Abstract
Krüppel-like factors (KLFs), of which there are currently 17 known protein members, belong to the Specificity-protein (Sp) family of transcription factors and are characterized by the presence of Cys2/His2 zinc-finger motifs in their carboxy-terminal domains that confer preferential binding to GC/GT-rich sequences in gene promoter and enhancer regions. While previously regarded to simply function as silencers of Sp1-transactivity, many KLFs are now shown to be relevant to human cancers by their newly identified abilities to mediate cross-talk with signaling pathways involved in the control of cell proliferation, apoptosis, migration, and differentiation. Several KLFs act as tumor suppressors and/or oncogenes under distinct cellular contexts, underscoring their prognostic potential for cancer survival and outcome. Recent studies suggest that a number of KLFs can influence steroid hormone signaling through transcriptional networks involving steroid hormone receptors and members of the nuclear receptor family of transcription factors. Since inappropriate sensitivity or resistance to steroid hormone actions underlie endocrine-related malignancies, we consider the intriguing possibility that dysregulation of expression and/or activity of KLF members is linked to the pathogenesis of endometrial and breast cancers. In this review, we focus on recently described mechanisms of actions of several KLFs (KLF4, KLF5, KLF6, and KLF9) in cancers of the mammary gland and uterus. We suggest that understanding the mode of actions of KLFs and their functional networks may lead to the development of novel therapeutics to improve current prospects for cancer prevention and cure.
Introduction
Endocrine-responsive cancers of female reproductive tissues constitute a complex set of pathologies that arise, in part, from aberrant levels and/or activity of the ovarian hormones estradiol (E) and progesterone (P) (Pasqualini 2007, Eliassen & Hankinson, 2008). While there are many factors that affect sex steroid hormonal profiles, foremost of which is the control of ovarian steroidogenic activity, numerous studies have shown that defects in steroid hormone signaling prominently underlie target cell resistance to the biological actions of E and P. Evidence for the latter is provided by the noted dysregulation of uterine and mammary functions consequent to the loss or aberrant expression of proteins involved in their respective steroid signaling cascades (Lydon et al. 1995, Curtis-Hewitt S et al. 2000, Mulac-Jericevic et al. 2000, Spears & Bartlett 2009). E and P actions are mediated by their cognate receptors, namely estrogen receptor (ESR) and progesterone receptor (PGR) in coordination with a whole host of functionally context-dependent coregulators, to stimulate or inhibit target gene transcription. The signal transduction pathways initiated by the binding of E and P to their respective receptors are the subject of excellent recent reviews (Beato & Klug 2000, Hall et al. 2001). Similarly, the biochemical and biological properties of ESR and PGR, each of which exists classically in two forms, designated ESR1 and ESR2 and PGR-A and PGR-B, respectively, have been well-described (Kastner et al. 1990; Katzenellenbogen BS et al. 2000). By contrast, there is yet an incomplete understanding of the pathways by which ESR and PGR specify, recruit, and functionally categorize their co-regulatory proteins to optimize target cell sensitivity (Lonard DM et al. 2007). In this review, we consider the emerging role of a subset of nuclear proteins, namely the Krüppel-like factors (KLFs) in the regulation of steroid hormone signaling leading to appropriate responses of mammary epithelial and uterine endometrial cells to E and P. We also review findings to support the concept that maintenance of appropriate KLF expression is tightly controlled in mammary and uterine tissues and that the consequence of deregulated KLF expression is aberrant cell proliferation and differentiation leading to pre-neoplasia and cancer.
Krüppel-like Factors (KLFs)
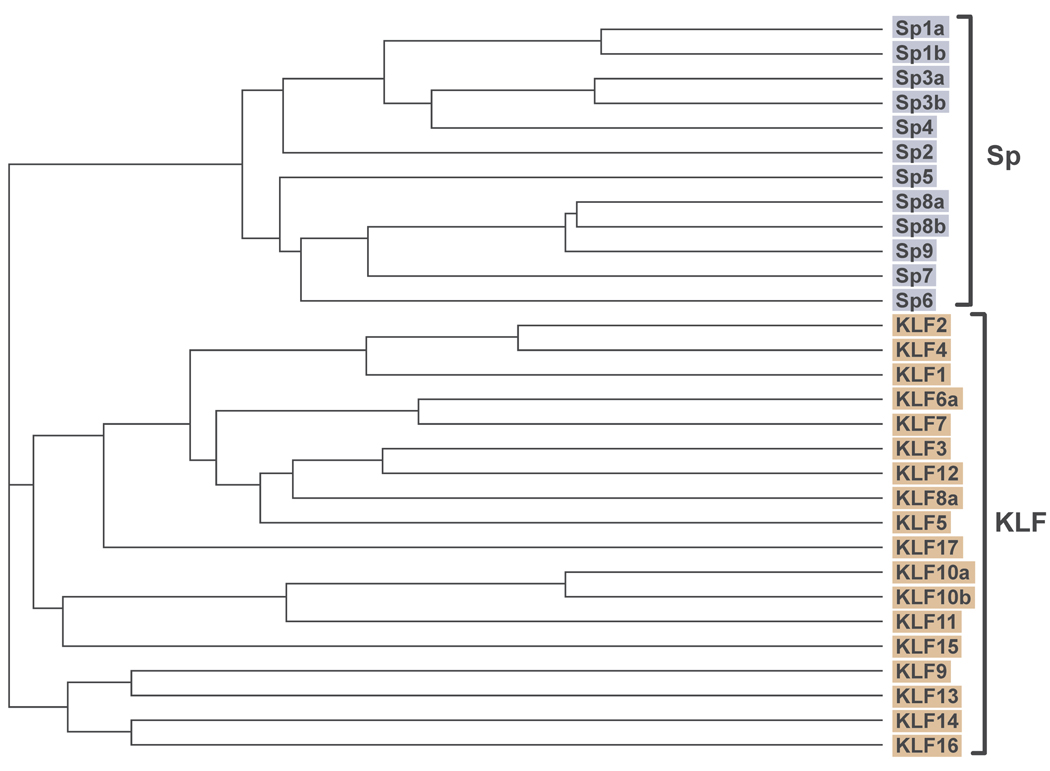
KLFs, so named for their similarity to the Drosophila segmentation gene product Krüppel (Preiss et al. 1985), belong to the evolutionary conserved Sp/KLF family of which there are currently 26 members (Fig.1). The Sp family is comprised of 9 members (Sp1-9), while the KLF family consists of 17 distinct members, a few of which (e.g., KLF6, KLF10, and KLF8) exhibit splice variants (Kaczynzki et al. 2003; Suske et al. 2005; Pearson et al. 2008). The family is characterized by a DNA-binding domain with conserved three tandem C2H2 type zinc-finger motifs at the carboxy-terminus and which recognizes the GT/GC box or CACCC element sites on promoter/regulatory regions. The phylogenetic relationship depicted in Fig. 1 is based on the similarity in sequences of their DNA binding domains. While Sp-family members are distinguished by glutamine and to a limited extent, serine-threonine-rich domains at the N-terminus, there is considerable diversity in the corresponding region among KLF members, which can display acidic, proline-rich, serine-rich, or hydrophobic transactivation domains. The highly variable amino-terminus confers functional specificity to KLF interactions with distinct nuclear proteins (Bieker 2001, Suske et al. 2005). Most KLFs are ubiquitously expressed, while others are found to be developmentally or temporally expressed in tissue- and cell-type specific manner; in recent years, however, the latter notion has been questioned with increasing evidence to the contrary (Kaczynski et al. 2001; Pearson et al. 2008). KLF members were previously designated as