Abstract
The UvrB protein is a central unit for damage recognition in the prokaryotic nucleotide excision repair system, which excises bulky DNA lesions. We have utilized molecular modeling and MD simulations based on crystal structures, mutagenesis, and fluorescence data, to model the 10R-(+)-cis-anti-B[a]P-N2-dG lesion, derived from the tumorigenic (+) anti-B[a]PDE metabolite of benzo[a]pyrene, at different locations on the inner and outer strand in UvrB. Our results suggest that this lesion is accommodated on the inner strand where it might translocate through the tunnel created by the β-hairpin and the UvrB domain 1B, and ultimately could be housed in the pocket behind the β-hairpin prior to excision by UvrC. Lesions that vary in size and shape may be stopped at the gate to the tunnel, within the tunnel or in the pocket when UvrC initiates excision. Common features of β-hairpin intrusion between the two DNA strands and nucleotide flipping manifested in structures of prokaryotic and eukaryotic NER lesion recognition proteins are consistent with common recognition mechanisms, based on lesion-induced local thermodynamic distortion/destabilization and nucleotide flipping.
Keywords: Nucleotide Excision Repair, UvrB, Lesion Recognition, Flip Out, Lesion Translocation
Damage to DNA occurs continuously in living organisms, either endogenously through reactive oxygen species (ROS) produced during normal metabolic processes or exogenously from sunlight, pollutants, dietary mutagens and other factors (1). To preserve the DNA as the reliable storehouse for genetic information, repair mechanisms play a vital role in maintaining DNA integrity (2,3). Nucleotide excision repair (NER) is the predominant pathway for repair of bulky DNA lesions (4-6).
In prokaryotes, the dual incisions induced by the NER apparatus involve the UvrABC repair system. The UvrABC system consists of three proteins: UvrA, UvrB, and UvrC (7,8). The incision pathway is initiated when UvrA and UvrB form a heterotrimeric (UvrA2B) (9) or heterotetrameric (UvrA2B2) (10) complex with the DNA lesion. This complex is believed to initially recognize the distortion / destabilization of the DNA duplex caused by bulky lesions. At this stage, the damaged DNA interacts primarily with the UvrA2 units (11). A recent crystal structure of UvrA identifies the UvrA dimerization interface that is regulated by ATP, and suggests the location of the DNA and UvrB binding domains (12). After an ATP hydrolysis cycle, the damaged DNA moves to the binding domain of the UvrB unit and the UvrA2 units leave (7). Recently it has been shown that DNA wrapping around UvrB, mediated by UvrA, is an early recognition event (13). Thus, UvrB is the central component of the bacterial NER system that recruits the nuclease UvrC to the complex. Most recently, the crystal structure of the complex between the interaction domains of UvrA and UvrB was reported (14). A structural model for the full length UvrA-UvrB assembly was constructed and two models for lesion recognition involving these two enzymes were proposed: the recruitment and the handoff model. In the recruitment model, lesion recognition is exclusively by UvrA, and UvrB binding occurs without lesion contact. In the handoff model, the damaged site is handed off to UvrB (14). The handoff model is consistent with earlier work suggesting that UvrB is directly involved in distinguishing damaged from undamaged DNA, and that it locates the position of the damage and guides the damaged DNA strand from the recognition to the repair stage (15). At the damaged site, the duplex DNA is separated by a UvrB β-hairpin protruding between the two strands; this generates an inner strand between itself and domain 1B (domains are defined in Figure 1A) of UvrB, and an outer strand on the exposed side (15). A change in the conformation of the UvrB / DNA complex triggers the recognition by the third component of the UvrABC system, UvrC. At this stage, the UvrB / DNA complex is in the pre-incision state, ready for processing by UvrC. In E. coli UvrABC, the UvrC carries out incisions at about the 5th phosphodiester bond on the 3′ side and about the 8th bond on the 5′ side of the damaged base; the exact length of the excised oligonucleotide can depend on the nature of the adduct and the sequence context (7,16-20).
