Abstract
The presence and biological importance of DNA secondary structures in eukaryotic promoters are becoming increasingly recognized among chemists and biologists as bioinformatics, in vitro, and in vivo evidence for these structures in the c-Myc, c-Kit, KRAS, PDGF-A, hTERT, Rb, RET and Hif-1α promoters accumulates. Nevertheless, the evidence remains largely circumstantial. This minireview differs from previous ones in that here we examine the diversity of G-quadruplex and i-motif structures in promoter elements and attempt to categorize the different types of arrangements in which they are found. For the c-Myc G-quadruplex and Bcl-2 i-motif, we summarize recent biological and structural studies.
Introduction
Although recent reviews on G-quadruplexes in telomeres have been published [1–3], in this minireview we focus on the increasingly observed complexity of G-quadruplex (G-rich strand) and i-motif (C-rich strand) folding patterns and structures in the promoter regions of oncogenes. Accompanying minireviews in this issue discuss other aspects of the biology of G-quadruplexes [4,5]. In previous bioinformatics searches [6,7], relatively simple algorithms have been used to examine promoter regions for G-quadruplexes, but it is likely that more-defined subcategory algorithm searches might yield more useful information on the relative distribution of different classes of G-quadruplexes present in promoter regions. Our minireview begins by examining the diversity of G-quadruplex structures associated with the six hallmarks of cancer and then makes a first attempt to categorize different types of G-quadruplexes and i-motifs that have been identified in promoter regions. We then select examples from two different types of G-quadruplex-containing promoters and discuss these in more detail to illustrate the different principles that we believe are important in considering how these G-quadruplexes and i-motifs function from a biological standpoint. Finally, we point to critical questions that need to be addressed for this exciting new area to be launched from a solid scientific basis.
Examples of G-quadruplex structures in oncogene promoters representing the six hallmarks of cancer
In a review to characterize the gene ontology of promoters that contained putative G-quadruplex-forming motifs, Eddy and Maizels discovered a significant enrichment of these motifs in oncogenes [8]. Consistent with this finding, G-quadruplex motifs within several oncogene promoters have been shown to transition to stable G-quadruplex structures. More importantly, altered expressions of these oncogenes are recognized as hallmarks of cancer. At the turn of the century, Hanahan and Weinberg proposed six vital cellular and microenvironmental processes that are aberrantly regulated during oncogenic transformation and malignancy [9]. These include self-sufficiency for growth signals, insensitivity to anti-growth signals, evasion of apoptosis, sustained angiogenesis, limitless replicative potential, and tissue invasion and metastasis. When each of these categories is examined, a critical protein or proteins can be found with a G-quadruplex in the core or proximal promoter (Fig. 1). This is especially significant when one realizes how young the G-quadruplex field is, and that new genes regulated by these structures are being continually identified. This observation led to our recent discussion of the G-quadruplexes of cancer [10], highlighting c-Myc, c-Kit and KRAS (self-sufficiency); pRb (insensitivity); Bcl-2 (evasion of apoptosis); VEGF-A (angiogenesis); hTERT (limitless replication); and PDGF-A (metastasis).
Fig. 1.
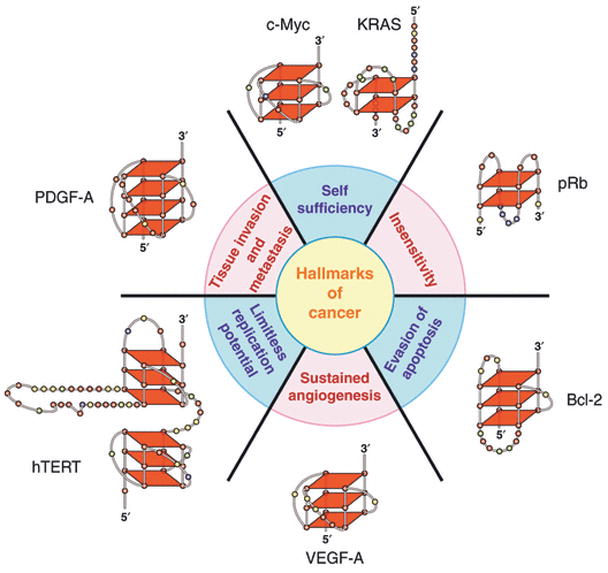
The six hallmarks of cancer [9] shown with the associated G-quadruplexes found in the promoter regions of these genes. As described in the text, the various G-quadruplexes differ by folding pattern, number of tetrads, loop size and constituent bases. In this a subsequent models, bases are colored as follows: guanine, red; cytosine, yellow; thymine, blue; adenine, green.