xKLF, where x refers to the tissue in which the gene was first identified (e.g., EKLF for erythroid KLF; GKLF for gut KLF). A single nomenclature system (KLF1, KLF2, etc.) has now been adopted by the scientific community to describe their order of discovery. Sp-family members Sp1–4 are highly related to KLF9, although Sp1 with 717 amino acids and a mol wt of 120 kDa is at least three times larger than KLF9 with 244 amino acids and a mol wt of 30 kDa. Other family members have molecular sizes in between. To date, only Sp1–4 and Sp7 among Sp-family members have documented cellular functions (Philipsen & Suske 1999, Waby et al. 2008). The expression patterns of Sp5, 6, 8 and 9 in various tissues have yet to be examined, and their transactivation potential relative to Sp1 remains relatively unknown. Among KLFs, several members, including KLF4, KLF5, KLF6, KLF8, KLF9, KLF10, KLF11, and KLF13, have been implicated in the regulation of a wide range of cellular functions including cell growth, differentiation, apoptosis, migration, and tumor formation (Black et al. 2001, Ghaleb et al. 2005, Wang et al. 2006, Pearson et al. 2008). Interestingly, family members can antagonize each other’s transcriptional activity, mostly because of physical competition between them in binding to cognate sequences in target gene promoters. In vivo, the physiologic functions of most KLF members have been validated by use of gene targeting technologies. This has been the subject of a recent review and will not be discussed here (Pearson et al. 2008). Suffice it to say that these studies have confirmed the pleiotropic actions of KLFs during embryonic and postnatal development in diverse tissue and cell types, and in adult tissues during distinct physiological states such as early pregnancy, parturition, and adipogenesis. The current review will focus on a subset of KLFs (KLF4, KLF5, KLF6, and KLF9) for which experimental evidence exists for their roles in growth control and in the pathobiology of uterine endometrial and breast cancers.
Figure 1.
Cladogram of the human Sp- and KLF-transcription factors. The 110-aa domain containing the buttonhead box (BTD)/zinc finger motifs was used for the multiple alignment with ClustalW, as described by Suske et al. 2005. *KLF6b, KLF6c, and KLF8b are truncated isoforms that contain deletion in the zinc finger motifs, and hence, were excluded in the alignment.
KLFs in the control of cell proliferation
The regulation of genes involved in cell cycle control and cell proliferation has surfaced as a major aspect of KLF action in diverse cell types (Black et al. 2001, Ghaleb et al. 2005). KLFs interact with different promoters and with other coregulators in their capacity to function as transcriptional activators, repressors, or both to influence cell growth regulation. This duality in functions is likely dependent on the architecture of the specific promoter (e.g., presence of single or multiple GC-rich motifs); the chromatin environment; and cellular co-expression of family members. KLF members mediate cell proliferation by attenuating or enhancing the transcription of anti-proliferative genes such as p21/wif1/cip1 (CDKNIA), p53 (TP53), and E-cadherin (CDH1) (Simmen et al. 2002, Yoon et al. 2003, Rowland et al. 2005, Wang et al. 2007) and of pro-proliferative genes such as those encoding cyclin E1 (CCNE1), cyclin D1 (CCND1), cyclin B1 (CCNB1), ornithine decarboxylase (ODC) and IGF-binding protein 2 (IGFBP2) (Shie et al. 2000, Chen et al. 2002, Simmen et al. 2002, Yoon et al. 2005, Evans et al. 2007). Several mechanisms involved in KLF transcriptional activation or repression have been described. KLFs can directly bind to GC-rich regions within target gene promoters to alter specific gene transcription. In this capacity, KLFs may bind alone or in complex (e.g. KLF4 and p53 in the CDKNIA promoter) with other proteins (Simmen et al. 2002, Yoon et al. 2003). By interfering with the recruitment of or competing with Sp1 for binding to recognition motifs within gene promoter regions, KLFs can suppress the well-recognized Sp1 induction of pro-proliferative gene transcription (Lomberk & Urrutia 2005). Finally, KLFs can selectively recruit negative co-regulators such as histone deacetylase-1 (HDAC-1) and mSin3A to gene regulatory regions to support transcriptional repression (Kaczynski et al. 2001). Recent studies, however, indicate that KLFs may also alter proliferative signaling pathways independent of binding to gene promoters. For example, KLF6 has been shown to interact with cyclin D1, thereby disrupting the phosphorylation of Retinoblastoma protein to promote cell cycle arrest (Benzeno et al. 2004). In addition, KLF4 was reported to inhibit Histone H4 acetylation by interacting with histone deacetylase 3 (HDAC-3) leading to transcriptional repression of proliferation-associated genes (Evans et al. 2007). Further, our group recently showed that KLF9 facilitates the recruitment of ESR1 to its own promoter, thus contributing to ESR1 auto-inhibition and decreased cell proliferation in the context of a high E2-environment (Velarde et al. 2007). Since the effect of KLF9 occurred without binding to DNA or ESR1, this suggests KLF9 interactions with other yet unknown nuclear proteins. Gene expression profiling has expanded the repertoire of KLF-induced or -repressed genes encoding cell cycle regulators in distinct cell types, revealing novel gene targets (Simmen RC et al. 2002, Goldstein et al. 2007, Simmen FA et al. 2008). It is unlikely that these up- or down-regulated genes all constitute direct targets of KLFs; however, data strongly suggest the depth and range of KLF involvement in growth signaling pathways.