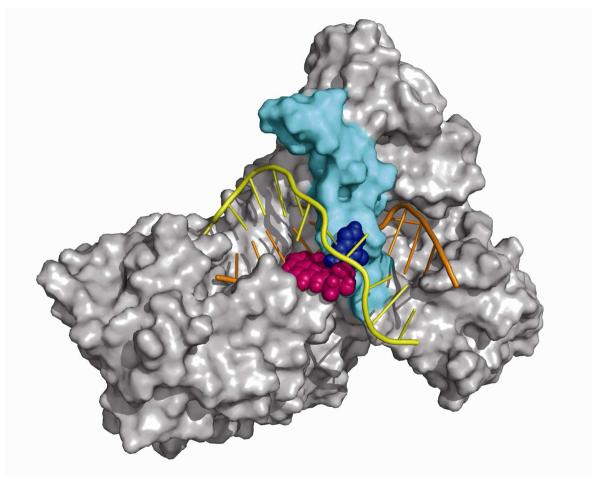
Figure 1.
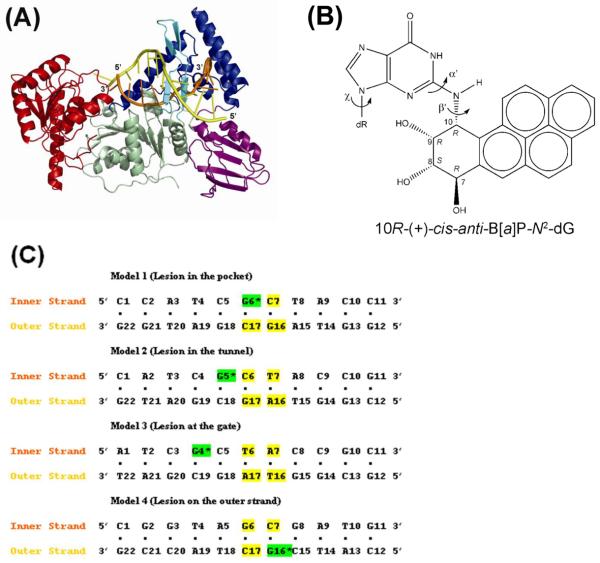
(A) UvrB-DNA model. The UvrB/DNA complex model is shown in cartoon. The domains of UvrB are color coded as Domain 1A (residues 1-151, 324-340, 379-411), green; Domain 1B (residues 252-323, 341-378), blue; Domain 2 (residues 151-251), purple; Domain 3 (residues 412-593), red. The β-hairpin which belongs to Domain 1A is in cyan. The DNA inner strand (located between the protein and the β-hairpin) is in orange, and the outer DNA strand is in yellow. (B) Structure of the 10R-(+)-cis-anti-B[a]P-N2-dG adduct. The absolute configurations of the four chiral atoms C7, C8, C9, and C10 are indicated. Torsion angles are defined as χ = O4′(dR)-C1′(dR)-N9-C4, α′ = N1-C2-N2-C10(B[a]P), β′= C2-N2-C10(B[a]P)-C9(B[a]P). (dR is deoxyribose.) (C) Base sequences and base numbering conventions of the four models considered. The B[a]P diol epoxide-modified guanine (G*) is indicated by the green background and is positioned on the inner strand in Models 1, 2, and 3, and on the outer strand in Model 4. The flipped out bases in the initial models are shown in yellow.
The intrinsic fluorescence of 2-aminopurine (2-AP) positioned on the damaged strand flanking a lesion on its 3′ side has been utilized to demonstrate that this base is flipped out of the duplex and is positioned in a hydrophobic region of the UvrB protein (21). Further fluorescence studies have recently shown that this flipped-out base helps stabilize the UvrB/DNA binding complex (22). In addition, studies with UvrB mutants have shown that stacking interactions occur between UvrB tyrosine residues and several flipped out bases around the damaged base, including one on the damaged strand and one on the partner strand (23-25).