Promoters in each of these oncogenes are able to form G-quadruplexes with vast diversity in their folding patterns and loop lengths, making them putatively amenable to specific drug targeting [10]. These G-quadruplexes include varying numbers of tetrads, most commonly three, but sometimes two or four. They also vary in their loop directionality, either parallel, antiparallel or mixed parallel/antiparallel. Most often the tetrads are continuously connected, but a snap-back configuration has been confirmed in at least one naturally occurring G-quadruplex formation, c-Kit. The greatest variability among these secondary structures is found in loop lengths and constituent bases. Although the G-tetrad stacks are almost exclusively formed from guanines, there are no such limitations on bases in the loops. Shorter loops, especially in double-chain reversals, help stabilize the G-quadruplex. However, loop lengths have been seen to vary from only 1 base (the minimum required) to as many as 26 (forming their own secondary loop–stem structure in the hTERT promoter) [11]. Most commonly the loops are 1–9 bases long. All of these variations, detailed in Brooks & Hurley [10] for the G-quadruplexes of cancer, lead to the formation of 3D structures with distinctive binding pockets that offer sites for specific targeting with drugs. This diversity expressed in different folding patterns (e.g. parallel vs. mixed parallel/antiparallel), loop sizes and base composition (e.g. one to seven and bases that have specific interactions), number of tetrads (i.e. two, three or four) and inter-quadruplex binding sites in c-Myb and hTERT represents opportunities for specific binding interactions. Some of these drug–G-quadruplex interactions have been addressed in recent reviews [12–14]. In addition to the unique G-quadruplex structures, the formation of i-motifs also provides even more potential for potent and specific drug targeting (see later).
Classes of G-quadruplex/i-motif complexes found in promoter elements
In a first attempt to categorize promoter G-quadruplex folding patterns and structures, we have identified four classes of quadruplexes (Fig. 2). These classes differ in the number of G-quadruplexes that can be formed (1 = Classes I and IV, 2 = Classes II and III). Classes I and IV differ in that Class IV can form multiple G-quadruplexes that overlap in a region containing multiple G-tracts. Classes II and III differ in their relative positions in the promoter, either distant, so that direct interaction is less likely to occur (Class II), or adjacent, so that they can have inter-quadruplex stacking interactions (Class III). We recognize that there are other possible means of classifying G-quadruplexes in promoter regions, such as by folding patterns or whether the biological function is suppression or activation of gene expression; however, for the purpose of thinking beyond a single G-quadruplex in a promoter element, we propose that this is an important starting point. We also suspect that as new promoter elements containing G-quadruplexes are characterized, we will need to expand and revise this initial classification, which is admittedly based on quite limited information.
Fig. 2.
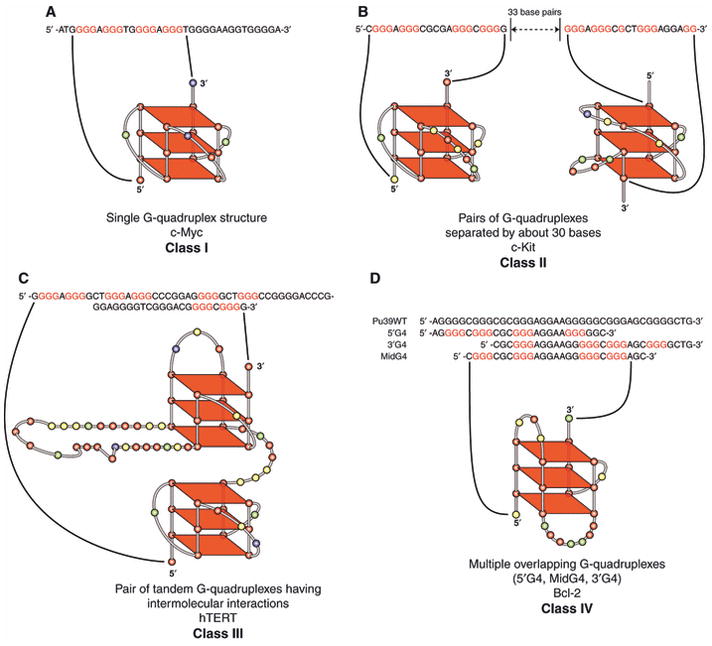
Proposed classes of unimolecular G-quadruplexes found in eukaryotic promoter elements. Class I (A) is represented by the single G-quadruplex found in the c-Myc promoter element. Class II (B) contains a pair of different G-quadruplexes separated by about three turns of DNA. Class III (C) is represented by the tandem G-quadruplexes from the hTERT promoter. Class IV (D) represents multiple overlapping G-quadruplexes. The example shown is from the Bcl-2 promoter and the G-quadruplex shown (MidG4) is the most stable of the three structures. A more complete description is found in the text.