KLFs and Endometrial Carcinoma
Endometrial carcinoma ranks as the fourth most frequent cancer among women in the Western world and causes significant morbidity and mortality in advanced stages (Jemal et al. 2008). The possible involvement of KLFs in uterine dysfunction as exemplified by endometrial carcinoma initially came from our group’s studies demonstrating cell type-dependent expression of KLF9 (previously designated Basic Transcription Element Binding Protein, BTEB-1; Imataka et al. 1992, Ohe et al. 1993) in uterine endometrium during pregnancy. We found KLF9 expression predominantly in endometrial stromal cells and to a lesser extent in glandular epithelial cells, with no or undetectable expression in luminal epithelial cells of normal cycling and early pregnant mice (Simmen et al. 2004, Velarde et al. 2005; Pabona et al. 2009). Importantly, we observed that null mutation of Klf9 by gene targeting in mice resulted in altered patterns of proliferation and apoptosis in all endometrial cell types, suggesting an essential role for largely stromal-derived KLF9 in uterine growth regulation (Velarde et al. 2005). Further, ovariectomized Klf9 null mutant mice were refractory to the proliferative effects of estradiol-17β (E2) in uterine cells, when compared to similarly treated ovariectomized wildtype counterparts (Pabona et al. 2009), documenting KLF9 involvement in E-mediated uterine proliferation. Using clonal sub-lines of HEC-1A human endometrial carcinoma cells that were stably transfected with sense and anti-sense Klf9 expression vectors, we found distinct cell phenotypes and gene expression patterns with KLF9 over- vs. under-expression (Zhang et al. 2001). KLF9 over-expressing cells displayed higher DNA synthesis and promoted G1/S progression of the cell cycle, concomitant with increased expression of CCND1, PCNA, CDKN1A, secretory leukocyte protease inhibitor (SLPI) and mitosin genes, all of which (with the exception of CDKN1A) are associated with increased proliferation status, relative to parent cells. Conversely, KLF9 under-expressing HEC-1A cells had lower expression levels of these genes, displayed lower mitotic index and interestingly, manifested increased ability to grow in multi-layers, the latter indicative of disruption in cell adhesion and cytoskeletal organization. Subsequent gene profiling of the same cell lines demonstrated regulation by KLF9 of gene transcripts encoding additional proteins associated with proliferation (e.g., brain-derived neurotrophic factor; KLF4); extracellular matrix (ECM) formation, motility and cell adhesion (e.g., integrin, beta 8; laminin gamma 2 protein; collagen type IV; versican); and signal transduction (e.g., mitogen-activated protein kinase-activated protein kinase 3; Wnt5b receptor) (Simmen et al. 2008). Collectively, these findings indicated that KLF9 levels are normally tightly regulated to maintain cellular homeostasis, and that inappropriate expression of KLF9 may lead to aberrant growth regulation and loss of epithelial-mesenchymal communication, contributing to endometrial carcinoma.
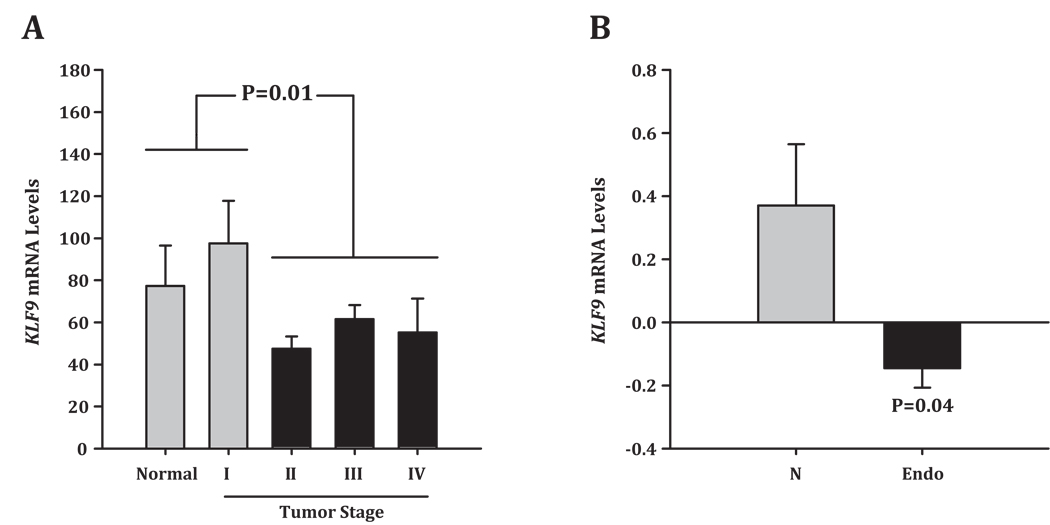
Two recent analyses suggested an association of KLF9 with human endometrial tumor pathology. In one study from our group, quantitative RT-PCR analyses of human endometrium and endometrial tumors using a normalized cDNA panel demonstrated a significant increase in KLF9 transcript levels in normal endometrium and stage I (more differentiated) endometrial tumors, when compared to tumors of more aggressive pathology (stages II, III, and IV) (Simmen et al. 2008; Fig. 2A). In the second study using the Cancer Microarray Data Mining Program (Oncomine.org; Compendia Biosciences, Ann Arbor, MI), we analyzed a published gene array database (Mutter et al. 2001) that compared the expression profiles of normal (from proliferative and secretory phases of the menstrual cycle) and malignant endometria, for KLF9 transcript levels. We found that levels of KLF9 transcripts were decreased in endometrial carcinoma tissues relative to normal endometria (Fig. 2B). Further studies using increased tumor sample sizes and at the level of the KLF9 protein for each tumor grade will be necessary to confirm this associational findings.