An X-ray crystallographic study (26) has revealed the structure of a B. caldotenax UvrB complex with double stranded DNA (part of a hairpin loop) containing a single stranded 3′ end overhang and a fluorescein-derived lesion. While this structure did not have the lesion positioned in the recognition site, it provided essential information on the mode of DNA binding and the relevant amino acid residues in UvrB that are important for damage recognition. In this structure, three bases on the single strand overhang behind the β-hairpin make multiple contacts with several amino acids including Tyr92 and Tyr93, and one base is flipped out and stabilized by stacking over Phe249 (26). However, exactly where the lesion is positioned remains uncertain: the bulky fluorescein adduct is not positioned in close contact with the β-hairpin motif that is believed to contain the lesion recognition region; instead it is adventitiously placed in the loop of the DNA hairpin, near the interface between domains 1A and 3 of UvrB. Based on the crystal structure and fluorescence data (21), a possible positioning of the damaged base on the inner strand was suggested by Truglio et al. (26) to be just outside the tunnel created by the β-hairpin, with the last base pair of the duplex making strong contacts with Tyr96, and the next 3′ base flipped out into a “shape-complementary (planar) pocket” created in part by Phe249. The possible positioning of the lesion on the inner strand at the gate to the tunnel was also suggested from a crystal structure of UvrB containing a single stranded dT 5mer oligonucleotide with a fluorescein adduct. In this case, two of the five dT residues were positioned behind the β-hairpin, and the large lesion was linked to a dT positioned at the β-hairpin / domain 1B interface; this structure suggested simple steric exclusion as inhibiting translocation of this very bulky lesion through the β-hairpin (27). Another position for the lesion would be inside the tunnel as suggested by Truglio et al. (26) who indicated that the damaged base at the tunnel gate would be the next one to be translocated behind the β-hairpin into the pocket created by Phe249. Finally, it is also possible that the damage may reside on the outer strand (7,26): a suggestion that this may occur, stems from the observation that DNA photolyase bound to a pyrimidine dimer in the dark stimulated incision by the UvrABC system (28,29) which would indicate a modified outer strand for this case.
The objective of the present work was to analyze possible lesion positioning mechanisms of the UvrB enzyme in complex with damaged DNA as in the handoff model (14), using molecular modeling and dynamics methods. We selected for investigation a bulky lesion derived from the reaction of the highly tumorigenic and mutagenic metabolically activated form of the environmental pollutant benzo[a]pyrene (30), (+)-7r,8t-dihydrodiol-t9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (+)-anti-B[a]PDE), with the exocyclic amino group of guanine in double-stranded DNA. Reaction between guanine and (+)-anti-B[a]PDE produces two stereoisomeric adducts, the 10S-(+)-trans-anti-B[a]P-N2-dG and the 10R-(+)-cis-anti-B[a]P-N2-dG adducts (31,32). While the 10S-(+)-trans-anti-B[a]P-N2-dG stereoisomer is the predominant reaction product, the 10R-(+)-cis-anti-B[a]P-N2-dG [(+)-cis-B[a]P-N2-dG] is more efficiently incised by the Bacillus caldotenax, Thermotoga maritima, and Escherichia coli UvrABC system (19,33) and hence was selected for the present study.
We built models of UvrB / DNA preincision complexes, based on available crystal structures of UvrB (26,34,35), and then explored four possible binding sites for positioning the lesion based on crystallographic data (26-28), fluorescence evidence (21), and mutation studies (23-25). Specifically, we considered [1] lesion placement on the inner strand in the UvrB shape-complementary (planar) pocket behind the β-hairpin; [2] lesion placement on the inner strand in the tunnel between the β-hairpin and domain 1B; [3] lesion placement on the inner strand at the gate before entering the tunnel; and [4] lesion placement on the outer strand in front of the β-hairpin. We carried out 5 ns molecular dynamics simulations and analyzed the resulting ensembles. Our results suggest that this lesion is housed on the inner strand where it might translocate through the tunnel between the β-hairpin and domain 1B, and could finally be located in the hydrophobic pocket behind the β-hairpin prior to excision by UvrC. Differently sized and shaped lesions may be halted at the gate to the β-hairpin tunnel, or become trapped in the tunnel prior to excision. The UvrB β-hairpin intrusion model bears a resemblance to the insertion of a hairpin between the two DNA strands at the lesion site by the yeast Rad4 NER protein, the orthologue of the human NER recognition factor XPC (36).