Class I (Fig. 2A) is seemingly the simplest case, in which a single G-quadruplex predominates, but there may be loop isomers, so although the same guanine runs are used, the loop sizes may vary. The c-Myc G-quadruplex is the prototypical member of this class, in which four contiguous guanine runs are used, producing four isomers having loop sizes of 5′-(1:2:1)-3′, 5′-(2:1:1)-3′, 5′-(1:1:2)-3′ and 5′-(2:1:2)-3′ [15]. Of these, the predominant loop isomer is the 5′-(1:2:1)-3′, in which the four 5′ guanine runs from the six guanine runs are utilized [16]. Unimolecular G-quadruplexes possessing an all-parallel folding pattern are found in the RET, Hif-1α, PDGF-A and VEGF promoters [17]. They differ in the central loop size, which can vary from two (c-Myc) to five (PDGF-A). The biological consequence of formation and stabilization of G-quadruplexes in these promoter elements is gene silencing [17]. For this class, the c-Myc system is the best characterized, and this is discussed later.
The second class is one in which there are two distinctly different G-quadruplexes separated by about three turns of DNA (Fig. 2B). There is only one known example here, the c-Kit [18–20]. For the c-Kit G-quadruplexes, NMR studies have shown that the downstream G-quadruplex has an unusual folding pattern in which a 2 + 1 discontinuity exists for one of the edges, but overall a parallel-stranded G-quadruplex exists [19]. The upstream G-quadruplex is an all-parallel structure having a 5′-(1:5:1)-3′ loop arrangement [21]. As is also the case for Class I, ligand stabilization of the G-quadruplexes results in inhibition of c-Kit gene expression [18,22]. The third class also includes a pair of G-quadruplexes, but they are sufficiently close that they have been shown to form tandem G-quadruplexes, and together these tandem structures are more stable than the individual G-quadruplexes. Thus there are intermolecular interactions between the two adjacent G-quadruplexes. The two examples are c-Myb [23] and hTERT [11] (Fig. 2C). The first example occurs in the c-Myb promoter, where there are three potential tandem G-quadruplexes, but only two co-exist at one time. For c-Myb, the heptad–tetrad is not stable under physiological conditions, but the interactions between the two heptads provide the additional stabilizing focus so that the tandem G-quadruplexes form a stable structure. The two linker sizes are either 4 or 19 bases. The second example of a tandem repeat is found in the hTERT promoter, which is proposed to have an unusual G-quadruplex with a large hairpin loop containing 25 or 26 bases (Fig. 2C). Unlike c-Myb, the two hTERT G-quadruplexes are dissimilar, with the upstream G-quadruplex forming a standard parallel structure having loop sizes of 5′-(1:3:1)-3′, whereas the downstream G-quadruplex most likely forms a mixed parallel/antiparallel structure with loop sizes of 5′-(3:26:1)-3′, similar to the folding pattern of the major Bcl-2 G-quadruplex (see later). The intermolecular G-quadruplex linker size of the hTERT is seven bases. In both cases, the duplex GC elements sequestered by the tandem G-quadruplexes contain multiple Sp1 binding sites [11,23]. For hTERT, stabilization of the tandem G-quadruplex complex leads to inhibition of gene expression, thus providing a direct mechanism to inhibit telomerase expression rather than by interaction with telomere G-quadruplexes [11].
The fourth class, in which multiple overlapping G-quadruplexes exist, is found in Bcl-2 [24] and platelet-derived growth factor receptor β (PDGFR-β) [25] (Fig. 2D). For Bcl-2, three equilibrating G-quadruplexes exist (5′G4, MidG4 and 3′G4), overlapping in a 39-base region containing six runs of three or more guanines. Of the three equilibrating G-quadruplexes, the MidG4 is the most stable and has been shown by NMR to have a mixed parallel/antiparallel folding pattern [26]. Recently, we have uncovered another complex G-quadruplex-forming region in the PDGFR-β promoter that covers 38 bases and contains four overlapping G-quadruplex-forming sequences (5′-end, mid-5′, mid-3′ and 3′-end) that appear to produce one or more unusual folding patterns [25]. These folded structures probably contain a 2 + 1 discontinuity, because dimethyl sulfate (DMS) footprinting shows isolated guanines that are protected as well as runs of two or four guanines that are also protected from DMS cleavage [25].
Altough there is less data on the i-motifs formed in promoter complexes, they also appear to belong to multiple classes (Fig. 3), which we have classified as small-loop (Class I) and large-loop (Class II) i-motif structures. Because slightly acidic pH values are required to stabilize the i-motifs formed from single-stranded DNA templates, the driving force for i-motif formation arises from maximizing the number of cytosine+–cytosine hemiprotonated base pairs [27]. Under negative supercoiling, the i-motif forms under physiological conditions, and in this case it is more likely that stabilizing capping interactions may drive the formation of a favored i-motif [16]. For example, in the case of the Bcl-2 i-motif, specific interactions between bases in the loops are believed to be responsible for the stability of the i-motif [28]. Fluorescence and mutational studies demonstrate the importance of these interactions in stabilizing the structure. Thus it is necessary to be cautious in drawing conclusions from experiments in which acidic conditions are used to drive i-motif formation. With this caveat in mind, the two classes of i-motifs shown in Fig. 3 can be identified. In Class I, the loop sizes are 5′-(2:3–4:2)-3′ with either four, five or six cytosine+–cytosine base pairs and members include VEGF, RET and Rb. In Class II, the loop sizes are 5′-(6–8:2–5:6–7)-3′, with Bcl-2 having the larger cumulative loop size (20). Only in the case of c-Myc have the conditions for formation of the i-motif relied upon negative superhelical stress, rather than acidic pHs [16].
Fig. 3.
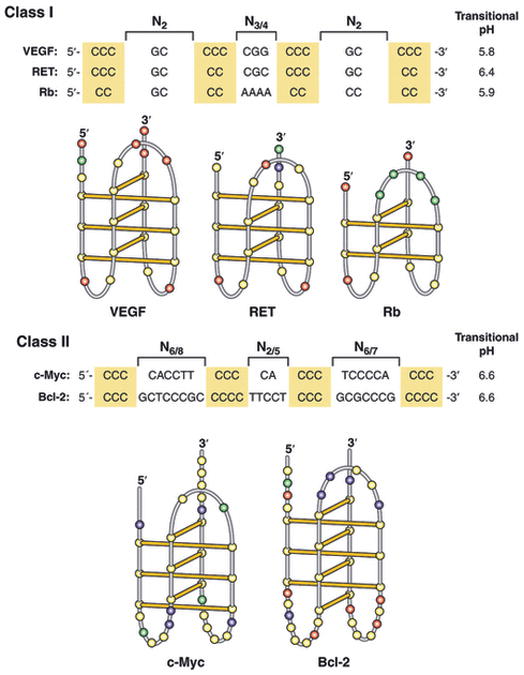
Sequences and folding patterns of i-motifs in the two proposed classes of i-motifs found in eukaryotic promoter elements. Class I, having small loop sizes, is found in the VEGF, RET and Rb promoter elements, and Class II, having larger loop sizes, is found in the c-Myc and Bcl-2 promoter elements. See text for additional details.
The role of negative supercoiling, NM23-H2 and nucleolin in the control of c-Myc gene expression via the nuclease hypersensitive element III1
There are two legitimate objections to the biological role of secondary DNA structures such as those described in this review: (a) how can these structures evolve from duplex DNA; and (b) once formed, how are they dissipated (at least in the case of the G-quadruplex, they can be very stable structures)? Indeed, the c-Myc G-quadruplex has a melting point in excess of 85°C. To address these issues directly, we set out to examine conditions such as supercoiling that might provide the torque necessary for conversion of duplex DNA to G-quadruplexes and to identify proteins that might serve to facilitate the formation and then resolve the G-quadruplex and i-motif structures in the nuclease hypersensitive element (NHE) III1 of the c-Myc promoter. We reasoned that if we could show that the G-quadruplexes and i-motifs could be formed under physiological conditions from duplex DNA, and if we could identify the proteins involved in the control of this process, then this would go a long way toward convincing skeptics that these ‘odd’ DNA structures are important components of eukaryotic transcriptional regulation. The experiments described below, taken from recent publications, provide this evidence. The importance of supercoiling and these proteins in modulating the effects of drugs on c-Myc transcription is described in more detail in a recent review [10]. A more complete description of the transcriptional factors and their role in the control of c-Myc via the NHE III1 are also described in a separate review [29].
The role of negative supercoiling in conversion of duplex DNA to G-quadruplex/i-motif structures in the NHE III1
Supercoiling has been known for many years to be an important factor in gene transcription in both eukaryotic and prokaryotic organisms [30,31]. Furthermore, it has been more recently shown that transcription itself can be a source of this supercoiling in eukaryotic cells [32]. We employed a system in which the negative supercoiling induced upstream of the transcription site is mimicked in a supercoiled plasmid [16]. Using this system, a wild-type and mutant sequence of the NHE III1 in the c-Myc promoter were inserted into a Del4 plasmid [16]. A comparison of chemical (DMS, KMnO4 and Br2) and enzymatic (S1 nuclease, DNase 1) footprinting on the wild-type and mutant inserts provided the evidence that supports the conclusions shown in Fig. 4. Figure 4A shows the equilibrium between duplex (i), locally unwound duplex (ii), single-stranded DNA (iii) and the G-quadruplex/i-motif structure (iv) formed as a consequence of negative supercoiling. Because the one-base mutant is unable to form a stable G-quadruplex, but is nevertheless a polypurine/polypyrimidine tract, it becomes locally unwound (i–iii) but is unable to form the G-quadruplex/i-motif complex that is evident with the wild-type sequence (i–iv). Figure 4B shows the asymmetric positioning of the G-quadruplex and i-motif in the NHE III1 deduced from the DMS and Br2 footprinting experiments.
Fig. 4.
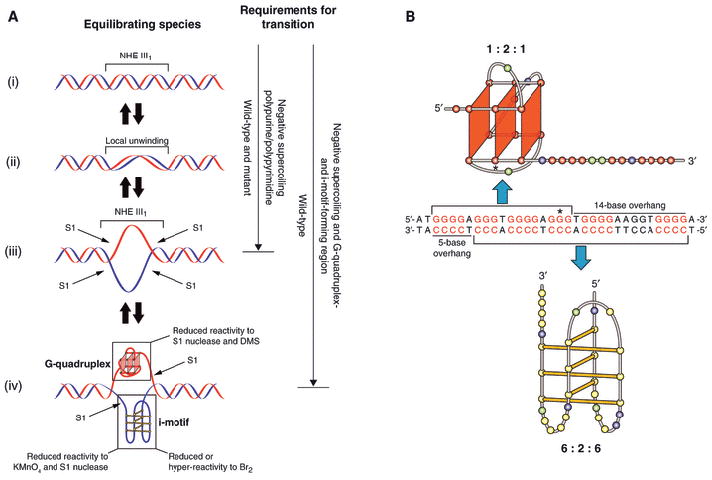
(A) Proposed equilibrating forms of the NHE III1 produced under negative supercoiling. The resistance/sensitivity to S1 nuclease, DMS, KMnO4 and Br2 of the various forms is shown in the left-hand panel. Requirements for transition to the single-stranded form or G-quadruplex/i-motif species are shown in the right-hand panel. (B) Asymmetric positioning of the DMS-protected G-quadruplex (top bracket) and Br2-protected i-motif (bottom bracket) together with 14- and 5-base overhangs. An asterisk marks the position of the G-to-A mutant in the G-quadruplex loop isomer [16].
The importance of NM23-H2 in transcriptional activation of c-Myc
The ubiquitous human non-metastatic 23 isoform 2 protein (NM23-H2) occurs as a hexamer and has been known for more than 15 years to be an important factor in c-Myc transcriptional activation [33]. However, until recently its precise role has remained controversial. This controversy centered around the identification of the favored DNA species for binding to NM23-H2 (duplex, single-stranded purine or pyrimidine strands) and whether enzymatic-induced cleavage of the NHE III1 occurred. It now appears that NM23-H2 binds to both the purine and pyrimidine strands of NHE III1 but not to duplex [34], and the purported DNA strand cleavage [35,36] was due to a contaminating protein that is either an accessory protein or a minor recombinant protein [34]. Studies show that an R88A mutation (arginine to alanine) in the nucleotide-binding site eliminates binding of the NM23-H2 to single-stranded DNA. Because NM23-H2 is a hexameric protein with six nucleotide-binding sites that favor purine residues [37], we propose that NM23-H2 sequentially traps out the single-stranded purine and pyrimidine strands as it unfolds the G-quadruplex and i-motif (Fig. 5A, B). Furthermore, the NM23-H2–DNA complex is highly reversible [34], so we propose that the transcriptional factors CNBP and hnRNP K readily displace the NM23-H2 to activate c-Myc transcription (Fig. 5A–C). Conditions in which the G-quadruplex is stabilized, such as with the compound TMPyP4 or the monovalent cation KCl, should inhibit NM23-H2 activation, and indeed this has been shown to be the case (Fig. 5A–E) [34].
Fig. 5.
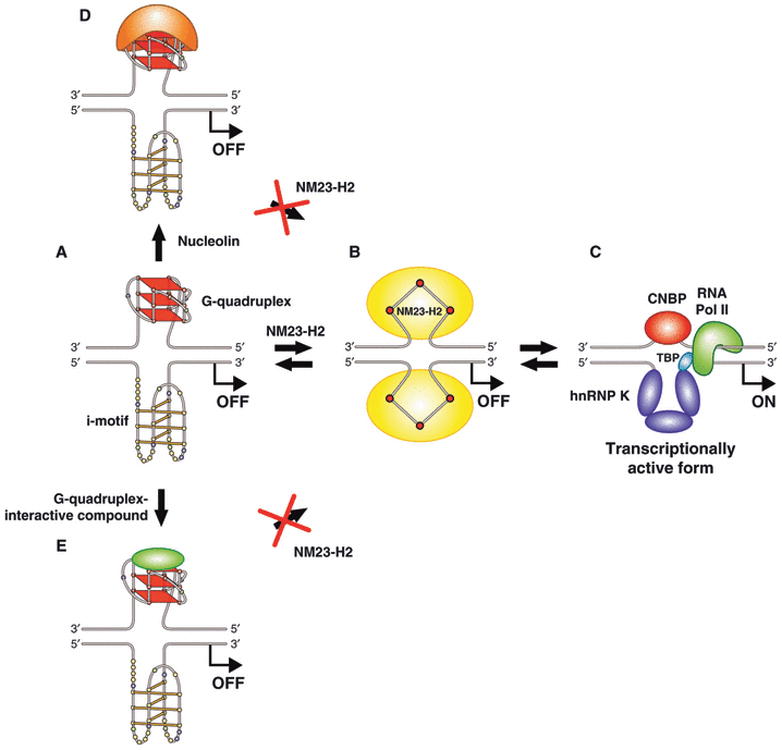
Cartoon showing the involvement of NM23-H2, nucleolin and a G-quadruplex-interactive compound in modulating the activation and silencing of the NHE III1 in the c-Myc promoter. (A) shows the G-quadruplex/i-motif form of the NHE III1, which is the silencer element. (A) to (C) via (B) illustrates the remodeling of the G-quadruplex/i-motif complex by NM23-H2, in which a stepwise unfolding of the secondary DNA structure is proposed to take place. Binding of nucleolin (A–D) or a G-quadruplex-interactive compound (A–E) to the silencer element prevents conversion by NM23-H2 to the transcriptionally active form of the NHE III1 (C) [10].
Identification of nucleolin as a c-Myc G-quadruplex binding protein
To identify potential c-Myc G-quadruplex-binding proteins, an affinity chromatography method was used followed by LC-MS/MS sequencing analysis [38]. Of the proteins identified, nucleolin was the most abundant, and many of the other proteins identified were known to bind to nucleolin. Subsequent studies with nucleolin showed that it facilitated the formation of the c-Myc G-quadruplex from the single-stranded purine-rich strand and then stabilized the resulting structure [25]. Furthermore, nucleolin bound more avidly to the c-Myc G-quadruplex than its previously suggested RNA substrate and had a specificity for this G-quadruplex over other promoter G-quadruplexes [38]. Chromatin immunoprecipitation analysis showed that nucleolin bound to the NHE III1 [38]. Furthermore, experiments with a nucleolin expression plasmid and using a luciferase reporter gene showed a dose-dependent decrease in c-Myc expression and inhibition of a Sp1-induced transcription [38]. Finally, inhibition of c-Myc transcription occurred preferably over VEGF and PDGF-A (unpublished results). The role of nucleolin in the inhibition of c-Myc gene expression is shown in Fig. 5(A–D).
The next logical series of experiments will examine the differential binding of Sp1, Pol II, CNBP, hnRNP K, nucleolin and NM23-H2 to the NHE III1 by chromatin immunoprecipitation analysis following activation or inhibition of c-Myc gene expression.
The Bcl-2 promoter element forms an i-motif with an unexpected 8:5:7 loop isomer opposite the multiple G-quadruplex-forming purine-rich strand
Similar to the c-Myc promoter region, within close proximity to the transcriptional start site (−46 to −28 base pairs upstream) in the Bcl-2 promoter there is a GC-rich element that has the potential for DNA secondary structure formation. However, in contrast to the c-Myc G-quadruplex-forming sequence, the Bcl-2 promoter G-rich element has been shown to adopt three different G-quadruplex structures [24]. Interestingly, the most stable G-quadruplex utilizes the middle four runs of guanines, because it requires the least amount of KCl for stabilization in comparison with the 5′- and 3′-end runs [24]. This raises the question as to the purpose of the additional guanine runs. Although an equilibrium between the three G-quadruplex structures may exist, recent studies involving the complementary strand suggest that the 5′- and 3′-end runs are necessary for providing the cytosines for i-motif formation. This has a similarity to the c-Myc G-quadruplex, in which the addition of two 3′-runs of guanines not used in G-quadruplex formation (Fig. 4B) are required because the i-motif on the opposite strand uses all six cytosine runs.
In contrast to the G-quadruplex, the i-motif may favor larger loop sizes for stability and therefore requires a longer sequence of nucleotides. Indeed, the complementary Bcl-2 C-rich promoter sequence has been shown to form a stable i-motif structure that requires the entire pyrimidine-rich element [28]. Studies similar to those using the G-quadruplex-forming sequence were performed using the Bcl-2 C-rich sequence; however, none of the truncated sequences (5′, middle or 3′ cytosine runs) displayed an i-motif with as high stability as the full-length sequence. Further analysis with the full-length sequence revealed that the most stable Bcl-2 i-motif consists of an 8:5:7 loop conformation requiring all six cytosine runs (shown in Fig. 3) [28]. Presumably these large loops enable capping structures to form and further stabilize the Bcl-2 i-motif, contributing to the significant stability that is reflected by the high transitional pH of ~ 6.6 [28]. Formation of the G-quadruplex and i-motif structures within the Bcl-2 promoter region may play a role in the complex transcriptional regulation of this oncogene. The majority of Bcl-2 transcription is driven by the P1 promoter, and to a lesser extent by the P2 promoter [39]. There are several negative and positive transcriptional response elements within the P1 promoter region with the double-stranded binding protein WT-1, a known repressor of Bcl-2 transcription and the most extensively studied. WT-1 has been shown to interact with the same GC-rich sequence that has the potential to form DNA secondary structures [39,40]. We propose that the formation of a G-quadruplex and i-motif upstream of the Bcl-2 P1 promoter prevents the binding of WT-1 and abrogates the transcriptional repression, thereby allowing activation of Bcl-2 transcription. Although a number of whole-genome studies have demonstrated a potential activating role for G-quadruplexes and i-motifs [7,41–43], if the hypothesis regarding the role of these nontraditional DNA secondary structures in the Bcl-2 promoter turns out to be true, this would be the first demonstration of an activating G-quadruplex in a specific promoter. Because a relatively small number of promoters containing G-quadruplexes have been studied, it would not be at all surprising to find activating G-quadruplexes and i-motifs, and they may even be relatively common.
Future important issues to be addressed
Although there is considerable circumstantial evidence from cellular and in vivo studies that G-quadruplexes and i-motifs are functionally relevant in promoter regions, some of which is summarized in this minireview, direct evidence for their existence in cells is still not available. This objective and other future important issues that need to be addressed are listed below.
Direct evidence for the existence of G-quadruplexes and i-motifs in the promoter regions of cells is the most important issue to be addressed.
Direct evidence for the interaction of G-quadruplex-interactive compounds with G-quadruplexes in promoter regions is needed.
The structure of composite G-quadruplex/i-motif assemblies is the next important structural objective.
Up to now the G-quadruplexes in promoter regions have been targeted for drug discovery; the next frontier is bringing i-motifs into focus as drug targets.
Acknowledgments
This research has been supported by grants from the National Institutes of Health (CA95060, GM085585, T32CA09213) and the Leukemia & Lymphoma Society (6225-08). We are grateful to Dr David Bishop for preparing, proofreading and editing the final version of the manuscript and figures.
Abbreviations
- DMS
dimethyl sulfate
- NHE
nuclease hypersensitive element
- NM23-H2
non-metastatic 23 isoform 2
- PDGFR-β
platelet-derived growth factor receptor β
References
- 1.Chaires JB. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 3.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 4.Huppert JL. Structure, location and interactions of G-quadruplexes. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Brosh RM., Jr G-quadruplex nucleic acids and human disease. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma A, et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 8.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 11.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arola A, Vilar R. Stabilisation of G-quadruplex DNA by small molecules. Curr Top Med Chem. 2008;8:1405–1415. doi: 10.2174/156802608786141106. [DOI] [PubMed] [Google Scholar]
- 13.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Zhao C, Chen Y, Fu M, Ren J, Qu X. DNA loop sequence as the determinant for chiral supramolecular compound G-quadruplex selectivity. J Med Chem. 2010;53:492–498. doi: 10.1021/jm9014795. [DOI] [PubMed] [Google Scholar]
- 15.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 16.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunaratnam M, et al. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J Med Chem. 2009;52:3774–3783. doi: 10.1021/jm900424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu ST, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J Am Chem Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J Am Chem Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor β (PDGFR-β) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-β expression. Biochemistry. 2010;49:4208–4219. doi: 10.1021/bi100330w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin KS, Shin SR, Ahn B, Rho Y, Kim SJ, Ree M. pH-dependent structures of an i-motif DNA in solution. J Phys Chem B. 2009;113:1852–1856. doi: 10.1021/jp808186z. [DOI] [PubMed] [Google Scholar]
- 28.Kendrick S, Akiyama Y, Hech SM, Hurley LH. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J Am Chem Soc. 2009;131:17667–17676. doi: 10.1021/ja9076292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González V, Hurley LH. The c-MYC NHE III1: function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 30.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travers A, Muskhelishvili G. DNA supercoiling – a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 32.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 33.Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 34.Dexheimer TS, et al. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1. Mol Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postel EH. Cleavage of DNA by human NM23-H2/nucleoside diphosphate kinase involves formation of a covalent protein–DNA complex. J Biol Chem. 1999;274:22821–22829. doi: 10.1074/jbc.274.32.22821. [DOI] [PubMed] [Google Scholar]
- 36.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc Natl Acad Sci USA. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agou F, Raveh S, Mesnildrey S, Véron M. Single strand DNA specificity analysis of human nucleoside diphosphate kinase B. J Biol Chem. 1999;274:19630–19638. doi: 10.1074/jbc.274.28.19630. [DOI] [PubMed] [Google Scholar]
- 38.González V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, Korsmeyer SJ. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2–Ig fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckman C, Mochon E, Arcinas M, Boxer LM. The WT1 protein is a negative regulator of the normal bcl-2 allele in t(14;18) lymphomas. J Biol Chem. 1997;272:19609–19614. doi: 10.1074/jbc.272.31.19609. [DOI] [PubMed] [Google Scholar]
- 41.Fernando H, Sewitz S, Darot J, Tavare S, Huppert JL, Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]


