Abstract
Development of cisplatin resistance in cancer cells appears to be a consequence of multiple epigenetic alterations in genes involved in DNA damage repair, proto-oncogenes, apoptosis, transporters, transcription factors, etc. In this work, we found that expression of the hypothetical transmembrane protein TMEM205 (previously known as MBC3205) is associated with cisplatin resistance. TMEM205 was first detected by functional cloning from a retroviral cDNA library made from human cisplatin-resistant (CP-r) cells. TMEM205 is predicted to be a transmembrane protein, but its expression, localization, and function have not previously been investigated. A polyclonal antibody directed to the TMEM205 protein was raised in our laboratory. Using this antibody, it was demonstrated that this protein is located at the cell surface. Its expression is increased in our cisplatin-selected CP-r cell lines, as demonstrated by immunoblotting, confocal examination and immuno-electron microscopy. Stable transfection of the TMEM205 gene confers resistance to cisplatin by approximately 2.5-fold. Uptake assays with Alexa Fluor-cisplatin showed reduced accumulation in CP-r KB-CP.3 and KB-CP.5 cells, and in TMEM205-transfected cells. Analysis of TMEM205 expression profiles in normal human tissues indicates a differential expression pattern with higher expression levels in the liver, pancreas, and adrenal glands. These results indicate that a novel mechanism for cisplatin resistance is mediated by TMEM205, and also suggest that overexpression of TMEM205 in CP-r cells may be valuable as a biomarker or target in cancer chemotherapy.
Keywords: TMEM205, cisplatin resistance
Introduction
Cisplatin (cis-Diamminedichloroplatinum II) revolutionized chemotherapy by improving treatment of a broad spectrum of solid tumors, and by facilitating the cure of metastatic testicular germ-cell cancer. However, despite the high efficacy of the compound, the ability of cancer cells to become resistant to the drug remains a significant impediment to successful chemotherapy. Intensive efforts have been made through biochemical characterization, cellular, and genetic approaches to determine the basis of resistance and define genes that are involved in acquisition of cisplatin resistance since multiple mechanisms of cisplatin resistance were described in murine leukemia cells two decades ago (Richon et al., 1987). Recent studies using gene knockout (Niedner et al., 2001), differential display (Francia et al., 2004), subtractive hybridization (Yasui et al., 2004), cDNA microarrays (Cheng et al., 2006; Roberts et al., 2005), and microRNA profiling (Yang et al., 2008) have documented that a large number of genes were either up-regulated or down-regulated in cisplatin-resistant (CP-r) cells, including genes that encode transcription factors, DNA damage-repair pathways, stress-response proteins, cell cycle checkpoints, apoptosis mediators, and transporters (reviewed in (Borst et al., 2007; Gottesman et al., 2002; Stewart, 2007; Wang and Lippard, 2005). Secondary mutations as a mechanism of cisplatin resistance have also been reported recently (Sakai et al., 2008).
To explore genes primarily involved in cisplatin resistance, we introduced a double-stranded cDNA into a retroviral expression vector, pLNCX2, from CP-r KB-CP.5 cells that were selected by a single step of cisplatin at 0.5 μg/ml. In our previous work, a ribosomal protein L36 and a heat shock protein HSP10 were found to be associated with cisplatin resistance by functional cloning and intermittent cisplatin selection (Shen et al., 2006). In this report, we have further determined that a novel hypothetical protein, TMEM205 (MBC3205) whose sequence was previously reported by Strausberg et al. (Strausberg et al., 2002) and listed as a putative secreted transmembrane protein using SPDI (Secreted Protein Discovery Initiative) strategies by Clark et al. (Clark et al., 2003) was able to confer cisplatin resistance. The development of cisplatin resistance has been known to result from reduced accumulation of cisplatin in many resistant cells (Andrews et al., 1988; Hall et al., 2008; Loh et al., 1992; Shen et al., 1998). Reduced accumulation of fluorescence-labeled cisplatin was also detected in the TMEM205-transfected stable clones. This is the first time, to our knowledge, that the hypothetical protein TMEM205 has been characterized and its ability to mediate cisplatin resistance has been documented. The results presented here demonstrate that the membrane secretory protein TMEM205 may play an important role in cisplatin resistance by reducing cisplatin accumulation.
MATERIALS AND METHODS
Cell lines and cell culture
Two populations of CP-r cell lines and their parental cell lines were studied: the human epidermoid carcinoma cell line KB-3-1 (a HeLa subclone) and its independent CP-r derivatives, KB-CP.3 and KB-CP.5, were selected in a single step at 0.3 and 0.5 μg cisplatin/ml respectively by two individuals in the lab (Liang et al., 2003; Shen et al., 1998). The KB-CP1 and KB-CP20, and the human liver carcinoma cell line BEL-7404 and its CP-r derivative 7404-CP20 were selected with stepwise increases to 20 μg of cisplatin/ml of medium as described previously. The mouse Balb/3T3 CP-r cell lines, A.6, A1.5 and A10 were isolated by stepwise increases from a single step of 0.6 μg/ml, up to 1.5, and 10 μg of cisplatin/ml of medium, respectively (Shen, DW & Gottesman, MM, unpublished data). All the CP-r cells were maintained in the presence of cisplatin, but cisplatin was removed from the medium 3 days prior to preparation of proteins and RNAs. The RetroPack PT67 cell line was purchased from Clontech (Palo Alto, CA). All cell lines were grown as monolayer cultures at 37°C in 5% CO2, using Dulbecco’s modified Eagle medium with 4.5 g/l glucose (InVitrogen, Carlsbad, CA), supplemented with L-glutamine, penicillin, streptomycin and 10% fetal bovine serum (BioWhittaker, Walkersville, MD). Cisplatin and other chemicals were purchased from Sigma (St. Louis, MO).
Retroviral cDNA library, expression vectors, and functional cloning
Double-stranded cDNA was synthesized from the CP-r cell line KB-CP.5 (maintained in medium containing 0.5 μg/ml cisplatin), using a BD SMART cDNA library construction kit (Clontech), as described by the manufacturer. Retroviral transduction and cisplatin selection were described in detail previously (Shen et al., 2006). Briefly, a library of cDNAs of 1.8 × 106 cDNA clones was constructed, and a pool of 93,000 Neo-r colonies was intermittently selected with cisplatin. Then a pool of 112 CP-r clones was sequenced individually, as detailed in our previous paper (Shen et al., 2006). TMEM205 (Accession no. BC091472), was obtained from the ATCC collection (Strausberg et al., 2002), and then inserted into the expression vector pcDNA5.1 (Invitrogen, Carlsbad CA), according to the manufacturer’s instructions.
Preparation of RNA and reverse transcription (RT) PCR
For determination of expression levels of genes of interest, RNAs were isolated from cells using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RT-PCR was performed using a GeneAmp kit (Applied Biosystems, Branchburg, NJ) as described by the manufacturer. Specific primers for tested genes were: TMEM205: 5′-CTT CCC CTT CTA CTT CCA CAT C-3′ (forward), and 5′-AGC GTA AGG CTC AGG AAC AG-3′ (reverse); Primers for GAPDH were purchased from InVitrogen (Carlsbad, Ca) for verification of the PCR performance.
Total RNA from human normal tissues was obtained from Clontech (Palo Alto, CA). The quality (purity and integrity) of the RNA samples was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). The RNA was quantitated using a spectrophotometer (Ultrospec 3100 pro, Amersham).
Real-time quantitative PCR
For quantitative measurement of expression levels of TMEM205 in human tissues, real-time quantitative PCR was performed using the ABI PRISM 7900HT (Applied Biosystems, Foster City, CA). Specific oligonucleotide primers and the TaqMan probe for TMEM205 (reference sequence; NM_198536) and GAPDH were obtained from Applied Biosystems (TaqMan Gene Expression Assay IDs of TMEM205 and GAPDH are Hs00414441_m1, Hs99999905_m1, respectively. For details of TaqMan Gene Expression Assay, see http://www.appliedbiosystems.com/). Synthesis of cDNA from total RNA samples was carried out using TaqMan Reverse Transcription Reagents (Applied Biosystems) with 1 μg total RNA/50 μL reaction volume. The cDNA (0.5 μL RT sample) was amplified using TaqMan Gene Expression Assays (1×) and TaqMan Universal PCR Master Mix Reagents (1×) (Applied Biosystems) in a total volume of 10 μL. The PCR mixture was pre-incubated at 50°C for 2 min, incubated at 95°C for 10 min, and amplified by 40 cycles at 95°C for 15 s and 60°C for 1 min. No-template (water) reaction mixtures were prepared as negative controls. During the PCR amplification, fluorescence emission was measured and recorded in real time. Crossing point values were calculated using the ABI PRISM 7900HT software package. The raw results were expressed as number of cycles to reach the crossing point. Crossing point values for TMEM205 were normalized to the respective crossing point values for the reference gene GAPDH.
Preparation of crude membrane and whole cell lysates
Crude membrane proteins of cells were prepared by homogenization in hypotonic solution. In brief, 1 × 107 cells from each cell line were homogenized in hypotonic solution [0.5 mM Na2HPO4, 0.1 mM EDTA, and protease inhibitor (Sigma, St. Louis, MO)], and the nuclei were separated after a spin at 1200 rpm × 5 min. The crude membrane pellets were collected by ultracentrifugation at 35,000 rpm × 40 min at 4°C, then re-suspended in 3 ml of lysis buffer (10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, and protease inhibitors) and stored at −80°C until use. For whole cell lysates, cells were lysed in a lysis buffer [50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, and 0.5% (v/v) Nonidet P-40, and protease inhibitor.
Generation of rabbit polyclonal anti-TMEM205 antibody
A purified rabbit polyclonal antibody directed to TMEM205 was generated by Rockland (Rockland Immunochemicals, Inc., Gilbertsville, PA) using a peptide of the sequence: Ac-CGPDPYRQLREKDPK-amide (amino acids 136–149). Rabbits were injected regularly, and bleeds were collected and screened for reaction with the peptide, and further confirmed by immunoblots for expression in cells. Positive anti-sera were affinity purified using the prepared peptide.
Immunoblotting and confocal analysis
SDS-PAGE immunoblotting was run as recommended by the manufacturer (Invitrogen, Carlsbad CA). Following electrophoresis, the gels were transblotted onto nitrocellulose membranes (Schleicher & Schuell, Keene, NH) at 4°C. Immuno-reaction was performed with desired primary antibodies and secondary HRP-conjugated antibodies. Pierce ECL reagents (Pierce Biotechnology, Rockford, IL) were used for developing signals, as described by the manufacturer. Specific primary antibodies, such as TGN markers and HRP-labeled secondary antibodies were purchased from BD Biosciences (San Diego, CA) and Jackson Immuno-Research Lab (West Grove, PA), respectively. For confocal analysis, cells were cultured in a Lab-Tek Chamber Slide (Nalge Nunc International, Naperville, IL), and fixed with 70% ETOH at −20°C for 15 min for visualization of intracellular expression and localization. Glutaraldehyde and triton X-100 fixation was applied for visualization of fluorescence distribution on the cell surface. The cells were fixed with either ETOH or glutaraldehyde reacted with the primary antibodies, followed by reaction with the secondary antibody labeled with Rhodamine red. For determination of accumulation and distribution of Alexa Fluor 488- or 546-labeled cisplatin (Invitrogen, Carlsbad CA), the fluorescence complex (F-CP) was dissolved in dimethylformamide (DMF) according to the manufacturer, then loaded into cells at a final concentration of 10 U/ml for 1 hour of incubation at 37°C. For efflux assays, cells were loaded with F-CP at a final concentration of 10 U/ml for 1 hour of incubation at 37°C, immediately washed with pre-warmed regular medium, and counted as the 0 min time point. The remaining cells were then re-incubated for 20 min and 60 min at 37°C. After incubation, cells were washed and fixed, and counter-stained with DAPI for nuclei staining. Images of cells were monitored under a laser scanning confocal microscope (Zeiss LSM 510 Laser Scanning Confocal Microscope) at 600 × magnification. The Zeiss AIM software LSM510 version 4.2 was used for semi-quantitative analysis on intensities of F-CP in cells. At least 25 cells were counted for each time point of the CP-s and CP-r cells.
Immunoelectron microscopy
Cells grown on Thermonax coverslips were slightly fixed in a mixture of 2.0% paraformaldehyde and 0.5% glutaraldehyde in PBS for 30 min followed by extensive washing in PBS. Cells were permeabilized with 0.5% triton X-100 for 30 min and blocked with 1% BSA, 1% normal goat serum and 0.1% fish gelatin in PBS plus 0.05% Tween-20 (PBST) for 1hr. After rinsing in PBST, cells were incubated overnight at 4°C in affinity-purified TMEM205 polyclonal antibody (1: 20), washed, and then incubated in a goat anti-rabbit 6 nm colloidal gold conjugate (Ted Pella, Redding, CA) for 90 min. Cells were post-fixed in 0.5% glutaraldehyde in PBS for 30 min and 0.5% osmium tetroxide in sodium cacodylate buffer before being dehydrated in a series of ethanol dehydrations and finally embedded with Epon-Aradite (Ted Pella, Redding, CA). Ultrathin sections (about 80nm) were cut on a Reichert Ultracut E (Leica, Bannockburn, IL) and loaded onto film-coated copper grids (200–300 mesh). Sections were counterstained with 2% (w/v) uranyl acetate for 5 min and lead citrate for 2 min, and examined at 100kV in a Philips CM 120 electron microscope.
Gene transfection and assays of resistance levels to cisplatin
Full-length cDNA for the genes encoding TMEM205 (cDNA clone: MGC110858), Accession, BC091472) was obtained from ATCC (Manassas, VA). The gene was re-inserted into a mammalian expression vector, pcDNA5.1 (Invitrogen, Carlsbad CA), as described by the manufacturer. Gene transfection was done with Lipofectin (Invitrogen, Carlsbad CA). Stable transfected clones were isolated after selection with G418, or dual selection with G418 and hygromycin B for double-transfection with TMEM205. Cell sensitivities to cisplatin were tested by two methods. Clonogenic assays were applied in Figure 4. Cells were seeded at 4 × 102 in 3 ml of medium per 35 mm dish, and then colonies were stained with methylene blue after 9–10 days of incubation, and counted. Drugs at a desired concentration were introduced into each well or dish prior to cell seeding. Control cells were transfected with insert-free vector only. The values are means of triplicate determinations.
Fig. 4.
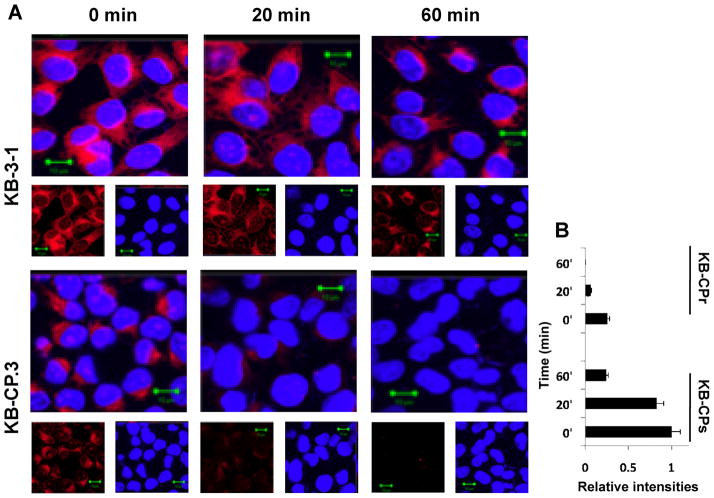
A: Confocal images showing reduced accumulation of Alexa Fluor 544- labeled cisplatin (F-CP) (red) in KB-CP.3 cells as compared to the parental KB-3-1 cells after 1 hour labeling (0 min), with a more rapid decrease in accumulation at time points 20 min and 60 min after depleting F-CP. B, a histogram showing faster F-CP clearance in KB-CP.3 cells than in KB-3-1 cells by measuring at least 25 cells using Zeiss AIM software LSM510, version 4.2 for semi-quantitative analysis on intensities of F-CP in CP-s and CP-r cells.
RESULTS AND DISCUSSION
Identification of TMEM205
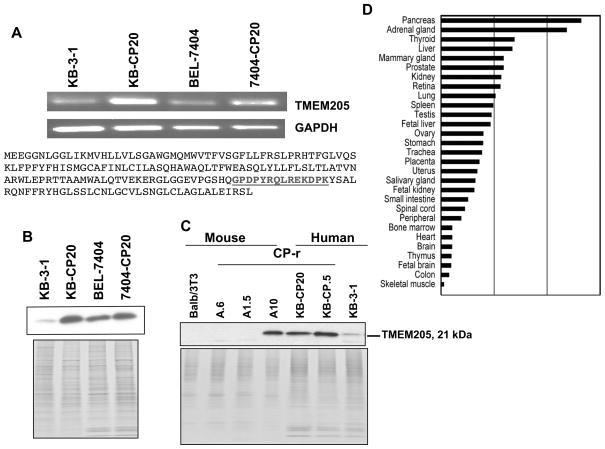
A pool of 112 CP-r clones was isolated from a retroviral cDNA library constructed from the single-step CP-r cell line KB-CP.5 by intermittent selection with cisplatin, as described in our previous paper (Shen et al., 2006). In addition to the previously reported 12 clones identified by functional cloning, another gene was identified in the screen named hypothetical protein TMEM205, which belongs to a group of secreted membrane proteins (Clark et al., 2003), and chosen for the present study. First, it was demonstrated by reverse transcription PCR that the gene was overexpressed in both CP-r cells, KB-CP20 and 7404-CP20, in comparison with their parent cell lines, KB-3-1 and BEL-7404 (Fig. 1A, upper panel). Second, we produced a polyclonal rabbit antibody (Rockland, PA) directed to TMEM205 using a peptide (amino acids 136–149) at the C-terminus of the sequence (AAQ89308) as shown in Fig. 1A, lower panel. After affinity-purification, the antibody was applied for further immunoblotting and confocal image analyses. Figure 1B shows that overexpression of the TMEM205 protein was detected at a molecular weight of ~21 kDa in both KB-CP20 and 7404-CP20 cells in comparison to their parent cell lines, KB-3-1 and BEL-7404, respectively, using the TMEM205 antibody as mentioned above. This result confirmed the determination by RT-PCR as seen in Figure 1A, upper panel.
Fig. 1.
A: RT-PCR analysis (upper panel) shows overexpression of TMEM205 in both cisplatin-resistant cell lines (KB-CP20 and 7404-CP20), compared with their parental cell lines (KB-3-1 and BEL-7404). The primers for TMEM205 and GAPDH are described in Materials and Methods. Peptide sequences of TMEM205 were used for generating polyclonal antibodies as highlighted (lower panel). B: an immunoblot showing increased expression of TMEM205 in KB-CP20 and 7404-CP20 cells using an affinity-purified antibody directed to TMEM205, and the lower panel showing the Coomassie blue stained gel as a loading control. C: increased expression of TMEM205 was detected by immunoblotting in human CP-r cell lines KB-CP.5 and KB-CP20, and mouse CP-r cell line, A10, and the lower panel showing a Coomassie blue stained gel as loading control. D: real-time PCR analysis to detect TMEM205 mRNA in normal human tissues, indicating a differential expression pattern with higher levels in tissues with secretory function, such as those of the pancreas, adrenal gland, thyroid, liver, and mammary gland.
To understand whether overexpression of TMEM205 is a common feature during development of cisplatin resistance, we compared the human cell lines with mouse Balb/3T3 cells by immunoblotting. The results demonstrate that the TMEM205 protein can be recognized in mouse cells by the antibody, and is significantly increased in mouse CP-r A10 cells (isolated after selection in 10 mg cisplatin/ml), as well as in human CP-r cell lines, KB-CP.5 and KB- CP20 (selected in cisplatin 0.5 and 20 μg/ml, respectively) as seen in Figure 1C, indicating that TMEM205 may play an important role in cisplatin resistance in human and mouse species. We have also demonstrated that acute exposure to cisplatin does not affect TMEM205 expression in the parental, sensitive (CP-s) cells (data not shown). Therefore, overexpression of TMEM205 in CP-r cells is likely a stable genetic change, rather than a stress response.
To obtain additional information about the potential role of TMEM205 in human tissues, Quantitative real-time PCR analysis of 29 different human tissue RNAs reveals a differential expression profile: higher expression levels in the pancreas, adrenal gland, liver, mammary gland, kidney, etc., as seen in Figure 1D. This expression profile reveals that TMEM205 is mainly present in tissues that are related to secretory function, indicating that this protein may play a significant role in secretion or vesicular trafficking. Such a function may also provide an explanation for its role in cisplatin resistance by involvement in vesicular trafficking or secretion of cisplatin, or by forming a cisplatin-protein complex that is effluxed from cells, resulting in a reduction of cisplatin accumulation in CP-r cells.
Localization of TMEM205
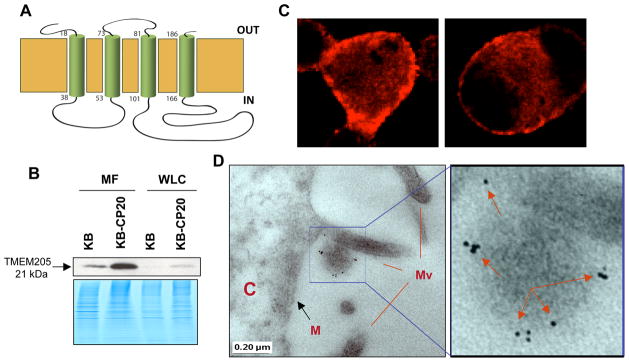
The protein is hypothetically a membrane protein, which has a four-transmembrane domain as shown in Figure 2A, based on the protein sequence analyzed using the PredictProtein Program (http://www.embl-heidelberg.de/predictprotein/predictprotein.html). Therefore, it was important to determine the localization and distribution of the protein in cells. We demonstrate here that TMEM205 is a membrane protein by the following evidence: i) much higher expression in the purified membrane fraction (MF) than in whole cell lysates (WCL) by immunoblotting (Fig. 2B); ii) expression on the cell surface as seen by strong fluorescent signals on the membrane, displayed in different regions of the cell surface of early-step KB-CP.3 cells (Fig. 2C); and, iii) immunoelectron microscopic observation further confirmed that the TMEM205 protein was largely located in microvilli of the cells, seen as dark grains which were labeled by the affinity-purified TMEM205 antibody and colloidal gold conjugate, as indicated by arrows (Fig. 2D). Therefore, the data presented in this work demonstrate that TMEM205 is, in fact, a membrane protein with molecular weight of ~21 kDa.
Fig. 2.
A: a predicted TMEM-205 topologic model, which has four transmembrane domains, based on the protein sequence analyzed using the PredictProtein Program (http://www.embl-heidelberg.de/predictprotein/predictprotein.html). B: an immunoblot showing increased expression of TMEM205 in KB-CP20 cells, and much stronger signals in an enhanced membrane fraction (MF) than in whole cell lysates (WCL) as indicated by an arrow. The lower panel shows a Coomassie blue stained gel as a loading control. C & D, KB-CP.3 cells were examined. C: confocal fluorescence images reveal cell surface distribution of TMEM205; D, immuno-electron microscopic images reveal location of TMEM205 in microvilli, C, Cell body; M, Membrane; Mv, Microvilli (the left panel), and an enlarged field is shown in the right panel, where the dark grains (labeled with TMEM205 antibody and colloidal gold) show the localization on the microvilli, indicated by arrows.
Correlation of TMEM205 expression with cisplatin resistance
Confocal images (Fig 3, the lower level) demonstrate that TMEM205 (labeled with Rhodamine red) is overexpressed in single-step low-level CP-r KB-CP.3 cells (selected with cisplatin 0.3 μg/ml), and shows stronger intensities in KB-CP.5 cells (independently selected with cisplatin 0.5 μg/ml) in comparison to their cisplatin-sensitive parental KB-3-1 cells. These results imply that expression levels of the TMEM205 protein are correlated with levels of cisplatin resistance, especially in the early stages of cisplatin resistance. In addition to the cell surface expression (Fig. 2), it was also found that the distribution pattern of TMEM205 in KB-CP.3 and KB-CP.5 cells appeared to be different, as shown in Figure 3, lower panel. The very early-step KB-CP.3 cells showed TMEM205 expression on the cell surface as well as in the cytoplasm from intracellular organelles or vesicles, whereas the KB-CP.5 cells presented an enriched pattern in the TGN region, near the nucleus, indicating a transition of TMEM205 distribution from the cell surface to intracellular regions that is associated with the level of cellular resistance to cisplatin.
Fig. 3.
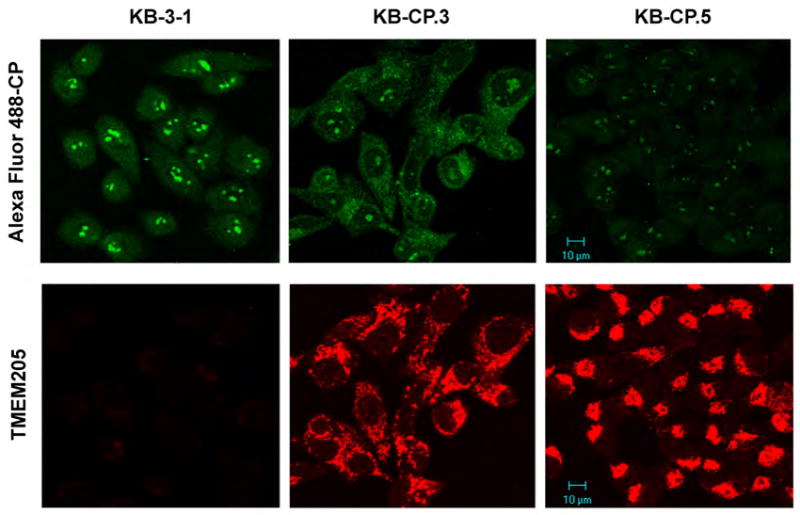
Confocal images. TMEM205 was overexpressed in CP-r cells, and distributed in the cell surface and intracellular compartment in KB-CP.3 cells, and translocated into the TGN region near the nucleus in KB-CP.5 cells. Decreased accumulation of fluorescence-labeled Alexa Fluor 488-cisplatin (green) was observed in human CP-r cells KB-CP.3 and KB-CP.5 compared to the parental cell line KB-3-1,
A prominent feature of cisplatin-resistant cells is reduced accumulation of cisplatin, due to either active efflux/secretion/sequestration, or defective uptake. As expected, when fluorescent Alexa-labeled cisplatin (F-CP) was used to trace intensities and distributions of a platinum compound in cells, it was found that the intensities of the F-CP (green) were significantly reduced in CP-r cells KB-CP.3 and KB-CP.5, as seen in Figure 3, the top panel, suggesting a close correlation between increased elevation of TMEM205 and reduced accumulation of F-CP in CP-r cells. The F-CP in KB-CP.3 cells appeared mostly in the cytoplasmic vesicles, with a small amount in the nucleoli, in comparison to the cisplatin-sensitive KB-3-1 cells, where there was a stronger green fluorescence in the nucleus, especially in nucleoli, with less in the cytoplasm. We further explored the possibility that the reduced accumulation of F-CP in KB-CP.3 cells was also associated with increased efflux of the compound. Figure 4, A and B demonstrate that reduced accumulation of F-CP is clearly seen after one hour incubation with F-CP (0 min). Then, the F-CP more rapidly decreased in the KB-CP.3 cells than in the parental KB-3-1 cells at the 20 min time point, and was almost undetectable at the 60 min time point after maintaining cells in F-CP-free medium, suggesting that reduced accumulation of cisplatin in CP-r cells is also involved in faster clearance of the compound. It should be noted that the Alexa-cisplatin, which is primarily used for labeling DNA in cells, may not precisely reflect the condition of cisplatin in cells. Nevertheless, it does reflect a correlation of increased TMEM205 and reduced F-CP accumulation in CP-r cells.
TMEM205 mediates cisplatin resistance
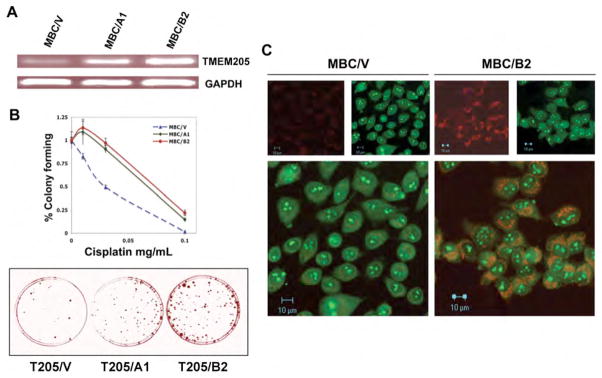
The next question we addressed was whether TMEM205 is directly related to cisplatin resistance or is a secondary feature during development of resistance to cisplatin. A full-length TMEM205 cDNA was inserted into the expression vector pcDNA5.1 with a neomycin selective marker. After transfection of TMEM205/pcDNA5.1 into cisplatin-sensitive KB-3-1 cells, with selection with G418, stable clones were further examined for expression levels of TMEM205 by reverse transcription PCR. Clones MBC (TMEM)/A1, and MBC (TMEM)/B2 were chosen for further drug sensitivity assays, as TMEM205 was well expressed with some difference in these two independent clones, as determined by RT-PCR (Fig. 5A). The results from clonogenic assays, described in Materials and Methods, showed approximately 2.2- to 2.5-fold more resistance in the TMEM205 (MBC3205)-transfected cells MBC/A1, and MBC/B2, respectively, than the control cells, which were transfected with vector only (Fig. 5B, upper panel). Figure 5B, the lower panel, shows methylene blue-stained colonies in this same test, indicating more colonies in TMEM205-transfected clones than the control, 9 days after exposure to cisplatin at 0.1 μg/ml. Figure 5C shows that F-CP was also reduced in the TMEM205-transfected cells, but not to the extent seen in cisplatin-selected KB-CP.5 cells (Fig. 3A). This result was expected, as the level of cisplatin resistance was much higher in KB-CP.5 cells (Liang et al., 2003) than the TMEM205-transfected cells. These results suggest that the gene could mediate cisplatin resistance, through reduction of cisplatin accumulation, by increased exclusion or “secretion” which was the role originally proposed for TMEM205. However, the precise mechanism of the TMEM205 in cisplatin resistance needs to be further elucidated. The major finding in this work of elevated expression of the hypothetical membrane secretory protein TMEM205 in CP-r cells, and the ability to confer cellular resistance to cisplatin may provide a novel mechanism for cisplatin resistance. TMEM205 may also be valuable as a biomarker, or as a target in clinical tumor chemotherapy.
Fig. 5.
A: RT-PCR determination of expression levels of two individual clones isolated from TMEM205 (MBC)-transfected, and G418-selected stable clones. MBC/V was a control clone transfected with vector only. GAPDH served as a control. B, resistance levels of the TMEM205 transfectants, MBC/A1 and MBC/B2, to cisplatin; the upper panel shows the killing curves, and the lower panel shows methylene blue-stained colonies after exposure to cisplatin 0.1 ug/ml for 9 days. Each point was a mean of triplicate determinations on clonogenic assays after 9–10 days of exposure to the compound. Confocal images shown in C demonstrate reduced accumulation of F-CP (green) in TMEM205-transfectants, MBC/B2, double-stained with Rhodamine red-conjugated antibody directed to the TMEM205 polyclonal antibody (red).
Acknowledgments
This research was funded by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute and the National Institute of Biomedical Imaging and Bioengineering. We would like to thank George Leiman for editorial assistance, and Susan Garfield, Poonam Mannan and Langston Lim in the Laboratory of Experimental Carcinogenesis (NCI) for assistance with confocal analysis.
Nonstandard abbreviations
- CP-r
cisplatin-resistant
- CP-s
cisplatin-sensitive
References
- Andrews PA, Velury S, Mann SC, Howell SB. cis-Diamminedichloroplatinum(II) accumulation in sensitive and resistant human ovarian carcinoma cells. Cancer Res. 1988;48:68–73. [PubMed] [Google Scholar]
- Borst P, Jonkers J, Rottenberg S. What makes tumors multidrug resistant? Cell Cycle. 2007;6:2782–2787. doi: 10.4161/cc.6.22.4936. [DOI] [PubMed] [Google Scholar]
- Cheng TC, Manorek G, Samimi G, Lin X, Berry CC, Howell SB. Identification of genes whose expression is associated with cisplatin resistance in human ovarian carcinoma cells. Cancer Chemother Pharmacol. 2006;58:384–395. doi: 10.1007/s00280-005-0171-8. [DOI] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee J, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand D, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia G, Man S, Teicher B, Grasso L, Kerbel RS. Gene expression analysis of tumor spheroids reveals a role for suppressed DNA mismatch repair in multicellular resistance to alkylating agents. Mol Cell Biol. 2004;24:6837–6849. doi: 10.1128/MCB.24.15.6837-6849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- Liang XJ, Shen DW, Garfield S, Gottesman MM. Mislocalization of membrane proteins associated with multidrug resistance in cisplatin-resistant cancer cell lines. Cancer Res. 2003;63:5909–5916. [PubMed] [Google Scholar]
- Loh SY, Mistry P, Kelland LR, Abel G, Harrap KR. Reduced drug accumulation as a major mechanism of acquired resistance to cisplatin in a human ovarian carcinoma cell line: circumvention studies using novel platinum (II) and (IV) ammine/amine complexes. Br J Cancer. 1992;66:1109–1115. doi: 10.1038/bjc.1992.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner H, Christen R, Lin X, Kondo A, Howell SB. Identification of genes that mediate sensitivity to cisplatin. Mol Pharmacol. 2001;60:1153–1160. [PubMed] [Google Scholar]
- Richon VM, Schulte N, Eastman A. Multiple mechanisms of resistance to cis-diamminedichloroplatinum(II) in murine leukemia L1210 cells. Cancer Res. 1987;47:2056–2061. [PubMed] [Google Scholar]
- Roberts D, Schick J, Conway S, Biade S, Laub PB, Stevenson JP, Hamilton TC, O’Dwyer PJ, Johnson SW. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer. 2005;92:1149–1158. doi: 10.1038/sj.bjc.6602447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Pastan I, Gottesman MM. Cross-resistance to methotrexate and metals in human cisplatin-resistant cell lines results from a pleiotropic defect in accumulation of these compounds associated with reduced plasma membrane binding proteins. Cancer Res. 1998;58:268–275. [PubMed] [Google Scholar]
- Shen DW, Liang XJ, Suzuki T, Gottesman MM. Identification by functional cloning from a retroviral cDNA library of cDNAs for ribosomal protein L36 and the 10-kDa heat shock protein that confer cisplatin resistance. Mol Pharmacol. 2006;69:1383–1388. doi: 10.1124/mol.105.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, Tomida A, Funato T, Yokomizo A, Naito S, Imoto I, Tsuruo T, Inazawa J. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.can-3263-2. [DOI] [PubMed] [Google Scholar]