METHODS
Molecular modeling of the preincision state of the UvrB/DNA adduct complex
Since there is no crystal structure of a UvrB/DNA complex with any adducts at its binding site, we created a model, based on three crystal structures: a UvrB/DNA complex (26) (PDB ID: 2FDC); a ternary complex containing UvrB, a polythymidine trinucleotide, and ADP (34) (PDB ID: 2D7D); and a UvrB in the ATP-bound form (35) (PDB ID: 1D9Z). The three UvrB crystal structures share considerable structural similarities. The 2FDC structure (chain B) from Bca was employed as the template structure since it is currently the only structure of a UvrB-DNA-duplex complex containing the β-hairpin motif with the inner strand behind the hairpin. However, the outer strand stops before reaching the β-hairpin. Also the ATP cofactor, needed for the preincision UvrB/DNA complex to bind UvrC (7), is missing. In addition, the adduct is not located near the β-hairpin region (21). Furthermore, 8 residues (residues 477 to 484) are missing in the 2FDC structure, causing the secondary structure from residues 485 to 489 to be misfolded as compared to the relatively complete structure 2D7D. Note that the UvrB C-terminal domain 4 is missing in most structures (only partially resolved in crystal structure 2D7D). UvrB domain 4 is an auto-inhibitory domain involved in UvrC binding, UvrB dimerization and inhibits DNA binding and ATP hydrolysis (10,37,38). Thus, we could not model domain 4 in our work. Further molecular modeling was needed to build the preincision UvrB/DNA complex, as described below.
The sequence of the 11 mer double stranded DNA investigated was selected because it has been used in previous experimental UvrABC DNA repair studies (Figure 1C) (19,33). The 2FDC crystal structure contains 3-base-pairs of double stranded DNA with a 2-base 3′ overhang, corresponding to 5′-A3-T4-C5-G6-C7-3′/3′-T20-A19-G18-5′ in Figure 1C, Model 1. We elongated the DNA duplex by modeling two nucleotides on the outer strand, G16 and C17, to wrap around the β-hairpin and then added 4 more base pairs 5′-T8-A9-C10-C11-3′ / 3′-A15-T14-G13-G12-5′ beyond the separated two strands emanating from the β-hairpin. The C17 on the outer strand was modeled to be flipped out based on the fluorescence data (21). The G16 on the outer strand was also modeled to be flipped out, stacking with Tyr95, based on mutation studies (24,25). We then added one base pair, T8•A15, which neighbors C7•G16 on the right side of the β-hairpin (Figure 2). The bases T8 and A15 on the inner and outer strands, respectively, were modeled to be base paired by manually adjusting the DNA backbone and glycosidic torsion angles. The remaining three base pairs were added using the NMR solution structure (39) of the 11mer duplex containing the B[a]P adduct, utilizing the last three base pairs 3′ to the lesion.
Figure 2.
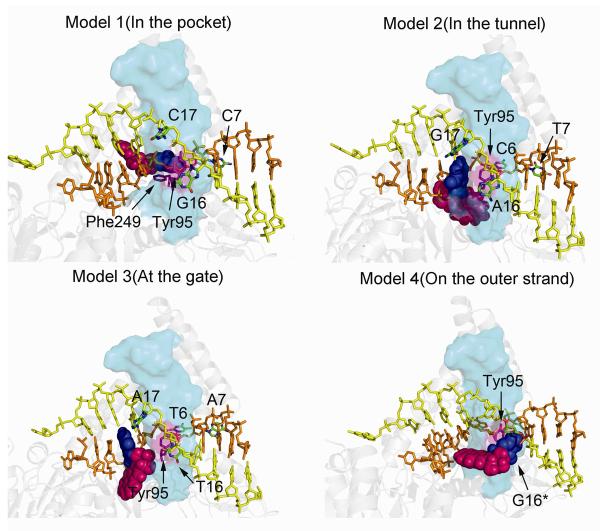
Initial models of the four UvrB/DNA complexes. Model 1 has the adduct positioned in the UvrB pocket, Model 2 has the adduct positioned in the tunnel between the β-hairpin and Domain 1B, Model 3 has the adduct positioned at the gate before entering the tunnel, and Model 4 has the adduct positioned on the DNA outer strand. The UvrB protein is rendered in cartoon, while the DNA is rendered in stick. The β-hairpin is rendered in cyan transparent surface. The B[a]P-dG is rendered in CPK model with G6* in blue and B[a]P in magenta. The DNA inner strand is shown in orange and the outer strand is shown in yellow. The flipped out bases and Tyr95 are labeled; these bases are colored by atom, and Tyr95 is in magenta. A stereo view is given in Figure S6.
