21 deletion suppresses tumor development In vivo
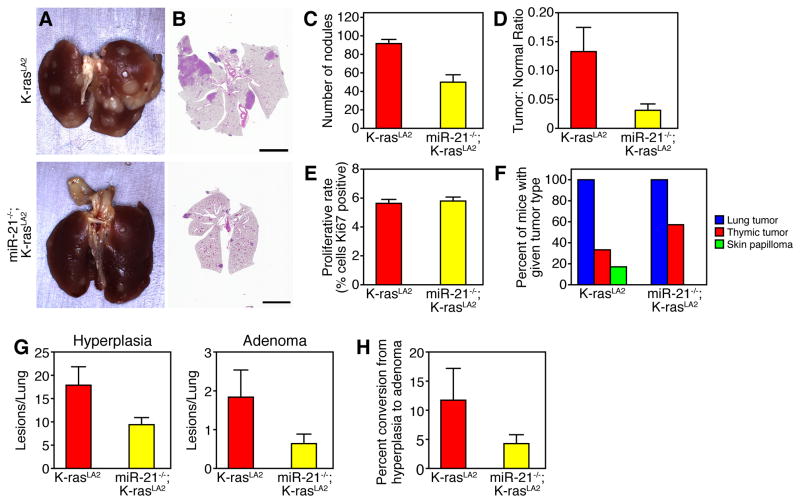
Given that high levels of miR-21 expression in patients with NSCLC serve as an independent negative prognostic factor and that miR-21-deficient MEFs form less colonies in soft agar, we hypothesized that genetic deletion of miR-21 in the K-rasLA2 model would alter tumorigenesis. To explore this possibility, we crossed the miR-21−/− allele into the K-rasLA2 NSCLC model. Significantly less tumors were present on the surface of the lungs of 20 week-old miR-21−/−;K-rasLA2 compound mutant mice compared to K-rasLA2 control mice (Figure 2A–C). Total tumor area as a proportion of total lung area was significantly decreased with miR-21 deletion (Figure 2D). MiR-21 deletion did not alter proliferation in tumors measured by Ki67 staining (Figure 2E). MiR21−/−;K-rasLA2 mice all develop lung tumors, but show an increased incidence of thymic lymphoma (Figure 2F). MiR-21−/−;K-rasLA2 mutant mice displayed a reduced incidence of hyperplasic lesions and adenomas compared to K-rasLA2 controls (Figure 2G). Although not statistically significant, there was a trend towards decreased conversion of hyperplasia to adenoma in the miR-21−/−;K-rasLA2 mice (Figure 2H). No adenocarcinomas were noted in any of the 14 miR-21−/−/K-rasLA2 animals evaluated. These results illustrate that miR-21 deletion suppresses tumor development In vivo.
Figure 2. miR-21 deletion suppresses tumorigenesis in K-rasLA2 model of NSCLC.
(A) Gross and (B) cross sectional H&E histology of lungs isolated from miR-21−/−;K-rasLA2 and K-rasLA2 mice at 20 weeks of age. Scale bar, 5 mm.
(C) Quantification of nodules grossly visible on the lung surface of miR-21−/minus;;K-rasLA2 (n = 5) and K-rasLA2 (n = 3) mice at 20 weeks of age. Results are mean ± SEM. P value 0.008 using two-tailed, unpaired Student’s t-test.
(D) Tumor burden measured as the ratio of total tumor area to total lung area of the lungs counted in (C). Results are mean ± SEM. P value 0.015 using two-tailed, unpaired Student’s t-test.
(E) Quantification of proliferating cells detected by Ki67 antibody immunostaining of lung tumors. Results are mean + SEM (n = 6). P value 0.4 by two-tailed, unpaired Student’s t-test.
(F) Tumor spectra in K-rasLA2 and miR-21−/−;K-rasLA2 mice.
(G) Lung H&E cross sections from miR-21−/−;K-rasLA2 (n = 14), K-rasLA2 (n = 6) mice were analyzed and all lesions were classified for tumor grade. Hyperplasia, P value 0.023. Adenoma, P value 0.057. All data represented as the mean ± SEM of the number of lesions per lung.
(H) Rate of conversion of hyperplasia to adenoma. Data represented as mean ± SEM, P value 0.089. See also Figure S2.
miR-21 targets multiple tumor suppressor genes
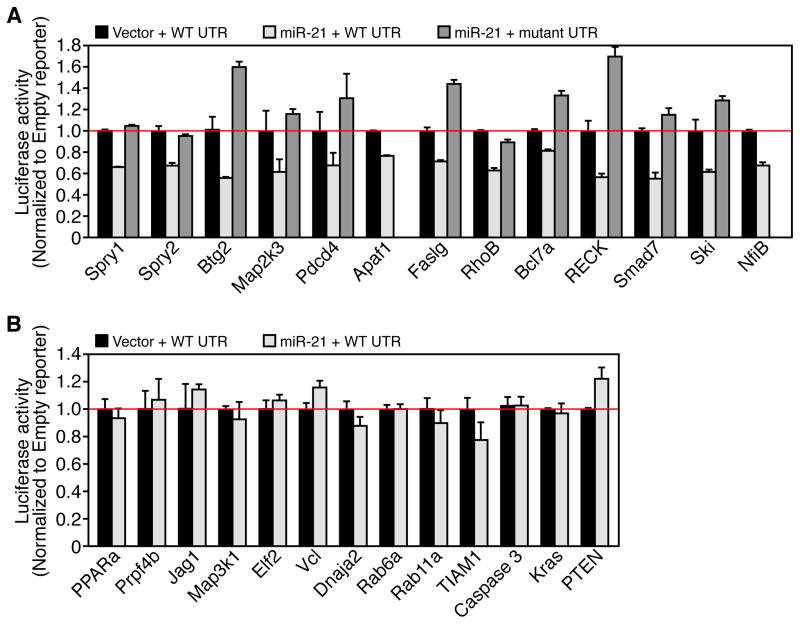
To identify miR-21 effectors responsible for phenotypes observed, we utilized two algorithms that predict the mRNA targets of miRNAs, TargetScan (Grimson et al., 2007) and PicTar (Krek et al., 2005). Approximately 180 mRNAs are predicted as miR-21 target genes based on the presence of miR-21 sites in their 3′ UTRs. Many of the miR-21 predicted targets have been reported to function as tumor suppressors and have been validated in vitro in previous studies (Selcuklu et al., 2009). We cloned the 3′ UTRs of 24 putative mouse miR-21 target genes into a luciferase reporter construct. Reporter assays in COS cells revealed miR-21 dependent repression of 13 of the 24 3′UTRs and mutation of the miR-21 site in the 3′ UTR abrogated the repression in 11 of the putative target gene 3′ UTRs (Figure 3A). Four of the validated miR-21 target genes are known negative regulators of the Ras/MEK/ERK pathway: sprouty 1 (Spry1), sprouty 2 (Spry2), B-cell translocation gene 2 (Btg2), and programmed cell death 4 (Pdcd4). Several pro-apoptotic genes were directly targeted by miR-21, apoptotic peptidase activating factor 1 (Apaf1), Fas ligand (Faslg), Pdcd4, and ras homolog gene family member B (RhoB). We show miR-21 dependent repression of the mouse Apaf1 3′ UTR reporter; however, the miR-21 site in the human APAF1 3′ UTR is conserved in primates but not conserved in the mouse. Eleven of the putative miR-21 target gene 3′ UTRs including phosphatase and tensin homolog (Pten) were not repressed by miR-21 (Figure 3B). Caspase 3 and K-ras do not contain miR-21 sites in their 3′ UTR and the 3′ UTR luciferase reporter activity is not suppressed by miR-21 (Figure 3B). These results suggest that in vitro miR-21 can repress multiple tumor suppressor genes that may participate in tumorigenesis.
Figure 3. miR-21 targets mRNAs encoding multiple tumor suppressors.
(A) Luciferase activity in COS cells co-transfected with miR-21 or control vector and miR-21 responsive target gene 3′ UTRs or miR-21 site mutant 3′ UTR luciferase reporters. Data represented as mean ± SEM and normalized to empty vector (no miR-21) control. n = 4.
(B) Luciferase activity in COS cells cotransfected with miR-21 or empty vector and indicated miR-21 unresponsive 3′ UTRs. Data represented as mean ± SEM and normalized to empty vector (no miR-21) control. n = 4.
Oncogenic Ras activation increases miR-21 expression In vivo
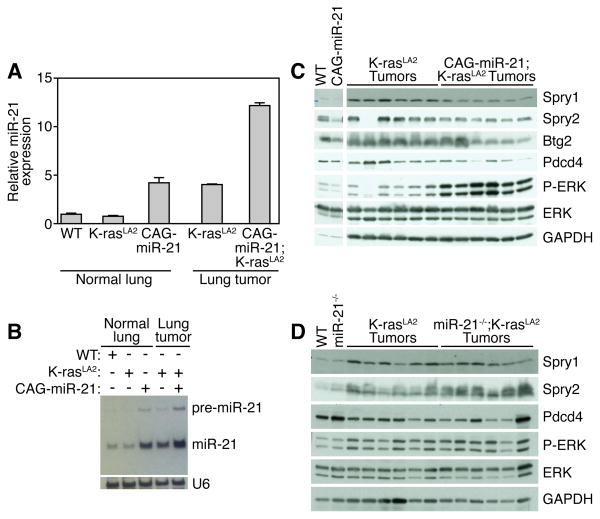
Previously, an autoregulatory loop connecting miR-21 and Ras activation through AP-1 was illustrated in a rat thyroid cell system (Talotta et al., 2009). miR-21 expression was induced by AP-1 in response to Ras activation, and miR-21 mediated Ras-dependent downregulation of Pdcd4 (Talotta et al., 2009), a known negative regulator of AP-1 transactivation (Goke et al., 2004; Jansen et al., 2005; Yang et al., 2003; Yang et al., 2001). As well, epidermal growth factor receptor (EGFR) and HER2/neu signaling positively regulates miR-21 expression in human lung and breast carcinoma cell lines, respectively (Huang et al., 2009; Seike et al., 2009). We confirmed this autoregulatory loop in vivo by analyzing miR-21 expression in normal lung and tumors of K-rasLA2 and CAG-miR-21;K-rasLA2 mutant mice. Normal lung tissue from K-rasLA2 mice (prior to K-ras activation) exhibits wild type expression levels of miR-21 (Figure 4A and 4B). K-ras activation in the tumors of K-rasLA2 mice increased miR-21 expression 4-fold similar to the expression level in the CAG-miR-21 transgenic mouse lung. In CAG-miR-21;K-rasLA2 tumors, miR-21 expression was more than additive compared to either CAG-miR-21 or K-rasLA2 mice alone. We speculate that by driving miR-21 expression in the transgenic mouse, we provided a “second hit” prior to K-ras activation, thus accelerating tumor initiation.
Figure 4. miR-21 induction by oncogenic K-ras represses negative regulators of the Ras/MEK/ERK pathway.
(A) MiR-21 expression in lung tumors from CAG-miR-21;K-rasLA2 and K-rasLA2 mice (after K-ras activation) compared to normal lung from wild-type, K-rasLA2 (prior to K-ras activation) and CAG-miR-21 by realtime PCR. Results are mean ± SEM.
(B) Northern blot analysis of miR-21 expression in lung tumors from CAG-miR-21;K-rasLA2 and K-rasLA2 mice (after K-ras activation) compared to normal lung from wild-type, K-rasLA2 (prior to K-ras activation) and CAG-miR-21.
(C) Western blot analysis of lung lysates from normal lung from wild-type (WT) and CAG-miR-21 mice and six isolated lung tumors from CAG-miR-21;K-rasLA2 and K-rasLA2 mice. Antibodies used are shown on right.
(D) Western blot analysis of lung lysates from wild-type (WT) and miR-21−/− mice and six isolated tumors from miR-21−/−;K-rasLA2 and K-rasLA2 mice. Antibodies used are shown on right. See also Figure S3.
miR-21 targets multiple negative regulators of Ras signaling
Multiple negative regulators of the Ras/MEK/ERK pathway were shown to be direct miR-21 target genes by luciferase reporter assays (Figure 3B). Although there have been reports of miR-21 target gene regulation in cancer cell lines, the constellation of genes regulated by miR-21 in NSCLC in vivo remains elusive. We hypothesize that increased miR-21 expression potentiates Ras signaling through inhibition of antagonists of the Ras pathway, such as Spry1, Spry2, Btg2, and Pdcd4 (Casci et al., 1999; Hanafusa et al., 2002; Lo et al., 2006).
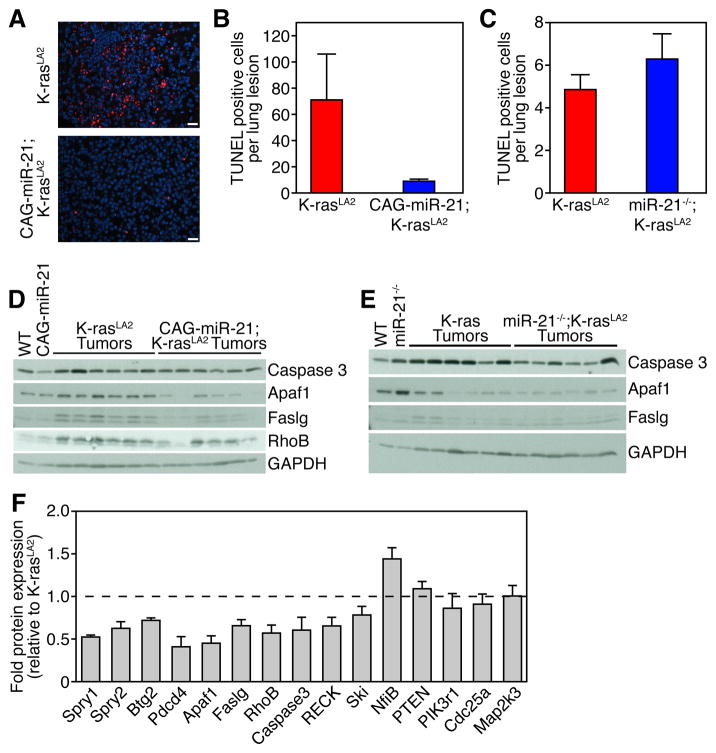
We isolated individual tumors from mice, made whole tumor lysates, and performed Western blots for putative miR-21 targets to address the effect of miR-21 expression on the protein levels of the miR-21 target genes in vivo within lung tumors. MiR-21 over-expression in CAG-miR-21;K-rasLA2 mutant tumors decreases Spry1, Spry2, and Btg2 protein expression compared to K-rasLA2 tumors (Figure 4C). The decrease in Spry1, Spry2, and Btg2 in CAG-miR-21;K-rasLA2 tumors correlates with increased Ras pathway activity, shown by increased ERK phosphorylation (Figure 4C). MiR-21−/−;K-rasLA2 mutant tumors did not show alteration in the regulation of ERK by Spry1, Spry2, and Btg2 (Figure 4D). Protein analysis of individually isolated tumors showed that Pdcd4 levels were decreased in CAG-miR-21;K-rasLA2 compound mutant mice compared to K-rasLA2 control tumors (Figure 4C). This regulation was absent in tumors from the miR-21−/−;K-rasLA2 compound mutant mice (Figure 4D). RECK, Cdc25a, Map2k3, NfiB, Trp63 and Ski have been shown to be miR-21 target genes in vitro, but only RECK showed decreased protein levels in the CAG-miR-21;K-rasLA2 tumors (Figure S3A). Several components of the PI3K pathway, including PTEN and PIK3R1, have been suggested as miR-21 target genes (Meng et al., 2007); however, miR-21 over-expression did not alter PTEN and PIK3R1 protein expression or Akt phosphorylation (Figure S3B). These data show that miR-21 over-expression decreases the protein expression of four miR-21 target genes that are tumor suppressors and negative regulators of the Ras/MEK/ERK pathway resulting in a concomitant increased ERK activity.
miR-21 suppresses apoptosis by targeting proapoptotic genes
Next, we evaluated the effect of miR-21 expression on apoptosis. MiR-21 over-expression in CAG-miR-21;K-rasLA2 mice reduced apoptosis in lung tumors as shown by TUNEL staining (Figure 5A and 5B). Deletion of miR-21 had no effect on the amount of apoptosis in the lung lesions as detected by TUNEL suggesting that lung tumors that develop in the background of the genetic deletion of miR-21 have escaped miR-21 regulatory effects (Figure 5C). Several miR-21 target genes are tumor suppressors involved in apoptosis including Apaf11, Pdcd4, RhoB, and Faslg. Lysates from individually isolated tumors from CAG-miR-21;K-rasLA2 mice showed decreased expression of Apaf1, Caspase 3, Pdcd4, Faslg and RhoB (Figures 4C and 5D). MiR-21−/−;K-rasLA2 mutant tumors did not show alteration in the regulation of Apaf1 or Faslg (Figure 5E). Apaf1, Pdcd4, Faslg, and RhoB were validated as direct miR-21 target genes by luciferase reporter assays; however, caspase 3 was not directly regulated by miR-21 (Figure 3B). Quantification of miR-21 target Western blots is shown in Figure 5F. These data illustrate that miR-21 over-expression decreases the protein expression of four miR-21 target genes that are involved in apoptosis thus promoting survival.
Figure 5. miR-21 reduces apoptosis through targeting multiple apoptotic modulators. (A) TUNEL staining of K-rasLA2 and CAG-miR-21;K-rasLA2 tumors. Scale bars 20 μm.
(B) Quantification of TUNEL positive nuclei per lung tumor. K-rasLA2 (n = 16) and CAG-miR-21;K-rasLA2 (n = 62). Results are mean ± SEM. P value 0.007 by unpaired, two-tailed Student’s t-test.
(C) Quantification of TUNEL staining in miR-21−/−;K-rasLA2 and K-rasLA2 controls. K-rasLA2 (n = 42) and miR-21−/−;K-rasLA2 (n = 22). Results are mean ± SEM. P value 0.28 using unpaired, two-tailed Student’s t-test.
(D) Western blot analysis of lung lysates from wild-type (WT) and CAG-miR-21 mice and six isolated lung tumors from CAG-miR-21;K-rasLA2 and K-rasLA2 mice. Antibodies used are shown on right.
(E) Western blot analysis of lung lysates from WT and miR-21−/− mice and six isolated lung tumors from miR-21−/−;K-rasLA2 and K-rsLA2 mice. Antibodies are shown on right. (F) Quantification of protein expression from Western blots shown in Figure 4C, Figure 5D, and Figure S3A and S3B. Bar values indicate the mean ± SEM of protein levels in six independent tumors from CAG-miR-21;K-rasLA2 mice after normalization to Gapdh and represented as fraction of the protein levels in the K-rasLA2 controls. Dashed line indicates the protein level in K-rasLA2 tumors.
miR-21 deletion sensitizes cells to DNA-damaging chemotherapy
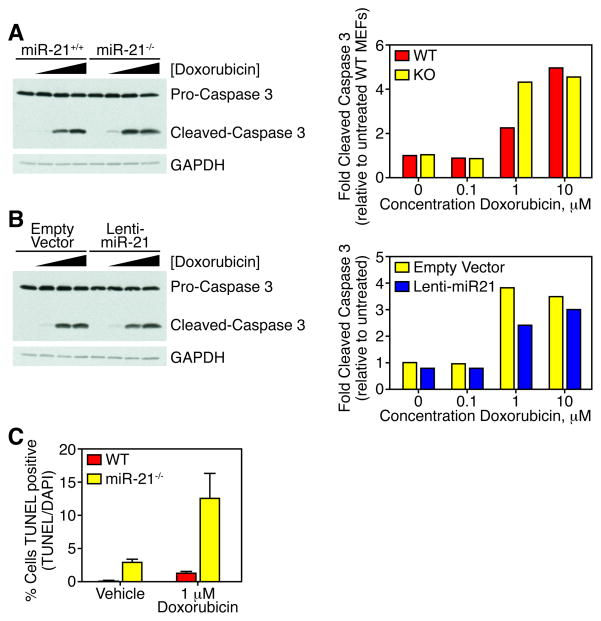
Recent studies illustrate that miRNAs modulate sensitivity of cell to chemotherapy (Blower et al., 2008). We utilized the miR-21−/− and wild type MEFs immortalized with TAg and transformed with RasV12 to tested the role of miR-21 in doxorubicin-induced apoptosis. MEFs were cultured with increasing concentrations of doxorubicin, whole cell lysates were isolated, and apoptotic activity was measured by immunoblotting for cleaved caspase 3. MEFs from miR-21−/− mice display increased sensitivity to doxorubicin-induced apoptosis measured by protein levels of cleaved caspase 3 (Figure 6A). miR-21−/− MEFs were transduced with a lentivirus expressing miR-21 to rescue the miR-21 deficiency (Figure S4). Lentiviral over-expression of miR-21 decreased cleaved caspase 3 levels compared to empty lentiviral transduced miR-21−/− MEFs. The increased sensitivity to doxorubicin-induced measure by cleaved caspase 3 levels was confirmed by TUNEL (Figure 6C). Therefore, miR-21 plays a significant role in inhibition of apoptosis thus allowing tumor survival.
Figure 6. miR-21 deletion sensitizes cells to doxorubicin-induced apoptosis.
(A) Western blot analysis (left panel) for cleaved caspase 3 in protein lysates from wild-type and miR-21−/− MEFs immortalized with T-Ag, transformed with H-rasV12, and treated with increasing concentrations of doxorubicin. Antibodies used are shown on right. Quantification of cleaved caspase 3 levels normalized to pro-caspase 3 and represented relative to untreated wild-type MEFs (right panel).
(B) Western blot analysis (left panel) for cleaved caspase 3 in protein lysates from miR-21−/− MEFs immortalized with T-Ag, transformed with H-rasV12, and transduced with lentivirus expressing miR-21 (lenti-miR-21) or empty vector, and treated with increasing concentrations of doxorubicin. Antibodies used are shown on right. Quantification of cleaved caspase 3 levels normalized to pro-caspase 3 and represented relative to untreated wild-type MEFs (right panel).
(C) TUNEL assay of wild-type and miR-21−/− MEFs treated with vehicle or 1 μM doxorubicin for 12 hours. Data represented as mean ± SEM, n = 6. See also Figure S4.
DISCUSSION
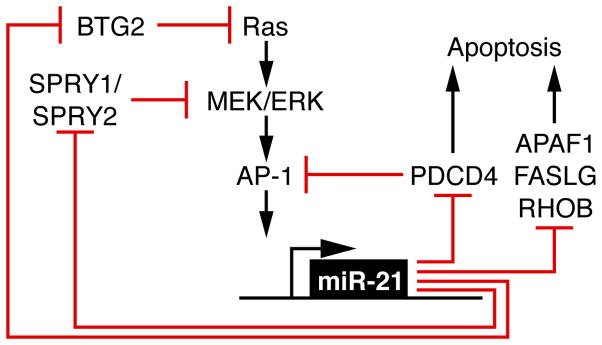
Our results show that miR-21 expression modulates tumor number, incidence and size in a mouse lung cancer model initiated by oncogenic K-rasG12D, consistent with miR-21 functioning as a tumor promoter. miR-21 was not sufficient in our model for tumorigenesis; however, under the control of other tissue-specific promoters, miR-21 could have cancer related phenotypes. miR-21 modulates several components critical to NSCLC pathogenesis through both relieving antagonism of the Ras pathway and reducing apoptotic effectors (Figure 7). By targeting antagonists of Ras/MEK/ERK signaling and pro-apoptotic genes, miR-21 enhances tumor proliferation and survival, two critical components of tumor promotion.
Figure 7.
Proposed model showing that miR-21 potentiates oncogenic Ras signaling and attenuates apoptosis through repression of multiple tumor suppressors, miR-21 target genes.
Our results define in vivo an autoregulatory loop between oncogenic Ras and miR-21 mediated by Spry1, Spry2, Btg2, and Pdcd4. PDCD4 is a pro-apoptotic tumor suppressor gene and has been verified as a miR-21 target in human colorectal and breast cancer cell lines (Afonja et al., 2004; Asangani et al., 2007; Bitomsky et al., 2008; Frankel et al., 2007; Lu et al., 2008; Zhang et al., 2006). Pdcd4 functions both as an inducer of apoptosis and a suppressor of AP-1 activity (Hwang et al., 2007). Loss of PDCD4 expression in human lung cancer correlates with higher grade and disease stage (Chen et al., 2003). MiR-21 targets SPRY2 in cardiomyocytes and SW480 colon cancer cells, enhancing cell migration (Sayed et al., 2008). SPRY2, in turn, negatively regulates Ras/MEK/ERK signaling and is a tumor suppressor in NSCLC in both mice (Minowada and Miller, 2004; Minowada and Miller, 2009; Shaw et al., 2007) and humans (Sutterluty et al., 2007). In laryngeal carcinoma, miR-21 targets BTG2 (Liu et al., 2009), a p53 target gene, recently shown to suppress oncogenic Ras activity by reducing the GTP bound, active state (Buganim et al., 2010). Although the reductions in protein levels of individual miR-21 targets appear modest with miR-21 over-expression, collectively this diminution of tumor suppressor activity allows for more robust tumor formation through relieving multiple nodes of inhibition of the Ras/MEK/ERK pathway, thus facilitating tumor proliferation.
Oncogenic Ras activation has been shown to increase the expression of Spry2, providing feedback inhibition of Ras activation (Shaw et al., 2007). We show that Ras activation increases miR-21 expression and in turn decreasing the expression of Spry2. The genetic deletion of miR-21 removes the miR-21 mediated down-regulation of Spry2 resulting in more Spry2-mediated Ras/MEK/ERK inhibition and tumor suppressor activity. We postulate that the tumors formed in the background of miR-21 deletion have developed escape mechanisms to circumvent the Spry2 tumor suppressor activity. The necessity to circumvent the lack of miR-21 pro-tumorigenic effects could explain the latency of tumor formation in the miR-21−/− mice.
Previous work in cancer cell lines has implicated miR-21 as a suppressor of apoptosis. Knock-down of miR-21 in glioblastoma cell lines triggers the activation of caspases and increases apoptosis (Chan et al., 2005). Zhang et al. showed in a human gastric cancer cell line that forced over-expression of miR-21 enhanced tumor proliferation and invasion and knock-down of miR-21 resulted in a marked reduction in proliferation and increase in apoptosis (Zhang et al., 2008). The present work illustrates in vivo that increased expression of miR-21 significantly reduces apoptosis in mouse lung tumors. MiR-21 over-expression resulted in reduced protein levels of Apaf1 a key component of the intrinsic, mitochondrial apoptotic pathway, as well as decreased expression of Faslg a key initiator of the extrinsic, death receptor apoptotic pathway. Faslg mRNA is expressed normally in human lung tumors; however, the Faslg protein is only detected in a small fraction (Badillo-Almaraz et al., 2003). miR-21 mediated translational inhibition could partially explain this finding. Over-expression of miR-21 correlated with a decrease RhoB protein levels. RhoB promotes growth inhibition and induces apoptosis in cancer cells (Kim et al., 2009), and RhoB expression is down-regulated in human NSCLC suggesting a role as a tumor suppressor gene (Sato et al., 2007). Relieving miR-21 inhibition of pro-apoptotic genes could provide a means to augment the effect of current chemotherapy.
Methods have been developed to manipulate miRNA function pharmacologically, facilitating the development of new cancer therapeutic strategies (Hutvagner et al., 2004; Krutzfeldt et al., 2005). Increased sensitivity to DNA-damaging agents in the miR-21−/− transformed MEFs suggests that inhibition miR-21 could provide a therapeutic strategy in NSCLC. In addition, inhibiting miR-21 potentially restores the activity of multiple tumor suppressors acting at various critical nodes of tumor development. Here we report an in vivo, functional role of miR-21 in non-small cell lung cancer.
EXPERIMENTAL PROCEDURES
Mouse Strains
K-rasLA2 mice in the B6.129S background were provided by T. Jacks (Massachusetts Institute of Technology) through the National Cancer Institute. Construction of CAG-Z-miR-21-EGFP transgenic mice detailed in Supplementary Methods. All mice used in these studies were of mixed genetic backgrounds and all comparisons were performed on littermate controls. The CAG-miR-21 transgenic mice (B6C3F1 background) were bred to K-rasLA2 mice to generate miR-21 over-expressing CAG-miR-21;K-rasLA2 compound mutant mice and control K-rasLA2 mice. The miR-21−/− mice (B6.129SvEv) were bred to K-rasLA2 mice to generate miR-21 deficient miR-21−/−;K-rasLA2 compound mutant mice and control K-rasLA2 mice. Mice were sacrificed at stated time points or when showing obvious tumor burden or distress and a full necropsy was performed. All experimental procedures involving animals in this study were reviewed and approved by the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center. For survival analysis, CAG-miR-21;K-rasLA2 and K-rasLA2 mice were observed daily from birth and sacrificed at the first sign of shortness of breath, reduced locomotion, or reduced body weight (>20% of total body weight).
RNA Purification, RT-PCR, and Real-time PCR
Total RNA was isolated from normal lung or lung tumors with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. miRNA levels were determined by Northern blot analysis and real-time PCR. RT-PCR was performed using the TaqMan microRNA reverse transcriptase kit (Applied Biosystems). Real-time PCR was performed using TaqMan probes on an ABI-PE Prism 7000 sequence detection system according to manufacturer’s protocol. The relative quantities of miRNA were determined using the CT method and were normalized to RNU6B. The recombination of the K-rasLA2 allele was determined by by RT-PCR. Total RNA was isolated from micro-dissected normal lung and lung tumors. Reverse transcription was performed using random hexamer primers and SuperScript III First-Strand Synthesis (Invitrogen). PCR primers used and conditions as previously described (Johnson et al., 2001).
Northern Blot Analysis
Ten micrograms of total RNA from different tissues or tumors was resolved on a 20% polyacrylamide (7.6M urea) gel in 1× TBE. RNA was then transferred onto a Zetaprobe GT membrane (Bio-Rad) in 0.5× TBE buffer at 80 V for 1 hour. Hybridization was performed at 39oC. 32P-labeled Star-Fire oligonucleotide probes (IDT) against the mature miR-21 and U6 were used in hybridization. U6 was used as a loading control.
Histology, Immunohistochemistry and TUNEL
To determine tumor incidence and grade, whole lungs were manually inflated with 10% neutral-buffered formalin, placed in fixative for 3 days, embedded in paraffin and sectioned at the level of the tracheal bifurcation at 5 μm intervals. H&E stains were performed using standard procedures. Lung and tumor areas were determined using ImageJ software. Tumor burden was expressed as the sum of the tumor area divided by the total lung area. Proliferation was assessed by immunohistochemistry using antibodies against Ki67 (Abcam). Paraffin-embedded sections were deparaffinized, heated in a microwave in 0.01 M sodium citrate buffer for antigen retrieval, treated with 3% H2O2 for 10 minutes, rinsed in H2O and PBS. Sections were blocked in 5% goat serum in PBS followed incubation with anti-Ki67 antibody (Abcam). Signals were detected with Vectastain ABC kit (Vector Laboratories) and 3,3′-diaminobenzidine (DAB) substrate (Vector Laboratories). Sections were counter-stained with hematoxylin and mounted. Ki67 positive cells were scored per high-power field from 3 separate animals, 4 different tumors per animal and each genotype and represented as the mean percentage of the total number of cells ± SEM. Apoptosis was evaluated by staining paraffin sections with In situ cell death detection kit, TMR red terminal deoxynucleotidyl-transferase-mediated dUTP nick-end labeling (TUNEL) system (Roche) according to manufacturer’s protocol. Slides were mounted with Vectashield mounting medium with DAPI (Vector Laboratories). The number of apoptotic cells per lung tumor were recorded and represented as the mean ± SEM.
Tumor Grading
All tumors from the survival cohort were analyzed on H&E sections and categorized as hyperplasia, atypical adenomatous hyperplasia, adenoma, adenoma with typia, and adenocarcinoma according to the Mouse Models of Human Cancer Consortium recommendations.(Nikitin et al., 2004)
Western Blot Analysis
Individual tumors were micro-dissected from the lungs and snap frozen in liquid nitrogen. Individual tumor and lung lysates were prepared in RIPA with protease inhibitors (Roche complete mini) and phosphatase inhibitor cocktail (Sigma) using a manual pestle homogenizer and clarified by centrifugation at 12,000 g for 10 minutes at 4oC. Protein concentration was determined by BCA assay (Themo Scientific) and equivalent amounts were resolved by SDS-PAGE, and immunoblotted by a standard protocol. Antibodies purchased from Abcam (Spry1, Spry2, Btg2, NfiB, Ski, and Trp63), Cell Signaling (phosphor-ERK, total ERK, total Caspase 3, Apaf1, RhoB, PTEN, PIK3R1, phosphor-Akt(Ser473), total Akt, RECK, Cdc25a, and Map2k3), Chemicon (Gapdh and Faslg), and Rockland (Pdcd4). Quantification of Western blots was performed by densitometry using NIH ImageJ software. Each sample was normalized to Gapdh and represented as a fraction of K-rasLA2 control tumors.
Reporter assays
The 466 base-pair genomic fragment encompassing miR-21 was amplified by PCR and ligated into pCMV6. Full-length 3′ UTRs of putative miR-21 target genes were cloned from mouse 129SvEv genomic DNA and subcloned into the pMiR-report vector (Ambion). Mutations in the putative miR-21 site in the 3′ UTRs were generated by QuickChange mutagenesis (Stratagene) to alter the 2nd and 3rd nucleotides of the targeting sequence. Cell culture, transfection, and luciferase reporter assays were performed as previously described(van Rooij et al., 2008).
Doxorubicin-induced Apoptosis
Wild-type and miR-21−/− MEFs immortalized with T-Ag and transformed with H-rasV12 were seeded in a 6-well plate at 1 × 105 cells per well. Cells were treated with vehicle control and increasing doxorubicin concentrations from 100 nM to 10 μM for 12 hours. Cells were lysed in RIPA and protein concentrations determined by BCA (Thermo Scientific). Western blotting of 50 μg of lysates was performed with antibodies directed against pro- and cleaved Caspase 3 (Cell Signaling) and Gapdh (Chemicon). Quantification of Western blots was performed by densitometry using NIH ImageJ software. Cleaved caspase 3 levels were expressed as a ratio of pro-caspase 3 and normalized to either WT untreated or empty vector untreated samples. For TUNEL assays, MEFs were seeded in a 6-well plate at 1 × 105 cells per well on cover slips. Cells were treated with vehicle control or 1 μM doxorubicin for 12 hours. TUNEL was performed using In situ cell death detection kit, TMR red terminal deoxynucleotidyl-transferase-mediated dUTP nick-end labeling (TUNEL) system (Roche) according to manufacturer’s protocol.
Statistical Analysis
Results are expressed as the mean ± SEM. We utilized a two-tailed, unpaired Student’s t-test for all pair-wise comparisons (GraphPad Prism version 5). P values less than 0.05 were considered significant.
SIGNIFICANCE.
MicroRNAs are small non-coding RNAs that are commonly dysregulated in human malignancies and play a substantial role in the pathogenesis and survival of many cancers. MiR-21 is over-expressed in the majority of human malignancies including non-small cell lung cancer (NSCLC). However, it is not clear whether miR-21 determines important aspects of lung cancer pathogenesis or is instead simply a marker for advanced disease. We show in vivo that miR-21 expression increases with oncogenic K-ras activation and modulates NSCLC tumorigenesis by targeting Spry1, Spry2, Btg2, and Pdcd4 which act as negative regulators of the Ras/MEK/ERK pathway, and Apaf1, Faslg, Pdcd4 and RhoB which promote apoptosis. MiR-21 deletion also sensitizes cells to DNA-damaging chemotherapy, suggesting that miR-21 inhibition could be of therapeutic value.
Supplementary Material
Acknowledgments
We are grateful to John McAnally for transgenic injection, Jose Cabrera for figure preparation, and Lillian Sutherland and John Shelton for experimental assistance. Work in Eric Olson’s laboratory was supported by grants from the National Institutes of Health, the Leducq Foundation, the Robert A. Welch Foundation, and the American Heart Association: Jon Holden DeHaan Foundation. E.V.R. was supported by grants from the American Heart Association. Mark E. Hatley is a Pediatric Scientist Development Program Fellow sponsored by the Eunice Shriver Kennedy National Institute of Child Health and Human Development (NICHD Grant Award K12-HD000850). E.N.O. and E.V.R. hold equity in miRagen Therapeutics, which is developing miRNA-based therapies for muscle disease.
Footnotes
Author Contributions
M.E.H. and E.N.O. developed hypotheses, designed experiments and wrote the manuscript. M.E.H. and R.B.D. designed animal protocols and experiments. M.E.H. and M.R.G. executed experiments. D.P. and E.V.R. developed and provided the miR-21 knock-out mouse. J.A.R. provided pathological review of histology.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene. 2004;23:8135–8145. doi: 10.1038/sj.onc.1207983. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Badillo-Almaraz I, Badillo-Salas C, Villalobos R, Avalos-Diaz E, Herrera-Esparza R. Defective expression of FasL and Bax in human lung cancer. Clin Exp Med. 2003;3:106–112. doi: 10.1007/s10238-003-0012-1. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bitomsky N, Wethkamp N, Marikkannu R, Klempnauer KH. siRNA-mediated knockdown of Pdcd4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene. 2008;27:4820–4829. doi: 10.1038/onc.2008.115. [DOI] [PubMed] [Google Scholar]
- Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Solomon H, Rais Y, Kistner D, Nachmany I, Brait M, Madar S, Goldstein I, Kalo E, Adam N, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70:2274–2284. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chen Y, Knosel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, Ozaki I, Petersen I. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–646. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2007 doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Mishina Y, Walker MP, DiAugustine RP. Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein. Mol Cell Biol. 2005;25:5270–5281. doi: 10.1128/MCB.25.12.5270-5281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1) Am J Physiol Cell Physiol. 2004;287:C1541–1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, Horiuchi D, Lebbink RJ, Mo YY, Goga A, McManus MT. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–18524. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SK, Jin H, Kwon JT, Chang SH, Kim TH, Cho CS, Lee KH, Young MR, Colburn NH, Beck GR, Jr, et al. Aerosol-delivered programmed cell death 4 enhanced apoptosis, controlled cell cycle and suppressed AP-1 activity in the lungs of AP-1 luciferase reporter mice. Gene Ther. 2007;14:1353–1361. doi: 10.1038/sj.gt.3302983. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kim DM, Chung KS, Choi SJ, Jung YJ, Park SK, Han GH, Ha JS, Song KB, Choi NS, Kim HM, et al. RhoB induces apoptosis via direct interaction with TNFAIP1 in HeLa cells. Int J Cancer. 2009;125:2520–2527. doi: 10.1002/ijc.24617. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–837. doi: 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- Lo TL, Fong CW, Yusoff P, McKie AB, Chua MS, Leung HY, Guy GR. Sprouty and cancer: the first terms report. Cancer Lett. 2006;242:141–150. doi: 10.1016/j.canlet.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic Value of Mature MicroRNA-21 and MicroRNA-205 Overexpression in Non-Small Cell Lung Cancer by Quantitative Real-Time RT-PCR. Clin Chem. 2008 doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minowada G, Miller YE. Sprouty 2 gene in mouse lung tumorigenesis. Chest. 2004;125:111S. doi: 10.1378/chest.125.5_suppl.111s. [DOI] [PubMed] [Google Scholar]
- Minowada G, Miller YE. Overexpression of Sprouty 2 in mouse lung epithelium inhibits urethane-induced tumorigenesis. Am J Respir Cell Mol Biol. 2009;40:31–37. doi: 10.1165/rcmb.2008-0147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Pawlish K, Gadgeel S, Demers R, Kalemkerian GP. Lung cancer in young patients: analysis of a Surveillance, Epidemiology, and End Results database. J Clin Oncol. 1998;16:651–657. doi: 10.1200/JCO.1998.16.2.651. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukui T, Taniguchi T, Yokoyama T, Kondo M, Nagasaka T, Goto Y, Gao W, Ueda Y, Yokoi K, et al. RhoB is frequently downregulated in non-small-cell lung cancer and resides in the 2p24 homozygous deletion region of a lung cancer cell line. Int J Cancer. 2007;120:543–551. doi: 10.1002/ijc.22328. [DOI] [PubMed] [Google Scholar]
- Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, Parisi T, Rajagopal J, Blank LJ, Bronson RT, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Soon YY, Stockler MR, Askie LM, Boyer MJ. Duration of chemotherapy for advanced non-small-cell lung cancer: a systematic review and meta-analysis of randomized trials. J Clin Oncol. 2009;27:3277–3283. doi: 10.1200/JCO.2008.19.4522. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, Corless CL, Fire AZ, van de Rijn M. MicroRNA expression signature of human sarcomas. Oncogene. 2007 doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- Sutterluty H, Mayer CE, Setinek U, Attems J, Ovtcharov S, Mikula M, Mikulits W, Micksche M, Berger W. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol Cancer Res. 2007;5:509–520. doi: 10.1158/1541-7786.MCR-06-0273. [DOI] [PubMed] [Google Scholar]
- Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene. 2009;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O’Brien C, Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R. Classifying microRNAs in cancer: the good, the bad and the ugly. Biochim Biophys Acta. 2007;1775:274–282. doi: 10.1016/j.bbcan.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.