Figure 2.
KLF9 and endometrial carcinoma. (A) A normalized cDNA panel of human endometrial tumors (OriGene Technologies, Inc.) was used to probe for KLF9 transcript levels by quantitative RT-PCR. Data were adapted from Simmen et al. 2008. For each stage, data from endometrioid and serous tumors were combined. Sample numbers (in parenthesis) for each tissue or tumors are: normal (6); I (9); II (8); III (19), and IV (6). (B) KLF9 expression levels from comparison of normal (N) and malignant endometria (Endo) obtained from Affymetrix Hu6800 GeneChip probe arrays, as reported by Mutter et al. 2001. The normalized values shown here were obtained using the Cancer Microarray Data Mining Program (Oncomine.org) and are presented in Oncomine graphical representations. Sample numbers for N and Endo are 4 and 10, respectively. Significant difference (P<0.05) between groups was determined by t-test.
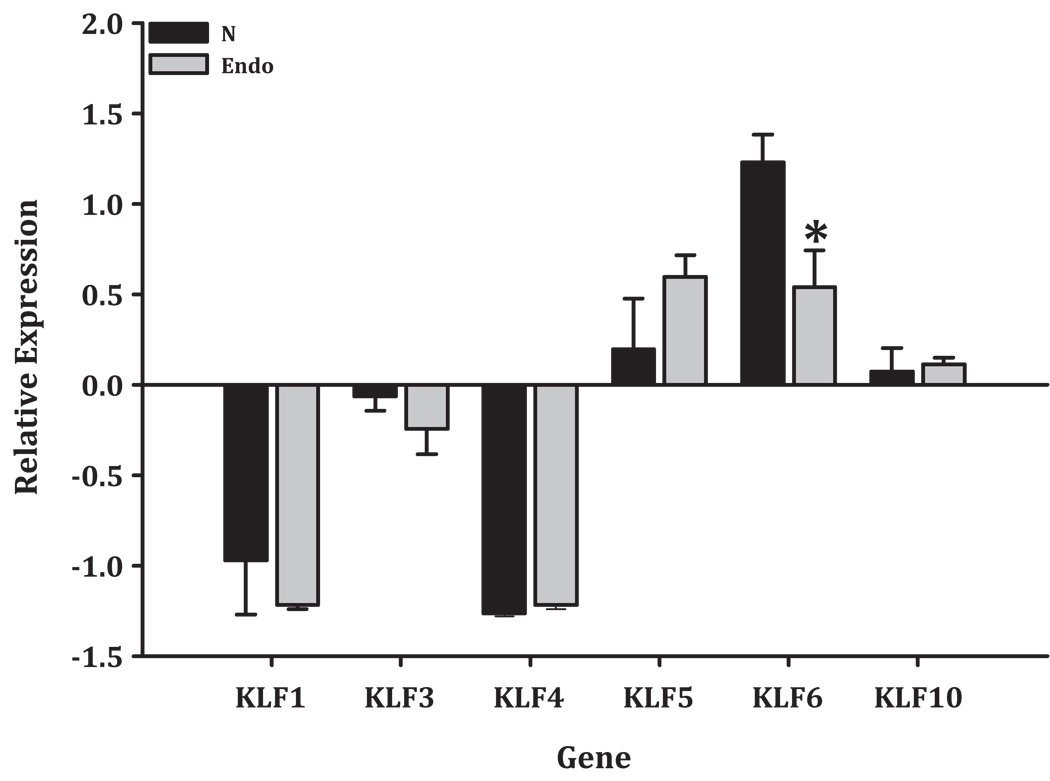
Given the observations that KLF4 expression was induced in HEC-1A cells over-expressing KLF9 (Simmen et al. 2008); that KLF13 can mimic KLF9 in transactivating genes in endometrial epithelial cells (Zhang et al. 2002; Zhang et al. 2003); and that KLF5 modulates the promoter of the uterine endometrial epithelial gene encoding lactoferrin (Shi et al. 1999), a protein with reported tumor-promoting activity (Albright & Kaufman, 2001), the expression of these and other KLFs in human normal endometrium (proliferative and secretory phases of the menstrual cycle) and endometrial carcinoma tissues were subsequently evaluated from a published gene array data by Mutter and colleagues (Mutter et al. 2001). Data mining indicated that the transcript levels of most KLFs were unaffected by malignant status (Fig. 3). The exceptions were KLF6 and KLF5, whose respective transcript levels were reduced and tended to increase, respectively in endometrial tumors, albeit not to the same magnitude as for KLF9 (Fig. 2B). These findings suggest that the physiological control of uterine epithelial proliferation may be limited to a small subset of KLFs. Moreover, since the expression of these KLFs has not been localized to specific cell types, it is not known whether the deregulated expression of these KLFs in tumors is directed from the stromal compartment or epithelium. Thus, a careful analysis of the adult uterine phenotypes of mice with conditional Klf null mutations will be required. Further studies will be also be needed to clarify whether in the context of normal vs. tumor cells, KLF members may have distinct, similar, or synergistic biological behaviors.
Figure 3.
Transcript levels of different KLF members in normal (N) and malignant (endo) endometria. The analysis was carried out using the same data set described by Mutter et al. 2001 and normalized values are presented in Oncomine graphical representations. Sample numbers for N and Endo are 4 and 10, respectively. Difference between groups was determined by t-test. *P=0.05.
KLFs and Breast Cancer
The potential loss of growth control mediated by distinct KLFs is well-studied and better documented in mammary epithelial cells than in endometrial cells, a fact likely related to the higher incidence and hence, more wide-spread and devastating consequences, of breast than endometrial cancers in the populace. In the USA alone, an estimated 180,000 new cases of breast cancer and 50,000 deaths from this disease are reported annually (Jemal et al. 2008). The linkage between breast cancer and KLFs is strongest for KLF4 and KLF5, although a consensus on whether these KLFs function as tumor suppressors or oncogenes in breast cancer is lacking. In support of a tumor suppressor function for KLF4, breast cancer cells were found to exhibit loss of KLF4 expression relative to normal mammary epithelial cells, and this was associated with markedly down-regulated expression of laminin B5, a component of the major ECM protein lamin α (Miller et al. 2001). However, KLF4 was also reported to be expressed at low levels in morphologically normal (uninvolved) breast epithelium adjacent to tumor cells, but displayed increased expression in neoplastic cells (Foster et al. 2000). Increased KLF4 expression in tumor cells was localized to the nucleus in the early stages of invasive ductal carcinoma of the breast, suggesting its prognostic potential for aggressive phenotype (Pandya et al. 2004). Similar to KLF4, KLF5 has also been reported to have a dual role as a tumor suppressor or as an oncogene. One study found that KLF5 is pro-proliferative, and the positive association between higher KLF5 expression coincident with increased expression of HER2/neu and Ki67 on the one hand, and shorter disease-free survival and limited overall survival time on the other hand, suggest the prognostic value of this KLF for patients with breast cancer (Tong et al. 2006). In another study, KLF5 was implicated in breast cancer progression by inducing the expression of fibroblast growth factor-binding protein, which is over-expressed in breast tumors and found to promote tumorigenesis (Zheng et al. 2008). In vitro, knockdown of KLF5 expression in the human mammary epithelial cell lines MCF-10A and BT20 resulted in induction of apoptosis (Liu et al. 2008). This effect was attributed to loss of KLF5-mediated inhibition of degradation of the pro-survival phosphatase MAPK-phosphatase-1 protein. In support of KLF5 as a tumor suppressor, elevated expression of KLF5 in non-neoplastic and normal human mammary tissues, in contrast to lower expression in breast cancer lines, has been reported (Chen et al. 2002). Recent studies, albeit limited, have also implicated KLF6 and KLF8 in breast cancer progression. KLF6 expression was found to be negatively associated with breast cancer status, suggesting a possible tumor suppressor function (Guo et al. 2007). By contrast, KLF8 is considered to be involved in the promotion of breast cancer based on its ability to increase epithelial-mesenchymal transition and to enhance motility as a consequence of its direct binding to the E-cadherin promoter to decrease this gene’s transcription (Wang et al. 2007). A role for KLF9 has not been specifically evaluated in normal mammary tissues or mammary tumors; however, we have found no gross morphological differences in mammary glands of young and adult Klf9 null and wildtype mice, and observed no spontaneous mammary tumor occurrence in older (∼1 year-old) Klf9 null mutants (Simmen RCM & Velarde MC, unpublished findings). It will be interesting to further evaluate the mechanisms of mammary tumor progression mediated by KLFs in mouse models of tumorigenesis (e.g. MMTV-Wnt transgenic mice), for example by a comprehensive study of the different KLFs during mammary tumor development and by an extensive analyses of the mammary phenotypes of specific Klf mutants crossed to Wnt-Tg mice.
KLFs and Steroid Hormone Signaling
Endometrial and ESR1-positive breast cancers arise from dysregulated E and/or P signaling. Given the experimental data that KLFs may promote or attenuate endocrine-responsive cancers, the possibility that KLFs exert their effects through cross-talk with ESR1 and PGR signaling pathways was anticipated (Zhang et al. 2002). Indeed, a subset of KLF family members have now been confirmed to function as co-activators of ESR1 and PGR based primarily on in vitro cell culture studies, but increasingly supported from analyses of in vivo mouse mutant models. The major evidence to date comes from analyses of KLF9 and its interaction with PGR in the regulation of PGR-dependent gene transcription in uterine endometrial cells. In the human endometrial carcinoma cell line Ishikawa which is of glandular epithelial cell origin, KLF9 was shown to physically interact with PGR-B and to promote the PGR-B dependent transactivation of P-responsive promoters (Zhang et al. 2003, Velarde et al. 2006). Interestingly, PGR-A isoform did not recapitulate PGR-B interactions with KLF9, suggesting the selective utilization of KLF9 by PGR-B as a co-regulator of its transactivity (Zhang et al. 2003). KLF13 can substitute for KLF9 as a PGR-B partner in this context (Zhang et al. 2003); this is likely due to the structural homology between KLF9 and KLF13, which exhibit the greatest similarities among all KLF members (Philipsen & Suske, 1999) (Fig. 1). In vivo, functional interactions between PGR and KLF9 were confirmed by comparison of Klf9 wildtype and null mutants for P-dependent gene expression; E+P-dependent cell proliferation and apoptotic status; and embryo implantation outcome, an E+P-dependent event (Simmen et al. 2004, Velarde et al. 2005). Similar to PGR signaling, ESR1 signaling may also involve the participation of KLF9. Evidence for this is provided by in vivo and in vitro studies describing: a) loss of responsiveness to E2-induced proliferation of endometrial cells with Klf9 null mutation, possibly mediated by loss of KLF9 inhibition of Repressor of Estrogen Receptor Activity (REA) expression (Pabona et al, 2009); b) increased Esr1 expression in peri-implantation stromal cells of Klf9 null mutants (Velarde et al, 2005); c) KLF9 transcriptional repression of ESR1 signaling in Ishikawa endometrial adenocarcinoma cells by promoting ligand-dependent ESR1 auto-downregulation (Velarde et al, 2007); and d) the negative association between Klf9 and Esr1 transcript levels in endometrial tumors (RCM Simmen, data not shown).