The ATP cofactor with a coordinated Mg2+ was modeled in the ATP binding site of 2FDC from the crystal structure1D9Z by superimposing the residues 11-46 of 2FDC and 10-45 of 1D9Z (Figure S1). These residues form the ATP binding pocket. To model the 8 missing residues, we superimposed residues 420-460 of 2FDC to residues 418-458 of 2D7D and then modeled residues 477-489 from 2D7D onto 2FDC (Figure S1).
The B[a]P base adduct coordinates (Figure 1B) were obtained from the NMR solution structure (40) and modeled into the hydrophobic pocket suggested by Truglio et al (26) by covalently linking the inner strand guanine (G6) N2 atom to the B[a]P C10 atom. In order to be housed in the pocket, the glycosidic conformation has to be syn. These strategies were utilized to generate Model 1 (Figure 2).
We also modeled the B[a]P adduct in the positions G5 (Model 2) and G4 (Model 3) to investigate the translocation of the adduct within the UvrB complex (Figure 2). Models 2 and 3 were constructed using Model 1 as a basis, but with the appropriate modification of the adduct site relative to the protein (Figure 1C). At both positions G4 and G5 the guanine must be in the syn conformation to minimize collisions with protein residues. At the position G5, the syn guanine is still stacked with Tyr96 of UvrB, consistent with observations from both crystallography (26) and experiments with UvrB mutants which showed that binding to DNA was abolished when Tyr96 was mutated to alanine (7,24,41).
In addition, we modeled the adduct on the outer strand utilizing the sequence shown in Figure 1C to produce Model 4. The B[a]P adduct was linked to the G16 base; the latter was modeled in a stacked conformation with Tyr95, as observed by Moolenaar et al and Zou et al (24,25). To stack with Tyr95 and position the adduct without collisions, the G16 must be in the syn conformation (Figure 2, Table S1).
Initial models were constructed to have minimal steric collisions by adjusting the (+)-cis-B[a]P-N2-dG torsion angles χ, α′, and β′ (Figure 1B) to locate the optimal positions. We also made an effort to visually optimize favorable interactions between the damaged base and the enzyme, such as hydrophobic and electrostatic interactions. All χ, α′, and β′ torsions for the models are shown in Table S1. A total of 4 models were created. Hydrogen atoms were added with the LEaP module in AMBER 8.0 (42). Each complex was then energy-minimized with implicit solvent (dielectric constant of 4.0r, while r is the distance between two atoms) for 400 steps of steepest descent (SD) followed by 600 steps of conjugate gradient (CG) minimization using the SANDER module of AMBER 8.0 (42).
Force field
Computations were carried out with the AMBER 8.0 suite of programs (42), the Cornell et al. force field (43), and the PARM99 parameter set (44). The force field for the (+)-cis-B[a]P-N2-dG adduct was parameterized consistent with the rest of the AMBER force field: partial charges were computed as described previously (45) and atom types were defined based on the PARM99 and GAFF (46) parameter sets. Missing parameters were assigned by analogy with chemically similar atom types already present in the force field. All of the added force field parameters, atom types, and topology assignments are listed in Tables S2-S4.
Molecular dynamics protocol
Full details of the MD protocol are given in Supplementary data.
Stability of the molecular dynamics simulation
Plots of the root-mean-square deviations (RMSD) of the current relative to the starting structure as a function of time are shown in Figure S2. The structures generally fluctuate in a stable manner after 2 ns, and our analyses were confined to the 2.0-5.0 ns time frame. We selected one best representative structure from each ensemble for graphic illustration. To pick the best representative structure, we extracted 100 structures at 30 ps intervals from the 2 -5 ns time frame for each model. Each extracted structure was superimposed to the remaining 99 structures and the RMSDs were computed and summed. The structure with the smallest sum RMSD was selected as the most representative structure (Table S6). This procedure essentially follows the philosophy in Moil-View (47), which could not be utilized due to the size of our system.
Structural analyses
Snapshots of the structures during the simulations and the average structures with solvent and counterions removed were obtained with the PTRAJ module of the AMBER 8.0 suite (42). PTRAJ was also employed to determine the time-dependence of the RMSD, and of the torsion angles α′, β′, and χ. These are given in Figure S3, which shows that these torsions occupy limited domains as previously delineated by molecular mechanical calculations (48). Detailed hydrogen bonding analyses were carried out with the CARNAL module of the AMBER 7.0 suite (49).
INSIGHT II and Discovery Studio from Accelrys, Inc., and PyMOL (50) from DeLano Scientific LLC. were employed for visualization and model building. Computations were carried out on our own cluster of Silicon Graphics Origin and Altix high performance computers.
RESULTS
The objective of this work was to systematically explore different structural hypotheses for the possible binding sites of the lesion in UvrB. Is the adduct positioned within the UvrB protein on the inner strand or the outer strand? If on the inner strand, is the lesion blocked at the UvrB β-hairpin steric gate or can it translocate through the tunnel behind the β-hairpin? For these purposes, we constructed four models, all based on available experimental data, of complexes containing UvrB and double stranded DNA with the (+)-cis-B[a]P-N2-dG adduct either on the inner or the outer strand (see Methods). The exact sequences and the base numbering schemes that were used in modeling the different translocation steps are shown in Figure 1C. The lesion was positioned on the inner strand in the pocket behind the β-hairpin (Model 1), in the “tunnel” between the β-hairpin and domain 1B (Model 2), at the entrance to the “tunnel” (Model 3), and on the outer strand contacting the β-hairpin (Model 4). Molecular dynamics simulations were performed in each case. The MD trajectories were evaluated, and the lesion positioning and the possibility of lesion translocation through the β-hairpin of the UvrB protein were examined.
Structures of the UvrB/DNA preincision complex
The initial model of the UvrB/DNA preincision complex was based mainly on the crystal structure 2FDC (26) of the Bca UvrB protein containing the modified DNA sequence context 5′-….CCATCG*CTACC…. with G* designating the modified guanine. The complex in the crystal structure has a DNA single strand region as well as a pocket with a flat floor motif, both located behind the β-hairpin; this pocket appears capable of accommodating the DNA adduct when the damaged site is located on the inner DNA strand between the hairpin and surface of the UvrB protein (26). The B[a]P residue was positioned into this pocket (see Methods) (Model 1 of the pre-incision complex) with minimal close contacts (Table S1). In order to study the feasibility of translocation of the adduct through the β-hairpin, the lesion was also positioned at G5 and G4 on the inner strand (Models 2 and 3, respectively). In all models, the sequence context in which the (+)-cis-B[a]P-N2-dG adduct is embedded is constant, and only the position of the adduct is different.
The feasibility of positioning the lesion on the outer DNA strand of the DNA duplex was investigated by modeling the lesion at G16 (Figure 1C) where the modified guanine is stacked with Tyr95 (Model 4), as suggested by experiments with mutants (24,25). In this case, G6 is positioned in the pocket.
Accommodation of the lesion in the pocket behind the β-hairpin
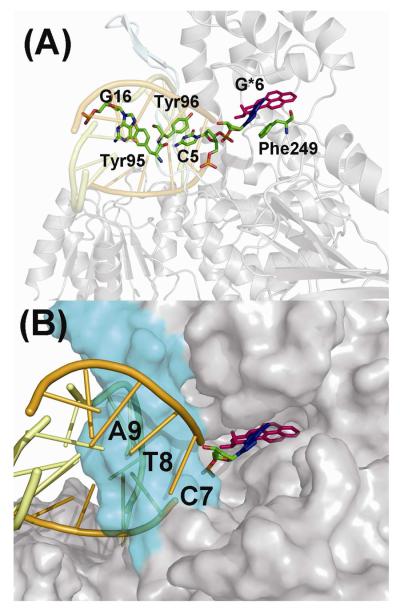
The bulky and hydrophobic B[a]P residue can be positioned in the spacious pocket located behind the β-hairpin, when the glycosidic bond of the modified guanine is in the syn conformation (Model 1). This lesion-containing pocket is surrounded by the following residues (within 5Å of any B[a]P atom): Tyr146, Gly147, Leu148, Gly149, Asn308, Ser310, Arg311, Thr312, Ser320, