Abstract
Residue 116 of major histocompatibility complex (MHC) class I heavy chains is an important determinant of assembly, that can influence rates of ER-Golgi trafficking, binding to the transporter associated with antigen processing (TAP), tapasin dependence of assembly, and the efficiency and specificity of peptide binding. Here we investigated assembly and peptide binding differences between HLA-B*3501(S116) and HLA-B*3503(F116), two alleles differing only at position 116 of the MHC class I heavy chain, that are associated respectively with normal or rapid AIDS progression. A reduced intracellular maturation rate was observed for HLA-B*3503 in HIV-infected and uninfected cells, which correlated with enhanced binding of HLA-B*3503 to TAP. No significant differences in the intrinsic efficiency of in vitro peptide binding by HLA-B*3501 and HLA-B*3503 were measurable with several common peptides or peptide libraries, and both allotypes were relatively tapasin independent for their assembly. However, thermostability differences between the two allotypes were measurable in a CD4+ T cell line. These findings suggest that, compared to HLA-B*3501, a reduced intracellular peptide repertoire for HLA-B*3503 could contribute to its slower intracellular trafficking and stronger association with rapid AIDS progression.
Keywords: MHC class I, tapasin, Antigen Presentation/Processing, TAP transporter, HLA-B*3501, HLA-B*3503, HLA-B*4402, HLA-B*4405, HLA-B*5701
INTRODUCTION
HIV/AIDS is a leading cause of infectious disease mortality worldwide. Among the multiple genetic factors that influence AIDS progression, the human leukocyte antigen (HLA) class I genes have the strongest effects identified to date (Carrington and O’Brien 2003). HLA-A, -B, and -C genes encode molecules that bind antigenic peptides, and present the peptides to CD8+ T cells, thereby initiating a cytotoxic T cell (CTL) response during infection. Extensive allelic polymorphisms are observed in the HLA-A, B and C genes, concentrated primarily among nucleotides that encode residues within the peptide binding grooves of the HLA class I molecules, which determine specificity for the associated peptide ligands (Bjorkman et al. 1987). Specific HLA alleles, primarily HLA-B alleles, are implicated in influencing AIDS progression rates (Kiepiela et al. 2004). Consistent associations include delayed progression to AIDS among individuals with HLA-B*27 or -B*57 (Kaslow et al. 1996; Migueles et al. 2000), and accelerated progression conferred by certain HLA-B*35 alleles (Gao et al. 2001; Itescu et al. 1992). In a study by Gao et al (Gao et al. 2001), accelerated AIDS progression among HLA-B*35-positive individuals was attributed to HLA-B*35-Px alleles (including HLA-B*3503) (Gao et al. 2001). HLA-B*35 subtypes were divided into two groups according to their peptide-binding specificities: (a) the HLA-B*35-PY group, composed of two closely related HLA-B*35 alleles (including the most common, HLA-B*3501), which bind peptides with proline in position 2 and tyrosine in position 9 (Falk et al. 1993); and (b) the HLA-B*35-Px group, which also binds peptides with proline in position 2, but with several amino acids excluding tyrosine at position 9 (Steinle et al. 1996). Although HLA-B*35-PY and HLA-B*35-Px groups differ in their preferences for tyrosine at P9, several other P9 residues are expected to be permissive for both groups.
HLA-B*3501 (PY) and HLA-B*3503 (Px) proteins differ by a single amino acid at position 116 of the heavy chain, which forms the floor of the peptide-anchoring F pocket, and influences specificity for peptide carboxy-terminal residues. Structural analyses have shown that residue 116 directly interacts with residue P9 of the bound peptide (Saper et al. 1991; Smith et al. 1996; Tynan et al. 2005). Recent studies have shown that residue 116 also profoundly influences intracellular assembly characteristics (Khanna et al. 1997; Turnquist et al. 2000; Williams et al. 2002; Zernich et al. 2004; Goodall et al. 2006; Thammavongsa et al. 2006). Allotypes that differ only at position 116, such as HLA-B*4402(D116) and HLA-B*4405(Y116), display markedly different assembly characteristics. HLA-B*4402 is highly dependent on the MHC class I assembly factor tapasin for its assembly, whereas HLA-B*4405 is relatively tapasin independent for its assembly (Williams et al. 2002; Zernich et al. 2004). Tapasin-independent peptide loading was found to be relatively inefficient for HLA-B*4402 compared to HLA-B*4405, and the rate of ER-Golgi trafficking was reduced for HLA-B*4402 compared to HLA-B*4405, even in tapasin-sufficient cells (Khanna et al. 1997; Thammavongsa et al. 2006). These assembly differences have been shown to impact the efficiency of induction of a CTL response, with CTL responses against HLA-B*4405 being less efficient compared to HLA-B*4402 (Khanna et al. 1997). The cell surface expression of HLA-B*4402 has also been found to be more susceptible to viral modulators of MHC class I assembly (Zernich et al. 2004). Together, these observations indicate that intracellular assembly differences could strongly impact infectious disease outcomes. Since HLA-B*3501 and HLA-B*3503 differ only at position 116 of their sequences, and have previously been shown to be differentially associated with the transporter associated with antigen processing (TAP) (Neisig et al. 1996), in the studies described here, we compared trafficking and assembly profiles of HLA-B*3501 and HLA-B*3503 with each other and with HLA-B*4402 and HLA-B*4405.
MATERIALS AND METHODS
Cell lines
CEM-SS lines were maintained in RPMI supplemented with 2 mM glutamine, 10% FCS, penicillin and streptomycin. The preparation of a CEM-SS T cell line stably expressing various MHC-I molecules has been previously described (Kasper and Collins 2003). For experiments using murine retroviral vectors, retroviral supernatants were prepared as previously described (Pear et al. 1993; Van Parijs et al. 1999) except that they were pseudotyped with VSV-G protein. 1 × 106 CEM T cells were spin-transduced with the retroviral supernatants as previously described for HIV infections (Kasper and Collins 2003). Insect SF21 cells were maintained in Grace’s insect media (Invitrogen), supplemented with 10% fetal bovine serum and 500 μg/ml gentamycin (Invitrogen).
DNA Constructs
Retroviral vectors
The construction of retroviral vectors encoding HA-tagged HLA-B*4402, HLA-B*4405 and tapasin have been previously published (Thammavongsa et al. 2006). For retroviral constructs encoding HA-tagged HLA-B*3501 and HLA-B*5701, cDNAs encoding B*3501, and B*5701 were isolated as follows: RNA was prepared from EBV-transformed B cell lines using RNeasy Midi Protocol for isolation of total RNA from animal cells (Qiagen). RT PCR was performed using Superscript First Strand Synthesis Kit (Gibco/Invitrogen). Primers used were as follows: forward primer 5′-GGGAATTCCTCAGAATCTCCTCAGACGCCGAG-3′ (this sequence is found just before the open reading frame of HLA-B sequences) and reverse primer 5′-GGCTCGAGTCAAGCTGTGAGAGACACATCAGA-3′. The RT PCR products were digested with Eco RI and Xho I and ligated into MSCV2.1 (cut with EcoRI and Xho I).
PCR was used to add a nine amino acid hemagglutinin (HA) epitope tag (YPYDVPDYA) to the N-terminus of each MHC-I molecule (just after the leader sequence cleavage site and before the Nae I site as previously described (Roeth et al. 2004)). As part of this construction, a second glycine-serine was added N-terminal to the tag at the signal peptide cleavage point to reproduce the cleavage site of the class I molecule. The primers used to amplify the HLA-B*3501 leader plus HA sequence were as follows: forward primer (same as above primer), 5′-GGGAATTCCTCAGAATCTCCTCAGACGCCGAG- 3′; reverse primer, 5′-TCCCGCCGGCATAGTCGGGTACG TCATACGGATAGGACCCGGCCCAGGTCTCGGT- 3′. The PCR product described above was digested with EcoRI and NaeI and subcloned into a shuttle vector (New England Biolabs, Inc.). The tag plus B3501 leader sequence was then isolated by EcoRI and NaeI digestion of the shuttle vector. The 3′ regions of HLA- B*5701 and HLA-B*3501 (encoding the portion of the protein downstream of the leader sequence) was isolated by digestion of MSCV2.1- HLA- B*5701 and HLA-B*3501 with NaeI and XhoI. Finally, the HLA-B*3501 and HLA- B*5701 NaeI-XhoI fragments and the EcoRI-NaeI fragment from the shuttle vector were ligated into MSCV 2.1 (cut with EcoRI and XhoI) in a three- way ligation to generate MSCV HA- B*5701 and HA-HLA-B*3501.
To make MSCV HA-HLA-B*3503 (B*3501S116F), B*3501 was used as a template for the following primers: forward primer 5′-TCCGCGGGCATGACCAGTTCGCCTACGACGGCAAGGA-3′; reverse primer 5′-TCCTTGCCGTCGTAGGCGAACTGGGTCATGCCCGCGGA-3′. HLA-B*3503 was then HA-tagged as described above for HA-HLA-B*5701 and HA-HLA-B*3501.
Baculovirus constructs
Construction of soluble histidine-tagged HLA-B*3501 has been described previously (Rizvi et al. 2004). Soluble histidine-tagged HLA-B*3503 was derived from full-length HLA-B*3503 using the following two PCR primers (5″-AAGGATCCGAATTCCCACCATGCGGGTCACGGCGCCC-3″) and reverse primer (5″-AAGGATCCCACGAGCTAATGATGATGATGATGATGCTCCCATCTCACGGTGAG-3″). This corresponded to a truncation after the LRWE sequence at residue 299 of the mature protein. The amplified PCR product was digested with BamH1 and ligated into the BamH1 site of a pACUW31 vector that had the chimp β2m cDNA ligated into the BglII site. The heavy chain region was sequenced.
Viruses and cell infections
Baculoviruses were generated using the BD Baculogold kit (BD Biosciences Pharmingen). HLA-B*35 heterodimers were purified from baculovirus-infected High-Five insect cell supernatants as described previously (Mancino et al. 2002).
Retroviruses were generated as previously described (Pear et al. 1993; Van Parijs et al. 1999) using Bosc cells, and used to infect CEM and the tapasin-deficient M553 (Belicha-Villanueva et al., 2008) cells. Cells were transduced with retroviruses encoding the class I molecules, and selected by treatment with 1 mg/ml G418 disulfate (Sigma-Aldrich), and maintained in 0.5mg/ml G418 disulfate. In M553 cells, after verifying MHC class I expression by flow cytometric analyses and by immunoblotting analyses of cell lysates, cells expressing HA-tagged class I were transduced with the tapasin retrovirus, and selected by treatment with 1 mg/ml puromycin. Cells were maintained in 0.5 μg/ml puromycin.
Intracellular trafficking and interactions analyses
Antibodies
Monoclonal antibody HC-10 recognizes free MHC class I heavy chains (Stam et al. 1990), monoclonal antibody anti-HA recognizes the Hemagglutinin epitope tag (YPYDVPDYA), monoclonal antibody W6-32 (Barnstable et al., 1978) recognizes correctly folded MHC class I/β2m heterodimers and anti-TAP1 is a rabbit polyclonal anti-sera which is specific for the C-terminus of TAP1 (Androlewicz et al. 1994).
Assessment of interactions of MHC class I molecules with TAP
Analyses of MHC class I – TAP interactions were performed as described previously using immunoprecipitations with anti-TAP1 (obtained from Dr. Matt Androlewicz), followed by immunoblotting analyses with anti-HA (Thammavongsa et al. 2006).
Pulse-chase analyses
For sequential immunoprecipitations with W6/32 and anti-HA, 8 × 107 CEM cells expressing various class I molecules were metabolically labeled for 10 minutes with [35S]-methionine and cysteine, and chased for the indicated times. Cells were lysed by incubation for 45 minutes on ice in a 1% Triton buffer (10mM Tris, 10mM phosphate, 130 mM NaCl, 1% Triton X-100, pH 7.5), and then centrifuged for 5 minutes at full speed in a microcentrifuge. Supernatants were collected and immunoprecipitated for 1 hour at 4 °C with 5 μL of W6/32 antibody and then with 20 μL of protein A/protein G beads. Beads were washed once with 1% Triton buffer, and boiled for 5 minutes in the presence of 100 μL of 1% SDS, 5 mM DTT in TBS. The supernatant was removed, and 1% Triton lysis buffer with 10 mM iodoacetamide was added to a volume of 1 mL. The supernatant was then immunoprecipitated with 5 μL of HA antibody and 20 μl of protein A/protein G beads for 1 hour at 4 °C. The beads were washed 3 times with RadioImmunoPrecipitation Assay (RIPA) buffer (1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate, 2mM EDTA, pH 7.2), boiled for 5 minutes in 1× denaturing buffer and treated with Endo H glycosidase as per the manufacturers protocol. The samples were then analyzed by 10% SDS-PAGE.
Trafficking rate assessments in HIV-infected cells
CEM cells were infected with HIV (NL-PIgagHXB) as previously described (Collins et al. 1998) and harvested 48 hrs after infection. Briefly, 15 × 106 cells per immunoprecipitation were pulse labeled for 30 minutes with [35S]-methionine and cysteine and chased for the indicated time. Immunoprecipitations were conducted as described above for uninfected cells.
Flow cytometric analyses
Cells were detached from plates using PBS/EDTA. 2×106 cells were washed three times with PBS containing 1% FBS. Cells were then incubated with anti-HA ascites (Covance Scientific) at a 1:250 dilution or with W6-32 or HC-10 antibodies for 1 hour on ice. Following this incubation, cells were washed three times with PBS + 1% FBS. Cells were then incubated for 1 hour on ice with FITC-conjugated goat anti-mouse IgG (Jackson Laboratories) at 1:250 dilution, followed by washing 3 times with PBS + 1% FBS. Cells were analyzed using a Becton FACS calibur cytometer and the cellquest software.
Peptide binding and thermostability assays
Thermostability assessments in insect and CEM cells
Thermostability assays were performed as described previously (Thammavongsa et al. 2006). Briefly, proteins in lysates from metabolically labeled insect cells were incubated at 4 °C or 37 °C for 12 minutes and then immunoprecipitated with the W6/32 antibody. Ratios of heavy chains recovered at 37 °C relative to 4 °C were quantified following SDS-PAGE. For thermostability assessments in CEM cells, a total of 4 × 107 CEM cells expressing HLA-B*3501 and HLA-B*3503 were incubated for 30 min in DMEM (lacking methionine/cysteine). Cells were metabolically labeled with 0.2 mCi [35S]methionine/cysteine for 10 min, lysed in 1% Triton X-100 lysis buffer (10 mM Tris, 10 mM phosphate, 130 mM NaCl (pH 7.5), and protease inhibitors) and split into two equal aliquots. One aliquot was incubated at 37°C or 50 °C for 12 min while the other was left on ice. Sequential immunoprecipitations were conducted with W6/32 (primary) and anti-HA ascites (secondary) as described. Following SDS-PAGE, H chain bands recovered under each condition were visualized by phosphorimaging analyses and quantified using ImageQuant.
Direct fluorescent peptide binding to HLA-B*35
Soluble HLA-B*35 heterodimers were purified from baculovirus-infected High-Five insect cell supernatants, using a W6/32 immunoaffinity column, as described previously for other class I molecules (Mancino et al. 2002). A fluorescent version of LPSSADVEF was obtained by cysteine substitution at position 4 of the sequence and labeling with iodoacetamidofluorescein as previously described (Arora et al. 2001). A fluorescent version of YPLHEQHGM was obtained by substituting the histidine residue at position 4 with a lysine residue labeled with 5-(and-6) carboxyfluorescein (C-1311) (Molecular Probes). The peptide was then synthesized using standard Fmoc Solid Phase Peptide Synthesis with the special lysine derivative incorporated and the labeled peptide purified using reverse phase HPLC. Proteins and the fluorescently labeled peptides were incubated under the indicated conditions in buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5), followed by native-polyacrylamide gel electrophoresis as described (Mancino et al. 2002; Rizvi et al. 2004). HLA-B*35/fluorescent peptide complexes were visualized by fluorescence imaging of the gels by using a FluorImager SI Fluorescence Scanner (Molecular Dynamics). Fluorescent bands were quantified and background subtracted using IMAGEQUANT software.
Effects of HLA-B alleles on HIV disease progression rates
Analyses of the effects of HLA on HIV disease progression and HLA typing were performed previously (Gao et al. 2001). Four AIDS-related outcomes were considered end points of survival analysis: a CD4 T-lymphocyte count of less than 200 per cubic millimeter, progression to AIDS according to the 1993 definitions of the Centers for Disease Control and Prevention (CDC), progression to AIDS according to the CDC’s more restrictive 1987 definition, and death from an AIDS-related cause. Data management and statistical analyses were performed with SAS 9.1 (SAS Institute, Cary, NC). PROC LIFETEST and PHREG were used for Cox model analyses.
RESULTS
ER export rates of HLA-B allotypes in CD4+ T cells
As described above, HLA-B allotypes have prognostic significance in HIV disease. To better understand the explanation for this observation, we sought to characterize the assembly and intracellular trafficking of a panel of relevant HLA-B allotypes in CD4+ T cells. To accomplish this goal, we created CD4+ CEM T cell lines expressing HA-tagged versions of each MHC-I allotype. The N-terminal HA-tag allowed the direct comparison of each HLA-B molecule of interest in a consistent manner and allowed them to be distinguished from endogenous MHC class I heavy chains. Previous studies have demonstrated that tagging HLA-A2 in this manner did not influence the intracellular trafficking of this molecule (Roeth et al. 2004). In analyses of surface expression we observed similar levels of cell surface expression of both HLA-B35 allotypes examined (Table I).
TABLE I. Comparison of stable class I molecules on the cell surface of CD4+ T cells.
CD4+ CEM cells expressing the indicated class I molecules were stained with HC-10, W6/32 or anti-HA antibodies and cell surface expression was measured by flow cytometric analyses. The mean fluorescence signals obtained with each antibody are indicated in columns 2–4. Mean fluorescence values from two independent FACS analyses are shown for all allotypes except HLA-B*4405 for which a single analysis was undertaken. The last two columns display HC-10/anti-HA and W6/32/anti HA mean fluorescence ratios.
| Anti-HA | W6/32 | HC-10 | W6/32/Anti-HA | HC-10/Anti-HA | |
|---|---|---|---|---|---|
| CEM | 11.830 10.660 |
182.550 263.580 |
14.970 15.440 |
20.075 +/− 4.650 | 1.377 +/− 0.112 |
| B*3501 | 391.740 515.660 |
1754.730 1192.000 |
63.050 49.730 |
3.396 +/− 1.084 | 0.129 +/− 0.033 |
| B*3503 | 310.790 487.560 |
1500.100 1002.820 |
26.830 44.490 |
3.442 +/− 1.385 | 0.089 +/− 0.002 |
| B*5701 | 243.870 223.960 |
1110.610 778.310 |
14.250 32.720 |
4.015 +/− 0.540 | 0.102 +/− 0.044 |
| B*4402 | 132.700 135.050 |
899.870 557.560 |
21.140 18.210 |
5.455 +/− 1.326 | 0.147 +/− 0.012 |
| B*4405 | 173.560 | 761.630 | 24.950 | 4.388 | 0.144 |
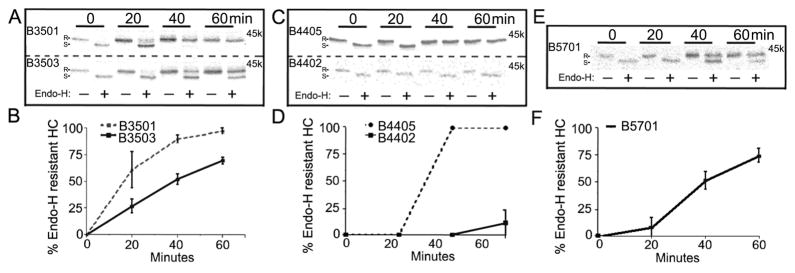
We assessed the rate of intracellular trafficking by pulse-chase and endo H digestion. Acquisition of resistance to endo H serves as an indication of migration into the medial Golgi apparatus where proteins are modified such that endo H is no longer active. Based on current models of the class I assembly pathway, the major steps involved in MHC class I assembly in the ER include (i) the formation of heterodimers (ii) peptide loading of heterodimers and (iii) dissociation of heterodimers from the TAP complex. The antibody W6/32 (Barnstable et al., 1978) which recognizes only assembled MHC class I heterodimers, was used to compare trafficking rates of HA-tagged molecules. HLA-B*3501(S116) and HLA-B*3503(F116) differ only at position 116 of their heavy chain sequences, as do HLA-B*4402(D116) and HLA-B*4405(Y116). Variations at position 116 are expected to impact peptide loading of heterodimers, and thus, pulse-chase comparisons with the W6/32 antibody allowed for comparisons of the rates of peptide loading within the two sets of closely-related allotypes.
Compared to HA-HLA-B*3501, a reproducible reduction in kinetics of HA-HLA-B*3503 ER exit was observed (Figs. 1A and 1B). The extent of the differences between HA-HLA-B*3501 and HA-HLA-B*3503 were smaller compared to that between HA-HLA-B*4402(D116) and HA-HLA-B*4405(Y116) (Fig. 1C and 1D). Among the five allotypes analyzed, HA-HLA-B*4402 and HA-HLA-B*4405 displayed the slowest and most rapid intracellular trafficking rates, respectively (Fig. 1). The trafficking rate of HA-HLA-B*4405 was closely matched with that of HA-HLA-B*3501 (Figs. 1A and 1B). HA-HLA-B*5701 and HA-HLA-B*3503 displayed intermediate rates of trafficking (Fig. 1A and 1E). The relative hierarchy of trafficking rates was found to be HA-HLA-B*4405=HA-HLA-B*3501>HA-HLA-B*5701=HA-HLA-B*3503>HA-HLA-B*4402.
FIG. 1.
Maturation rates of folded MHC class I heterodimers. CEM T cells were metabolically labeled for 10 minutes and chased for the indicated times in fresh media. Lysates were immunoprecipitated with mAb W6/32 followed by a secondary anti-HA immunoprecipitation then digested with Endo H or left undigested, and analyzed by SDS-PAGE and phosphorimaging analyses. Upper panels: Representative gels for indicated HA-HLA-B trafficking. Lower panels: Percent Endo H resistant heavy chain bands (Endo H resistant band/Endo H resistant + Endo H sensitive) were quantified from gels such as those shown in the upper panels. Data is the average of three independent measurements for HA-HLA-B*4402 and two independent measurements for HA-HLA-B*4405, HA-HLA-B*3501, HA-HLA-B*3503 and HA-HLA-B*5701. R indicates Endo H resistant MHC class I heavy chain band, and S indicates Endo H sensitive MHC class I heavy chain band.
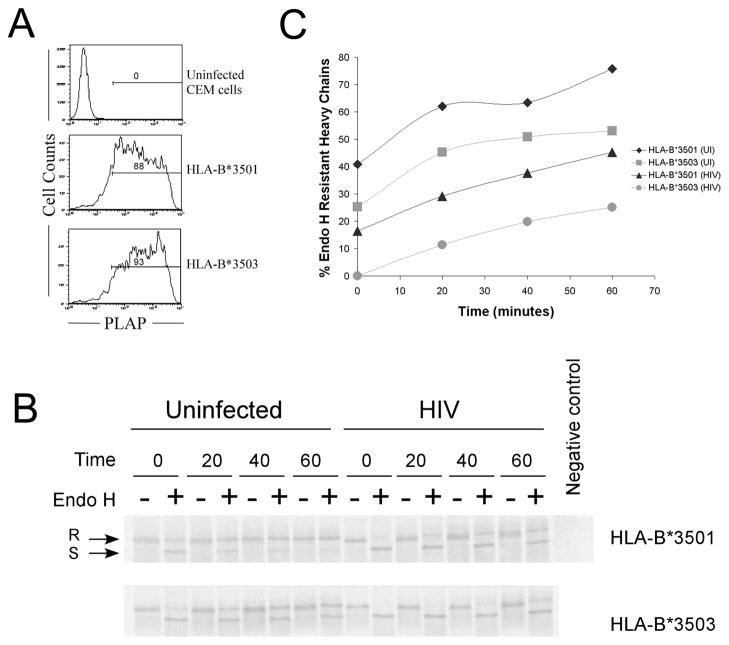
We asked whether the differences in the rate of transport of HLA-B*3501 and HLA-B*3503 observed in uninfected CEM cells (Fig. 1A) were also maintained in HIV-infected CEM cells. The transport rates of both HA-HLA-B*3501 and HA-HLA-B*3503 were found to be reduced in HIV-infected cells compared to uninfected cells (Fig. 2B and 2C; these experiments yielded lower signals and therefore used a longer pulse than those shown in Fig. 1, which resulted in higher levels of Endo H resistant molecules at the zero time point). Slowing of HLA-B*35 trafficking out of the ER in HIV-infected cells compared to uninfected cells may be related to previously described observations that HIV infection inhibits TAP activity and induces a reduction in rate of Endo H resistance acquisition of MHC class I molecules (as assessed by immunoprecipitation with W6/32 (Kutsch et al. 2002)). Despite the slowing of the overall HLA-B*35 transport rate that was induced by HIV infection, the differences in rates of transport between HA-HLA-B*3501 and HA-HLA-B*3503 were maintained in HIV infected cells as in uninfected cells (Fig. 2C).
FIG. 2.
Intracellular trafficking of HA-HLA-B*35 during HIV infection. (A) HIV infection: CEM cells expressing HA-HLA-B*3501 and HA-HLA-B*3503 were infected with HIV-1 encoding placental alkaline phosphatase (PLAP). Percentage infected cells are indicated. (B) CEM cells from part A or uninfected cells were pulse la(beled, chased for the indicated time in minutes, immunoprecipitated with W6/32, followed by a secondary immunoprecipitation with anti-HA, and then digested with Endo H or left undigested, and analyzed by SDS-PAGE and phosphorimaging analyses. (C) Percentage Endo H resistant heavy chains were quantified from the gels shown in the (B). Similar trends as in C were observed in a second experiment which had a lower percentage of infected cells (~50%).
Differences in steady-state TAP binding by HLA-B*3501 and HLA-B*3503
The slower rate of trafficking of HA-HLA-B*3503 compared to HA-HLA-B*3501 allowed detection of TAP1 complexes with HA-HLA-B*3503 (Fig. 3, lower panel, lanes 2 and 7). The slower rates of trafficking of HA-HLA-B*5701 and HA-HLA-B*4402 also allowed for the detection of TAP complexes with these allotypes (Fig. 3, lower panel, lanes 3 and 4). The amounts of each of these allotypes recovered in complex with TAP (Fig. 3, lower panel) correlated with the amounts of class I detected in the lysates (Fig. 3, upper panel). However, the allotypes that showed more rapid rates of ER export (HA-HLA-B*3501 and HA-HLA-B*4405) were not detectable in association with TAP at steady state (Fig. 3, lower panel, lanes 5 and 6). Thus, an overall trafficking rate resembling that of HA-HLA-B*3501 must be just above the threshold where visualizing TAP binding in the steady state is no longer possible.
FIG. 3.
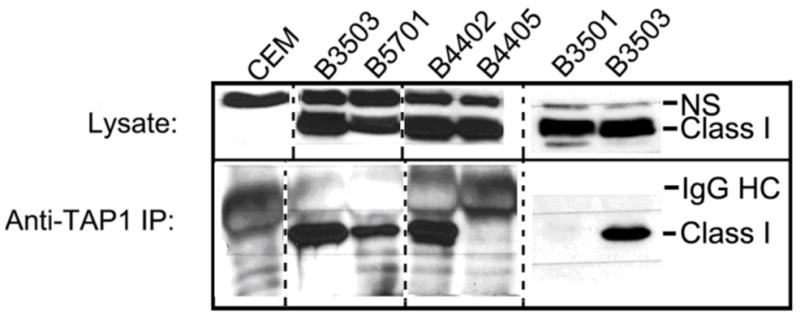
TAP binding profiles of HA-HLA-B allotypes in CEM cells. Lysates from CEM cells expressing the indicated class I were directly loaded (top panel), or proteins first immunoprecipitated with anti-TAP1 antisera (lower panel), separated by SDS-PAGE, and immunoblotting analyses undertaken with an anti-HA antibody. NS denotes a non-specific band. Data is representative of at least two independent sets of analyses for each class I. The dotted line indicates where different lanes from the immunoblots have been juxtaposed. The solid line indicates where different immunoblots were juxtaposed.
Similar tapasin-dependence profiles of HLA-B*3501 and HLA-B*3503
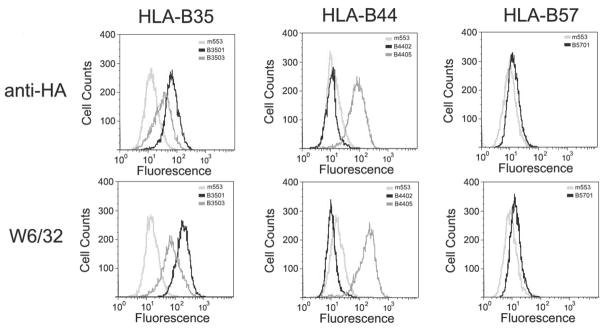
We next examined cell surface expression levels of various HLA-B alleles in the human tapasin-deficient melanoma cell line, M553 (Belicha-Villanueva et al., 2008). This cell line was chosen for analyses of tapasin dependence of the HLA-B assembly since tapasin’s effect on MHC class I assembly (rather than TAP stabilization) appears to be the predominant function of tapasin in this cell line (Belicha-Villanueva et al. 2008). Flow cytometric analyses were used to compare tapasin dependencies of the different HLA-B allotypes analyzed (Fig. 4). Following infection with each HLA-B-encoding virus and selection with neomycin, surface expression of each allotype in the surviving population of cells was assessed using flow cytometric analyses with an anti-HA antibody. In these analyses, surface expression of HA-HLA-B*4402 was essentially undetectable. By contrast, high levels of HA-HLA-B*4405 cell surface expression was observed in the absence of tapasin. These observations are consistent with the reported tapasin dependence and independence respectively, of HLA-B*4402 and HLA-B*4405 (Williams et al. 2002; Zernich et al. 2004). Significant levels of HA-HLA-B*3501 were detectable in the absence of tapasin, and HA-HLA-B*3503 was also detectable at significant levels in the absence of tapasin. Low levels of HA-HLA-B*5701 were detectable, consistent with the recent report of the low expression level of the endogenous HLA-B*5701 of M553 cells in the absence of tapasin (Belicha-Villanueva et al. 2008). Tapasin dependence as measured by the anti-HA antibody largely correlated with tapasin dependence assessed by the heterodimer-specific w6/32 antibody (Fig. 4). As recently reported for the endogenous HLA class I molecules of M553 cells (Belicha Villanueva et al. 2008), increasing TAP expression (by IFN-γ treatment) had only small effects on increasing HA-HLA-B expression in the absence of tapasin (data not shown).
FIG. 4.
Surface expression of indiacted HA-HLA-B allotypes in the tapasin deficient cell line M553. (A) M553 cells expressing the indicated HLA-B allotypes were stained with anti-HA (top panels) or with mAb W6/32 (lower panels) followed by FITC conjugated secondary antibody staining and analyzed by FACS.
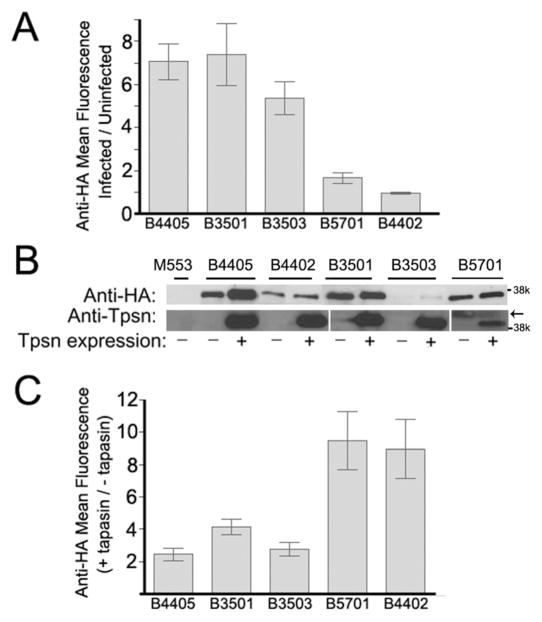
The mean fluorescence of cells infected with viruses encoding the indicated HA-tagged class I molecules were measured and plotted as a ratio relative to the mean fluorescence of the parent uninfected M553 cells (Fig. 5A), and representative immunoblots from one set of infected cells are shown in Fig. 5B. The immunoblotting analyses showed different steady-state levels of HLA-B allotypes, with the level of expression of HA-HLA-B*3503 being the lowest of all five allotypes compared (Fig. 5B). Despite the lower expression of HA-HLA-B*3503 compared to HA-HLA-B*4402 and HA-HLA-B*5701, higher levels of cell surface HA-HLA-B*3503 was detectable, indicating that HA-HLA-B*3503 is relatively tapasin-independent for assembly and cell surface expression. The intrinsic assembly efficiency of HA-HLA-B*3503 must be higher than those of HA-HLA-B*4402 and HA-HLA-B*5701, which likely accounts for higher tapasin-independent surface expression phenotype of HA-HLA-B*3503. HA-HLA-B*3503 was expressed at slightly lower levels on the surface of tapasin-deficient cells compared to HA-HLA-B*3501 (Fig. 4 and 5A). Total HA-HLA-B*3503 expression in lysates was also lower compared to HA-HLA-B*3501 (Fig. 5B). This result was observed in three separate infections of M553 cells with viruses encoding HA-HLA-B*3501 and HA-HLA-B*3503, raising the possibility that the observed differences in steady-state expression levels reflect intrinsic properties of the proteins. Together these findings indicate that HLA-B*3501 and HLA-B*3503 are both relatively tapasin-independent for their cell surface expression.
FIG. 5.
Tapasin dependence of the cell surface expression of HA-HLA-B allotypes. (A and C) M553 cells expressing the indicated class I molecules alone, co-expressing both tapasin and the indicated class I molecule or uninfected M553 control cells were stained with anti-HA and FITC-conjugated secondary antibodies followed by flow cytometric analyses to assess MHC class I cell surface expression. (A) Cell surface expression of indicated class I in the absence of tapasin were represented as ratios of mean fluorescence values relative to the parent uninfected M553 cells (following staining with anti-HA and FACS analyses). Data are the average of 8 independent analyses for HA-HLA-B*4405 from two separate infections, 5 independent analyses of a single infection for HA-HLA-B*4402, 8 independent analyses of three separate infections for HA-HLA-B*3501 and HA-HLA-B*3503, and 5 separate analyses from a single infection for HA-HLA-B*5701. (B) Cells used for the analyses in panels A and C were lysed and immunoblotting analyses was undertaken with anti-HA in the top panel and anti-tapasin in the lower panel. For the anti-HA immunoblots all cells were compared in the same set of blots; for the anti-tapasin analyses, separate blots are indicated by spaces. The arrow indicates the location of a non-specific band. (C) Tapasin induced expression of the indicated class I were represented as ratios of the mean fluorescence of anti-HA staining in the presence of tapasin/anti-HA staining in the absence of tapasin. Data are the average of 5 independent analyses from two separate infections for HA-HA-HLA-B*4405, HA-HLA-B*4402, HA-HLA-B*3501, and HA-HLA-B*3503, and two independent analyses from two separate infections for HA-HLA-B*5701. In A and C, error bars represent the standard error of the mean ratios.
M553 cells expressing the different HLA-B allotypes were next infected with a tapasin-encoding retrovirus that conferred puromycin resistance. Following selection with puromycin, total and surface class I expression was assessed by immunoblotting and FACS analyses, respectively, with anti-HA (Fig. 5B and 5C), and tapasin expression was verified by immunoblotting analyses (Fig. 5B). In general, higher levels of each class I were detectable in cell lysates in the presence of tapasin, indicating that tapasin influenced the assembly/stability of all allotypes to some extent (Fig. 5B). For a given allotype, the extent of tapasin-induced surface expression was highly dependent on tapasin expression levels (data not shown), as reported in other tapasin-deficient cells (Everett and Edidin 2007); thus, relative surface expression in the absence of tapasin (Fig. 4 and 5A) provided a better measure of tapasin dependence than relative tapasin-mediated induction (Fig. 5C). In general, however, at comparable tapasin concentrations, the smallest tapasin-induced enhancement was observed for allotypes that were most highly expressed in the absence of tapasin (HA-HLA-B*4405, HA-HLA-B*3501, and HA-HLA-B*3503), and the most significant enhancements were observed for those allotypes that were poorly expressed in the absence of tapasin (HA-HLA-B*5701 and HA-HLA-B*4402). However, some degree of tapasin-induced surface expression was observed for all the HLA-B allotypes that were analyzed.
Soluble forms of HLA-B*3501 and HLA-B*3503 bind to several peptides with similar intrinsic efficiencies
The data shown in Figures 1–2 indicated differences in rates of trafficking between HA-HLA-B*3501 and HA-HLA-B*3503 both in uninfected and HIV-1-infected CEM cells. To further understand whether those differences may reflect differences in the intrinsic efficiency of peptide loading as was observed for HLA-B*4402 and HLA-B*4405, we examined differences in peptide loading between soluble forms of the two allotypes in insect cells, or as purified proteins. Insects lack an adaptive immune response, and insect cells therefore do not express components of the MHC class I antigen processing machinery, including TAP and tapasin. Thus, MHC class I molecules expressed in insect cells are largely peptide-deficient, allowing for comparisons of intrinsic efficiencies of loading of closely-related allotypes, as previously described (Thammavongsa et al. 2006). To broadly compare efficiencies of loading of a large number of peptides by HLA-B*3501 and HLA-B*3503, we synthesized two random nonamer peptide libraries; one which was completely random at each position (xxxxxxxxx) and the other containing a proline at position 2, which is critical for binding to both HLA-B*3501 and HLA-B*3503 (xPxxxxxxx) (Falk et al. 1993; Steinle et al. 1996). A thermostability assay was first used to compare the abilities of soluble HLA-B*3501 and HLA-B*3503 to bind the peptides (Fig. 6A and B). The levels of heavy chain stabilization in the presence of the completely random peptide (xxxxxxxxx) were similar for both HLA-B*3501 and HLA-B*3503 at all of the peptide concentrations tested, with higher thermostability being observed at higher concentrations of exogenous peptide (Fig. 6A). As expected, compared to the completely random peptide library, the random peptide library with proline at position 2 induced enhanced stabilization of both HLA-B*3501 and HLA-B*3503 (Fig. 6B). However, no significant differences in stabilization conferred to HLA-B*3501 and HLA-B*3503 were observed. This result is in contrast to results obtained with similar experiments undertaken with HLA-B*4402 and HLA-B*4405, which revealed significant differences in stabilization efficiencies of those two allotypes by appropriate random peptide libraries in thermostability assays undertaken at 37 °C (Thammavongsa et al. 2006).
FIG. 6.
Intrinsic peptide loading efficiencies of soluble HLA-B*3501 and HLA-B*3503 in insect cells or as purified proteins. Sf21 cells were infected with baculoviruses encoding the indicated soluble HLA-B*35/β2m constructs, metabolically labeled, and thermostability analyses conducted in the absence (−) or presence of (A) a randomized nonamer library at the indicated concentrations, (B) randomized nonamer library with a proline at position 2 at the indicated concentrations, (C) randomized 11mer library with a proline at position 2 and methionine at position 11 at the indicated concentrations, (D) 0, 10, or 100 μM LPSSADEVF as indicated, (E) 0, 10 or 100 μM LPSSADEVFCL as indicated, (F) 0 or 100 μM YPLHEQHGM, or YFLHEQHGM as indicated. Lysates were incubated at 4° or 37° degrees for 12 minutes, followed by immunoprecipitation analyses with w6/32. Samples were separated by SDS-PAGE and proteins visualized by phosphorimaging analyses. (Upper panels) Representative images of heavy chains recovered at 4 °C and 37 °C in the absence or presence of peptide, following SDS-PAGE. (Lower panels) Quantifications of ratios of heavy chains recovered at 37 °C relative to 4°C under the indicated condition. Error bars represent the standard error of the mean (SEM) percentages. Data are the average of 3, 2, 4, 2, 2, and 2 experiments respectively for (A–F). Purified HLA-B*35 (2 μM) and (G) LPSCFADVEF (0.2 μM) or (H) YPLKFEQHGM (H; 0.2 μM) or (I and J) XPXKFXXXXM at the indicated concentrations were incubated at 37 °C for (I) 1 or (J) 2 hr. Mixtures were separated by native-PAGE and protein-peptide complexes were quantified by fluorimaging analyses of gels. Representative fluorescent scans of native gels are shown in the upper panels and lower panels display quantifications of fluorescent bands as percentage fluorescence relative to the maximum fluorescence within each experiment. Data is the average of two independent analyses and the error bars represent the standard error of the mean percentage fluorescence.
Recent reports have shown that HLA-B*35 molecules are able to bind peptides of up to 14 amino acids in length (Green et al. 2004; Tynan et al. 2005). The extended peptide has been shown to be bound to HLA-B*35 in a distinct conformation; with the amino and carboxyl termini of the peptide anchored into the peptide binding groove and the additional amino acids creating an outward “bulge” in the center of the peptide (Tynan et al. 2005). We thus examined whether peptides with extended lengths were also able to bind to HLA-B*3503, and whether there were differences in the abilities of HLA-B*3501 and HLA-B*3503 to recognize long peptide sequences. Such a difference could significantly impact the number of peptides that are able to bind to each allotype, and influence peptide repertoires during infection. We synthesized an 11-mer peptide containing a proline at position 2 of the peptide. This peptide also contained a fixed methionine at the C-terminus, as a C-terminal methionine appears to be a preferential anchor for both HLA-B*3501 and HLA-B*3503 (Steinle et al. 1996). When the thermostabilities of HLA-B*3501 and HLA-B*3503 were analyzed in the presence of this 11-mer peptide library (xPxxxxxxxxM), thermostability of HLA-B*3503 was enhanced to levels similar to that of HLA-B*3501 at two peptide concentrations tested (Fig. 6C). Thus, similar to HLA-B*3501, HLA-B*3503 is capable of efficiently loading long peptides.
To further examine loading of specific peptides onto HLA-B*3501 and HLA-B*3503, three known HLA-B*35 specific peptides (LPSSADVEF, LPSSADVEFCL and YPLHEQHGM) were used in the thermostability assays. These peptide sequences have been shown to elicit HLA-B*3501 and HLA-B*3503 specific T cell responses (Lee et al. 1995; Morel et al. 1999). Compared to the random peptides, these specific peptides were more efficient stabilizers of HLA-B*35, with slightly higher thermostability conferred to sluble HLA-B*3501 compared to HLA-B*3503 (Fig. 6D–F). The importance of a proline residue at position 2 of the peptide is exemplified in Figure 6F, as the peptide YFLHEQHGM (P→F at position 2) does not stabilize either HLA-B*3501 or HLA-B*3503 (compare stabilization of HLA-B*35 in the presence of YPLHEQHGM vs YFLHEQHGM).
To more precisely examine differences in the intrinsic kinetics of peptide loading onto HLA-B*3501 and HLA-B*3503, we examined binding of fluorescently labeled versions of LPSCFADVEF, YPLKFEQHGM, as well as a random xPxKFxxxxM library to purified soluble forms of HLA-B*3501 and HLA-B*3503. As with the thermostability measurements, no significant differences in the intrinsic efficiency of loading of these peptides to HLA-B*3501 and HLA-B*3503 were apparent with the fluorescent peptide-based binding experiments, after normalizing for total protein amounts. Thermostability assays were, however, sufficient to detect intrinsic peptide loading efficiency differences between HLA-B*4405 and HLA-B*4402 (Thammavongsa et al. 2006), consistent with the more dramatic differences in trafficking rates between HLA-B*4402 and HLA-B*4405 compared to those between HLA-B*3501 and HLA-B*3503 (Fig. 1).
Thermostability differences between HLA-B*3501 and HLA-B*3503 in CEM cells suggest a reduced ER peptide repertoire for HLA-B*3503
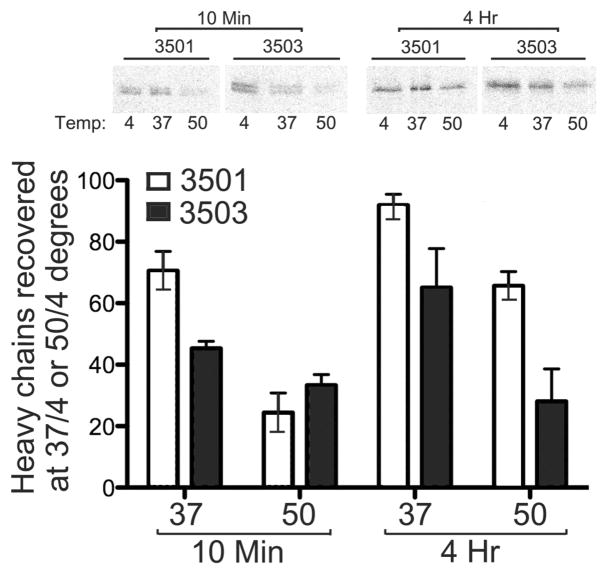
The results described in Figure 6 suggested that there were no differences in the intrinsic kinetics of loading for a large number of peptides of common specificities for both allotypes. There are, however, known specificity differences between the two allotypes (Steinle et al. 1996), and an over-representation of HLA-B*3501-specific peptides in the ER could account for the observed differences in rates of trafficking between HA-HLA-B*3501 and HA-HLA-B*3503. While quantitative differences in the endogenous peptide concentrations are hard to measure directly, thermostability differences between the two allotypes could be reflective of variations in peptide availability. To investigate whether such differences were observable, thermostability analyses were undertaken in CEM/B35 cells, by primary and secondary immunoprecipitations with w6/32 and anti-HA respectively, following heating of the lysates to 37 °C or 50 °C, and quantifying percent heated/untreated heavy chains recovered for each allotype over multiple independent analyses. As shown in Figure 7, higher levels of thermostable HA-HLA-B*3501 heavy chains were observed at early (10 minutes) and late (4 hour) time points post-syntheses, when lysates were heated to 37 °C. While the amount of thermostable heavy chains increased for both allotypes at the late time-point, differences in levels of thermostable heavy chains between HA-HLA-B*3501 and HA-HLA-B*3503 were maintained even at the late time-point. When these analyses were undertaken following incubations of lysates at 50 °C, no significant differences in thermostable heavy chains were observable at the early time point when neither repertoire is expected to be completely optimized, whereas significant differences between HA-HLA-B*3501 and HA-HLA-B*3503 were observed at the late time-point. Taken together with the analyses of Figure 6, these findings are consistent with the possibility that an increase in peptide availability for HLA-B*3501 accounts for the faster rate of trafficking of HLA-B*3501 compared to HLA-B*3503 in CEM cells.
FIG. 7.
Peptide loading efficiencies of HA-HLA-B*3501 and HA-HLA-B*3503 in CEM cells: Upper panel: CEM cells expressing HA-HLA-B*3501 or HA-HLA-B*3503 were metabolically labeled for 10 minutes and, lysed immediately or chased with fresh media for 4 hours, and lysates incubated at 4°, 37°, or 50° degrees for 12 minutes, followed by sequential immunoprecipitations with primary W6/32 and secondary anti-HA antibodies. Samples were separated by SDS-PAGE and proteins visualized by phosphorimaging analyses. Lower panel represents the quantification of percentage heavy chains recovered at 37 °C or 50 °C relative to 4 °C. Data are averages of 2 independent analyses with error bars representing the standard error of the mean.
DISCUSSION
The CTL response against HIV disease is important for controlling viremia. Recent epidemiological data suggests that antigen presentation of CTL epitopes by different HLA-B allotypes is an important determinant of disease prognosis. In this study we characterized the relative rates of intracellular trafficking and assembly of a panel of HLA-B allotypes that have prognostic significance in HIV-infected people. The trafficking profiles of the allotypes investigated in this study can be classified as (a) Group A (HA-HLA-B*4405 and HA-HLA-B*3501), with rapid assembly rates, undetectable TAP binding in the steady-state, and tapasin-independent surface expression (b) Group B (HA-HLA-B*5701 and HA-HLA-B*3503), with intermediate trafficking rates, detectable TAP binding in the steady state, and variable tapasin dependencies and (c) Group C (HA-HLA-B*4402), the slow-assembling, highly tapasin-dependent allotype, with readily detectable TAP binding in the steady state. Thus, observations in the literature relating to TAP association, tapasin dependence, and rates of ER exit appear, to some extent, to be interrelated. Either the formation of heterodimers, the peptide loading of heterodimers or the availability of peptides could be a rate-limiting step in the assembly of HLA-B molecules that display slow intrinsic assembly rates. HLA-B molecules with slow intrinsic assembly rates are in prolonged ER residence, thus making TAP interactions with such allotypes also detectable in the steady state. Rapid rate of trafficking of HA-HLA-B*4405 and HA-HLA-B*3501 correlated with tapasin independence, and the slowest trafficking allotype HA-HLA-B*4402 was strongly tapasin-dependent (Figs. 4A and 5), consistent with tapasin’s assembly-promoting functions (Chun et al. 2001; Everett and Edidin 2007; Lehner et al. 1998; Lybarger et al. 2001; Zarling et al. 2003). The tapasin dependence of allotypes with the intermediate rates of trafficking were harder to predict, with HA-HLA-B*5701 being more strongly tapasin-dependent than HA-HLA-B*3503 (Figs 4 and 5A). It is possible that HA-HLA-B*5701 has a slower intrinsic rate of peptide loading compared to HA-HLA-B*3503, which results in a stronger tapasin dependence of HLA-B*5701. On the other hand, the rate-limiting step in HA-HLA-B*3503 assembly may relate to peptide availability rather than reduced intrinsic kinetics of peptide loading (Figs. 6 and 7), making HLA-B*3503 relatively tapasin-independent for its assembly. Indeed, the tapasin dependencies of HA-HLA-B*3501 and HA-HLA-B*3503 showed only small differences (Figs 4 and 5A), consistent with similarities in their in intrinsic peptide loading efficiencies (Fig 6).
It has been shown that HLA-B*4405+ virus carriers showed a high frequency of CTL precursors for an Epstein Barr virus (EBV) epitope, whereas HLA-B*4402+ virus carriers consistently showed a weaker response. HLA-B*4405+ target cells were recognized more efficiently than HLA-B*4402+ target cells, regardless of whether the CTLs were derived from HLA-B*4405+ individuals or from HLA-B*4402+ individuals (Khanna et al. 1997). Thus, allotypes such as HLA-B*4405 that assemble and exit the ER more rapidly, may be advantageous for the CTL response. To further understand possible mechanisms that could underlie differential AIDS progression rates associated with HLA-B*3501 and HLA-B*3503, we compared assembly, trafficking and peptide loading of these two allotypes.
In CEM cells, the maturation and transport of HA-HLA-B*3503 was consistently slower relative to HA-HLA-B*3501 (Fig. 1–2). Consistent with the slower maturation rate, enhanced association of HA-HLA-B*3503 with the TAP transporter was also observed in the CEM T cell line (Fig. 3), as previously observed in PBLs (Neisig et al. 1996). The slower rate of trafficking did not correlate with significantly reduced rates of intrinsic in vitro peptide loading of HLA-B*3503 compared to HLA-B*3501, with epitopes and peptide libraries that were common to both allotypes, indicating comparable kinetics of assembly with epitopes that are common to both allotypes (Fig. 6). However, peptide loading of HA-HLA-B*3503 was reduced compared to that of HA-HLA-B*3501 in CEM cells (Fig. 7). It is likely that the known specificity differences between HLA-B*3501 and HLA-B*3503 (Falk et al. 1993; Steinle et al. 1996) contribute to an over-representation of HLA-B*3501 specific peptides in the ER, which in turn could contribute to the observed intracellular trafficking differences between HLA-*3501 and HLA-B*3503. HLA-B*3503 is associated with rapid AIDS progression for all AIDS outcomes (absolute CD4 T cell count, mortality, and the CDC classification of AIDS in 1987 and 1993), whereas HLA-B*3501 is associated with rapid AIDS progression only for the AIDS 1993 outcome ((Fig. 8) and (Gao et al. 2001)). If the HIV peptide repertoire of HLA-B*3503 in the ER is inherently more restrictive than that of HLA-B*3501 by the exclusion of all epitopes containing C-terminal tyrosines, a reduced diversity of the HLA-B*3503-specific CTL response could contribute to the stronger association of HLA-B*3503 with rapid AIDS progression. Indeed the presence of a C-terminal tyrosine on a peptide, a peptide sequence permissive for HLA-B*3501 but not HLA-B*3503, is strongly favored for transport by TAP (Uebel et al. 1997).
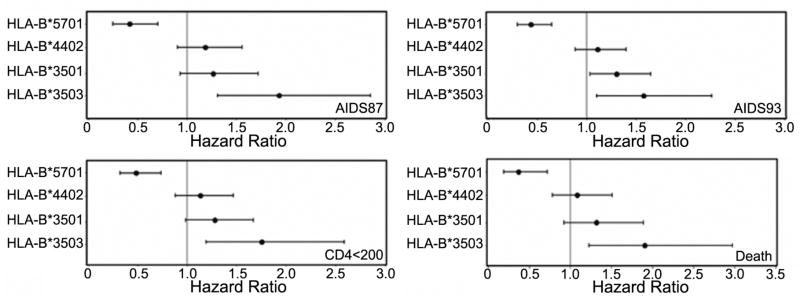
FIG. 8.
Effect of HLA Genotypes (HLA-B*5701, HLA-B*3501, HLA-B*3503 and HLA-B*4402) on progression to four AIDS-Related End Points. For each indicated outcome, relative hazards (HR), confidence intervals (CI), and significance were calculated for a dominant model as previously described (Gao et al. 2001; O’Brien et al. 2001). The ranges in sample size and p values were as follows: B*5701 (N = 75–78), p = 0.00003–0.003 for the four outcomes indicated (significantly associated with slow AIDS progression for all AIDS outcomes); B*4402 (N = 128–137), p = 0.22–0.62 for the four outcomes indicated; B*3501 (N = 117–123), p = 0.07 (CD4<200), 0.13 (AIDS 87), 0.03 (AIDS 93) and 0.13 (Death) (significantly associated with rapid AIDS progression only by the AIDS 93 criteria); B*3503 (N = 40–43) p = 0.004 (CD4<200), 0.001 (AIDS 87), 0.01 (AIDS 93) and 0.004 (Death) (significantly associated with rapid AIDS progression for all four AIDS outcomes). HLA-B*4405 is a very rare allele in the study cohort, and its effect on AIDS progression could not be studied.
A ranking of relative hazard ratios for AIDS progression for HLA-B allotypes considered in this study was HLA-B*3503>HLA-B*3501>HLA-B*4402>HLA-B*5701 (Fig. 8 for high resolution HLA typing and (O’Brien et al. 2001) for low-resolution typing); whereas the ranking for intracellular trafficking rates was HLA-B*3501>HLA-B*3503=HLA-B*5701>HLA-B*4402 (Fig. 1), indicating no correlations between the two sets of measurements. HLA-B*3503, which is associated with more rapid AIDS progression matures with an efficiency similar to that of HLA-B*5701, a highly protective allotype. Additionally, HLA-B*3503 is not expected to sequester the TAP complex components more significantly than HLA-B*5701 (Figures 3–5). Slow tapasin-dependent assembly such as that observed with HLA-B*5701 may confer a protective advantage by allowing sufficient ER residence time for pathogen-specific epitopes to be consistently sampled by MHC class I and presented to CTL. Conversely, rapid tapasin-independent trafficking could result in inconsistent loading/presentation of some pathogen-specific epitopes. In line with this possibility, a recent study by Altfeld et al described that CD8+ T cell responses to particular HIV epitopes presented by HLA-B*57 were observed at high frequencies in a study cohort of HIV-infected individuals who were HLA-B*57-positive (Altfeld et al. 2006). In the same study, the frequencies of responses to several HLA-B*35-specific HIV epitopes were low, and of the tested epitopes, a large number are expected to be specific for HLA-B*35(PY), due to the presence of a C-terminal tyrosine. The advantage of the slower trafficking of HLA-B35(Px) group may be overridden by a more restrictive peptide HIV-specific peptide repertoire of the HLA-B35(Px) group. That not every slow-trafficking tapasin-dependent allotype is protective is also exemplified by the association of the slowest-trafficking allotype (HLA-B*4402) with normal AIDS progression (Fig. 8 for high resolution HLA typing and (O’Brien et al. 2001) for low-resolution typing); protection undoubtedly also correlates with the availability of optimal pathogen-derived peptides specific for a given MHC class I molecule, and it is also possible that beyond a threshold, high assembly inefficiency results in inefficient CD8+ T cell responses.
Acknowledgments
This work was supported by an NIH grant (AI44155) to MR and an American Heart Association pre-doctoral fellowship to VT. The authors wish to thank Dr. Matt Androlewicz for anti-TAP1 antisera. We also thank the University of Michigan Biomedical Research core facilities for peptide synthesis and purification. We thank Dr. Syed Monem Rizvi for assistance with Figure 6G–6J.
Footnotes
Publisher's Disclaimer: DISCLAIMER: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, Yu XG, Meier A, Rockstroh JK, Allen TM, Jessen H, Rosenberg ES, Carrington M, Walker BD. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androlewicz MJ, Ortmann B, van Endert PM, Spies T, Cresswell P. Characteristics of peptide and major histocompatibility complex class I/beta 2-microglobulin binding to the transporters associated with antigen processing (TAP1 and TAP2) Proc Natl Acad Sci U S A. 1994;91:12716–20. doi: 10.1073/pnas.91.26.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Lapinski PE, Raghavan M. Use of chimeric proteins to investigate the role of transporter associated with antigen processing (TAP) structural domains in peptide binding and translocation. Proc Natl Acad Sci U S A. 2001;98:7241–6. doi: 10.1073/pnas.131132198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, Bangia N. Differential contribution of TAP and tapasin to HLA class I antigen expression. Immunology. 2008;124:112–120. doi: 10.1111/j.1365-2567.2007.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–12. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- Chun T, Grandea AG, 3rd, Lybarger L, Forman J, Van Kaer L, Wang CR. Functional roles of TAP and tapasin in the assembly of M3-N-formylated peptide complexes. J Immunol. 2001;167:1507–14. doi: 10.4049/jimmunol.167.3.1507. [DOI] [PubMed] [Google Scholar]
- Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- Everett MW, Edidin M. Tapasin increases efficiency of MHC I assembly in the endoplasmic reticulum but does not affect MHC I stability at the cell surface. J Immunol. 2007;179:7646–52. doi: 10.4049/jimmunol.179.11.7646. [DOI] [PubMed] [Google Scholar]
- Falk K, Rotzschke O, Grahovac B, Schendel D, Stevanovic S, Jung G, Rammensee HG. Peptide motifs of HLA-B35 and -B37 molecules. Immunogenetics. 1993;38:161–2. doi: 10.1007/BF00190906. [DOI] [PubMed] [Google Scholar]
- Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, O’Brien SJ, Carrington M. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- Goodall JC, Ellis L, Hill Gaston JS. Spondylarthritis-associated and non-spondylarthritis-associated B27 subtypes differ in their dependence upon tapasin for surface expression and their incorporation into the peptide loading complex. Arthritis Rheum. 2006;54:138–47. doi: 10.1002/art.21552. [DOI] [PubMed] [Google Scholar]
- Green KJ, Miles JJ, Tellam J, van Zuylen WJ, Connolly G, Burrows SR. Potent T cell response to a class I-binding 13-mer viral epitope and the influence of HLA micropolymorphism in controlling epitope length. Eur J Immunol. 2004;34:2510–9. doi: 10.1002/eji.200425193. [DOI] [PubMed] [Google Scholar]
- Itescu S, Mathur-Wagh U, Skovron ML, Brancato LJ, Marmor M, Zeleniuch-Jacquotte A, Winchester R. HLA-B35 is associated with accelerated progression to AIDS. J Acquir Immune Defic Syndr. 1992;5:37–45. [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O’Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kasper MR, Collins KL. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J Virol. 2003;77:3041–9. doi: 10.1128/JVI.77.5.3041-3049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Burrows SR, Neisig A, Neefjes J, Moss DJ, Silins SL. Hierarchy of Epstein-Barr virus-specific cytotoxic T-cell responses in individuals carrying different subtypes of an HLA allele: implications for epitope-based antiviral vaccines. J Virol. 1997;71:7429–35. doi: 10.1128/jvi.71.10.7429-7435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kutsch O, Vey T, Kerkau T, Hunig T, Schimpl A. HIV type 1 abrogates TAP-mediated transport of antigenic peptides presented by MHC class I. Transporter associated with antigen presentation. AIDS Res Hum Retroviruses. 2002;18:1319–25. doi: 10.1089/088922202320886361. [DOI] [PubMed] [Google Scholar]
- Lee SP, Morgan S, Skinner J, Thomas WA, Jones SR, Sutton J, Khanna R, Whittle HC, Rickinson AB. Epstein-Barr virus isolates with the major HLA B35.01-restricted cytotoxic T lymphocyte epitope are prevalent in a highly B35.01-positive African population. Eur J Immunol. 1995;25:102–10. doi: 10.1002/eji.1830250119. [DOI] [PubMed] [Google Scholar]
- Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line. 220. Immunity. 1998;8:221–31. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- Lybarger L, Yu YY, Chun T, Wang CR, Grandea AG, 3rd, Van Kaer L, Hansen TH. Tapasin enhances peptide-induced expression of H2-M3 molecules, but is not required for the retention of open conformers. J Immunol. 2001;167:2097–105. doi: 10.4049/jimmunol.167.4.2097. [DOI] [PubMed] [Google Scholar]
- Mancino L, Rizvi SM, Lapinski PE, Raghavan M. Calreticulin recognizes misfolded HLA-A2 heavy chains. Proc Natl Acad Sci U S A. 2002;99:5931–6. doi: 10.1073/pnas.092031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel S, Ooms A, Van Pel A, Wolfel T, Brichard VG, van der Bruggen P, Van den Eynde BJ, Degiovanni G. A tyrosinase peptide presented by HLA-B35 is recognized on a human melanoma by autologous cytotoxic T lymphocytes. Int J Cancer. 1999;83:755–9. doi: 10.1002/(sici)1097-0215(19991210)83:6<755::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Neisig A, Wubbolts R, Zang X, Melief C, Neefjes J. Allele-specific differences in the interaction of MHC class I molecules with transporters associated with antigen processing. J Immunol. 1996;156:3196–206. [PubMed] [Google Scholar]
- O’Brien SJ, Gao X, Carrington M. HLA and AIDS: a cautionary tale. Trends Mol Med. 2001;7:379–81. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–6. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SM, Mancino L, Thammavongsa V, Cantley RL, Raghavan M. A polypeptide binding conformation of calreticulin is induced by heat shock, calcium depletion, or by deletion of the C-terminal acidic region. Mol Cell. 2004;15:913–23. doi: 10.1016/j.molcel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Roeth JF, Williams M, Kasper MR, Filzen TM, Collins KL. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J Cell Biol. 2004;167:903–13. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- Smith KJ, Reid SW, Stuart DI, McMichael AJ, Jones EY, Bell JI. An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity. 1996;4:203–13. doi: 10.1016/s1074-7613(00)80429-x. [DOI] [PubMed] [Google Scholar]
- Stam NJ, Vroom TM, Peters PJ, Pastoors EB, Ploegh HL. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int Immunol. 1990;2:113–25. doi: 10.1093/intimm/2.2.113. [DOI] [PubMed] [Google Scholar]
- Steinle A, Falk K, Rotzschke O, Gnau V, Stevanovic S, Jung G, Schendel DJ, Rammensee HG. Motif of HLA-B*3503 peptide ligands. Immunogenetics. 1996;43:105–7. doi: 10.1007/BF00186615. [DOI] [PubMed] [Google Scholar]
- Thammavongsa V, Raghuraman G, Filzen TM, Collins KL, Raghavan M. HLA-B44 polymorphisms at position 116 of the heavy chain influence TAP complex binding via an effect on peptide occupancy. J Immunol. 2006;177:3150–61. doi: 10.4049/jimmunol.177.5.3150. [DOI] [PubMed] [Google Scholar]
- Turnquist HR, Thomas HJ, Prilliman KR, Lutz CT, Hildebrand WH, Solheim JC. HLA-B polymorphism affects interactions with multiple endoplasmic reticulum proteins. Eur J Immunol. 2000;30:3021–8. doi: 10.1002/1521-4141(200010)30:10<3021::AID-IMMU3021>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Tynan FE, Borg NA, Miles JJ, Beddoe T, El-Hassen D, Silins SL, van Zuylen WJ, Purcell AW, Kjer-Nielsen L, McCluskey J, Burrows SR, Rossjohn J. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. 2005;280:23900–9. doi: 10.1074/jbc.M503060200. [DOI] [PubMed] [Google Scholar]
- Uebel S, Kraas W, Kienle S, Wiesmuller KH, Jung G, Tampe R. Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc Natl Acad Sci U S A. 1997;94:8976–81. doi: 10.1073/pnas.94.17.8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–8. doi: 10.1016/s1074-7613(00)80103-x.Retracted in Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Immunity. 2009 Apr;30(4):611. doi: 10.1016/j.immuni.2009.04.002.
- Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–20. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- Zarling AL, Luckey CJ, Marto JA, White FM, Brame CJ, Evans AM, Lehner PJ, Cresswell P, Shabanowitz J, Hunt DF, Engelhard VH. Tapasin is a facilitator, not an editor, of class I MHC peptide binding. J Immunol. 2003;171:5287–95. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, Tait BD, Holdsworth R, Brooks AG, Bottomley SP, Beddoe T, Peh CA, Rossjohn J, McCluskey J. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 2004;200:13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]