The recent generation of Klf13 null mutants which are not embryo-lethal (Zhu et al. 2007) will now allow parallel comparison of KLF13 effects on ligand-dependent PGR transcriptional and biological events, to those of KLF9. More importantly, such studies would provide confirmation on the ability of KLF13 and KLF9 to compensate/substitute for each other’s function in the uterine endometrium. Nevertheless, since initial analyses of endometrial tumor samples indicated undetectable KLF13 expression in human endometrium and endometrial tumors (Mutter et al. 2001, Fig. 3), context-dependent functions of KLF13 are likely. Although no data is available regarding the participation of KLF6 and KLF5 in steroid hormone signaling, perturbations in their expression under a pathological E2-dominated environment (endometrial carcinoma) hint of potential linkages. However, given that null mutations of Klf4, Klf5, and Klf6 result in embryonic or perinatal lethality (Pearson et al. 2008), it is not currently possible to utilize knockout mice for evaluation of respective uterine and mammary gland phenotypes; such studies await the generation of mammary- and uterine-targeted gene mutations.
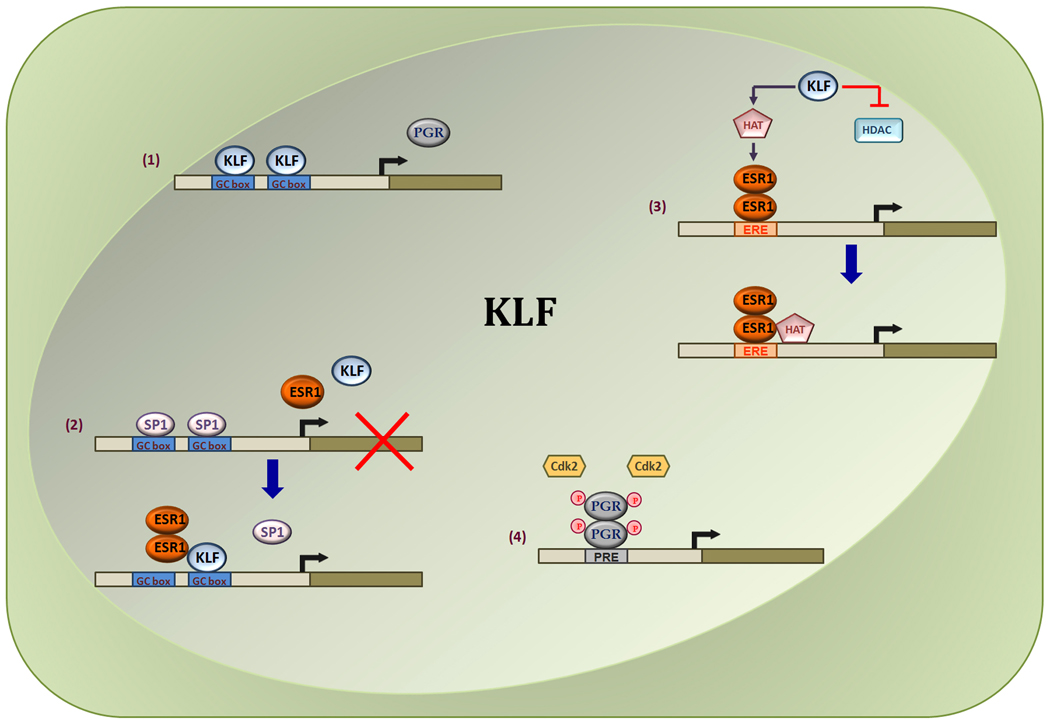
How may KLFs participate in steroid hormone signaling? Limited mechanistic data are available to fully describe KLF involvement, albeit insights gleaned from our data (Zhang et al. 2002, 2003, Velarde et al. 2005, 2006, 2008, Pabona et al. 2009) and those described for family member Sp1 (Khan et al. 2007, Wu et al. 2009) provide some directions. Given that KLFs are transcription factors, family members may modulate E- and/or P-sensitivity of target cells by: a) regulating ESR1 and PGR expression; b) by facilitating recruitment of PGR and ESR1 to steroid hormone-responsive promoters which lack canonical P-responsive elements (PRE) or E-responsive elements (ERE), through their direct binding to GC/GT boxes in gene promoters and by competing with SP factors to promote or inhibit transcription; c) by interacting with chromatin modifiers such as HAT, HDAC, and mSin3A to induce or repress recruitment of nuclear PGR/ESR1 co-regulators and components of the RNA pol II enzyme; and d) by post-translational modifications (e.g. phosphorylation) of nuclear receptors or their co-factors through control of expression and/or activity of specific kinases that modify these proteins. The latter possibility, while speculative, comes from findings that PGR phosphorylation is important for its transcriptional activity (Clemm et al. 2000, Knotts et al. 2001) and that CDK2, which has been implicated in PGR-A and PGR-B phosphorylation is a KLF9-induced gene in the human endometrial carcinoma cell line HEC-1A (Simmen et al. 2002). Clearly, the potential importance of KLFs in mediating multiple events (summarized in Fig. 4) necessitates a thorough understanding of the specific family member(s) involved in these and other similar yet unknown, regulatory processes.
Figure 4.
Postulated model for KLF involvement in ESR and PGR transcriptional pathways. KLF members may mediate transcriptional activities of steroid hormone receptors by regulating their levels of expression (1), and/or transactivities by interfering with Sp1 binding to gene promoters (2); promoting the recruitment of nuclear co-regulators (3); and influencing post-translational modifications (e.g., phosphorylation) of nuclear receptors or co-regulators through transcriptional regulation of kinase cascades (4). ESR1, estrogen receptor-α; PGR, progesterone receptor A/B; HAT, histone acetyl transferase; HDAC, histone deacetylase; Sp1, specificity protein-1.
Conclusions
Recent studies have documented KLF family members in the control of cell proliferation, differentiation, and apoptosis in steroid-responsive mammary and uterine endometrial cells. Since these processes are well-recognized as critical events regulated by ESR1 and PGR signaling, and loss of this regulation partly underlies endocrine-responsive cancers, the further understanding of the cross-talk between KLF-regulated pathways and those orchestrated by ligand-activated ESR1 and PGR may lead to the identification of common and possibly novel, gene targets that will facilitate the development of agents for the treatment of hormone-responsive cancers. Albeit Sp/KLF family member Sp1 has significant headway in the mechanistic understanding of its participation in growth control, current information predicts that KLFs may have far greater consequences on progression to neoplasia given their duality in functions (tumor suppressor or promoter) under distinct contexts even in the same target tissue. Further, given the fact that many other types of cancer (e.g. prostate cancer, colon and intestinal cancers, leiomyoma) have an endocrine component underlying their molecular pathologies, and some of these have been recently associated with loss of KLF expression (e.g., colorectal cancer and KLF9) (Kang et al. 2008), it is reasonable to assume that the development of effective therapies for a broad range of cancers may be well-served by further analyses of KLF signaling. In this regard, the increasing data in support of the involvement of multiple KLFs (KLF2, KLF4, KLF5) in the regulation of stem cell renewal and maintenance (Jiang et al. 2008, Chan et al. 2009) open new possibilities for the use of KLFs and signaling components to target cancer stem cells that drive tumor growth (Zhang & Rosen, 2006).
Acknowledgments
Funding
Research cited here from the Simmen’s laboratories was supported by funds from the National Institutes of Health grant no. HD21961, and the Arkansas Children’s Hospital Research Institute/Arkansas Biosciences Institute.
Footnotes
“Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Journal of Endocrinology, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the Society for Endocrinology accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at [insert DOI link]. © [insert year of publication] Society for Endocrinology.”
Declaration of interest
The authors declare that there are no conflicts of interest that could be perceived as prejudicing this report.
References
- Albright CD, Kaufman DG. Lactoferrin: a tamoxifen-responsive protein in normal and malignant human endometrial cells in culture. Exp.Mol.Pathol. 2001;70:71–76. doi: 10.1006/exmp.2000.2354. [DOI] [PubMed] [Google Scholar]
- Beato M, Klug J. Steroid hormone receptors: an update. Hum.Reprod.Update. 2000;6:225–236. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D&, Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J.Biol.Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J.Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Chan KK, Zhang J, Chia NY, Chan YS, Sim HS, Tan KS, Oh SK, Ng HH, Choo AB. KLF4 and PBX1 directly regulate nanog expression in human embryonic stem cells. Stem Cells. 2009 doi: 10.1002/stem.143. (In press) [DOI] [PubMed] [Google Scholar]
- Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J.Biol.Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- Clemm DL, Sherman L, Boonyaratanakornkit V, Schrader WT, Weigel NL, Edwards DP. Differential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site-specific monoclonal antibody. Mol.Endocrinol. 2000;14:52–65. doi: 10.1210/mend.14.1.0413. [DOI] [PubMed] [Google Scholar]
- Curtis HS, Couse JF, Korach KS. Estrogen receptor transcription and transactivation: Estrogen receptor knockout mice: what their phenotypes reveal about mechanisms of estrogen action. Breast Cancer Res. 2000;2:345–352. doi: 10.1186/bcr79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliassen AH, Hankinson SE. Endogenous hormone levels and risk of breast, endometrial and ovarian cancers: prospective studies. Adv.Exp.Med.Biol. 2008;630:148–165. [PubMed] [Google Scholar]
- Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J.Biol.Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am.J.Physiol Gastrointest.Liver Physiol. 2007;292:G1784–G1792. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- Guo H, Lin Y, Zhang H, Liu J, Zhang N, Li Y, Kong D, Tang Q, Ma D. Tissue factor pathway inhibitor-2 was repressed by CpG hypermethylation through inhibition of KLF6 binding in highly invasive breast cancer cells. BMC Mol Biol. 2007;8:110. doi: 10.1186/1471-2199-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J.Biol.Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J.Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Zhang JS, Ellenrieder V, Conley A, Duenes T, Kester H, van Der BB, Urrutia R. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J.Biol.Chem. 2001;276:36749–36756. doi: 10.1074/jbc.M105831200. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Lu B, Xu J, Hu H, Lai M. Downregulation of Kruppel-like factor 9 in human colorectal cancer. Pathol.Int. 2008;58:334–338. doi: 10.1111/j.1440-1827.2008.02233.x. [DOI] [PubMed] [Google Scholar]
- Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Montano MM, Ediger TR, Sun J, Ekena K, Lazennec G, Martini PG, McInerney EM, Delage-Mourroux R, Weis K, Katzenellenbogen JA. Estrogen receptors: selective ligands, partners, and distinctive pharmacology. Recent Prog.Horm.Res. 2000;55:163–193. [PubMed] [Google Scholar]
- Khan S, Wu F, Liu S, Wu Q, Safe S. Role of specificity protein transcription factors in estrogen-induced gene expression in MCF-7 breast cancer cells. J.Mol.Endocrinol. 2007;39:289–304. doi: 10.1677/JME-07-0043. [DOI] [PubMed] [Google Scholar]
- Knotts TA, Orkiszewski RS, Cook RG, Edwards DP, Weigel NL. Identification of a phosphorylation site in the hinge region of the human progesterone receptor and additional amino-terminal phosphorylation sites. J.Biol.Chem. 2001;276:8475–8483. doi: 10.1074/jbc.M009805200. [DOI] [PubMed] [Google Scholar]
- Liu R, Zheng HQ, Zhou Z, Dong JT, Chen C. KLF5 promotes breast cell survival partially through fibroblast growth factor-binding protein 1-pERK-mediated dual specificity MKP-1 protein phosphorylation and stabilization. J.Biol.Chem. 2009;284:16791–16798. doi: 10.1074/jbc.M808919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem.J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr.Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, Weitzman SA. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J.Biol.Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- Mulac-Jericevic B, Lydon JP, DeMayo FJ, Conneely OM. Defective mammary gland morphogenesis in mice lacking the progesterone receptor B isoform. Proc.Natl.Acad.Sci.U.S.A. 2003;100:9744–9749. doi: 10.1073/pnas.1732707100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutter GL, Baak JP, Fitzgerald JT, Gray R, Neuberg D, Kust GA, Gentleman R, Gullans SR, Wei LJ, Wilcox M. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol. Oncol. 2001;83:177–185. doi: 10.1006/gyno.2001.6352. [DOI] [PubMed] [Google Scholar]
- Ohe N, Yamasaki Y, Sogawa K, Inazawa J, Ariyama T, Oshimura M, Fujii-Kuriyama Y. Chromosomal localization and cDNA sequence of human BTEB, a GC box binding protein. Somat.Cell Mol.Genet. 1993;19:499–503. doi: 10.1007/BF01233255. [DOI] [PubMed] [Google Scholar]
- Pabona JM, Velarde MC, Zeng Z, Simmen FA, Simmen RC. Nuclear receptor co-regulator Kruppel-like factor 9 and prohibitin 2 expression in estrogen-induced epithelial cell proliferation in the mouse uterus. J.Endocrinol. 2009;200:63–73. doi: 10.1677/JOE-08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chieng DC, Grizzle WE, Engler JA, Krontiras H, Bland KI, LoBuglio AF, Lobo-Ruppert SM, Ruppert JM. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin.Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int.J.Biochem.Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat.Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27:4807–4815. doi: 10.1093/nar/27.24.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen FA, Su Y, Xiao R, Zeng Z, Simmen RC. The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod.Biol.Endocrinol. 2008;6:41. doi: 10.1186/1477-7827-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen RC, Zhang XL, Michel FJ, Min SH, Zhao G, Simmen FA. Molecular markers of endometrial epithelial cell mitogenesis mediated by the Sp/Kruppel-like factor BTEB1. DNA Cell Biol. 2002;21:115–128. doi: 10.1089/104454902753604998. [DOI] [PubMed] [Google Scholar]
- Simmen RC, Eason RR, McQuown JR, Linz AL, Kang TJ, Chatman L, Jr, Till SR, Fujii-Kuriyama Y, Simmen FA, Oh SP. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Kruppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J.Biol.Chem. 2004;279:29286–29294. doi: 10.1074/jbc.M403139200. [DOI] [PubMed] [Google Scholar]
- Spears M, Bartlett J. The potential role of estrogen receptors and the SRC family as targets for the treatment of breast cancer. Expert.Opin.Ther.Targets. 2009;13:665–674. doi: 10.1517/14728220902911509. [DOI] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin.Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- Velarde MC, Geng Y, Eason RR, Simmen FA, Simmen RC. Null mutation of Kruppel-like factor9/basic transcription element binding protein-1 alters peri-implantation uterine development in mice. Biol.Reprod. 2005;73:472–481. doi: 10.1095/biolreprod.105.041855. [DOI] [PubMed] [Google Scholar]
- Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor alpha signaling in Ishikawa endometrial adenocarcinoma cells. Mol.Endocrinol. 2007;21:2988–3001. doi: 10.1210/me.2007-0242. [DOI] [PubMed] [Google Scholar]
- Waby JS, Bingle CD, Corfe BM. Post-translational control of Sp-family transcription factors. Curr.Genomics. 2008;9:301–311. doi: 10.2174/138920208785133244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Zheng H, Castell C, Debussche L, Multon MC, Wasylyk B. Inhibition of the Ras-Net (Elk-3) pathway by a novel pyrazole that affects microtubules. Cancer Res. 2008;68:1275–1283. doi: 10.1158/0008-5472.CAN-07-2674. [DOI] [PubMed] [Google Scholar]
- Wu F, Ivanov I, Xu R, Safe S. Role of SP transcription factors in hormone-dependent modulation of genes in MCF-7 breast cancer cells: microarray and RNA interference studies. J.Mol.Endocrinol. 2009;42:19–33. doi: 10.1677/JME-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J.Biol.Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang XL, Michel FJ, Blum JL, Simmen FA, Simmen RC. Direct interaction of the Kruppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology. 2002;143:62–73. doi: 10.1210/endo.143.1.8590. [DOI] [PubMed] [Google Scholar]
- Zhang M, Rosen JM. Stem cells in the etiology and treatment of cancer. Curr Opin Genet Dev. 2006;16:60–64. doi: 10.1016/j.gde.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Simmen FA, Michel FJ, Simmen RC. Increased expression of the Zn-finger transcription factor BTEB1 in human endometrial cells is correlated with distinct cell phenotype, gene expression patterns, and proliferative responsiveness to serum and TGF-beta1. Mol.Cell Endocrinol. 2001;181:81–96. doi: 10.1016/s0303-7207(01)00536-6. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC. Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J.Biol.Chem. 2003;278:21474–21482. doi: 10.1074/jbc.M212098200. [DOI] [PubMed] [Google Scholar]
- Zheng H, Zhou Z, Chen C. Krupple-like factor 5 promotes breast cancer proliferation through activating fibrobalst growth factor-binding protein transcription. Cancer Res (Suppl) 2008;69 271s. [Google Scholar]
- Zhou M, McPherson L, Feng D, Song A, Dong C, Liu SC, Zhou L, Shi X, Ahn YT, Wang D, Clayberger C, Krensky AM. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J.Immunol. 2007;178:5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]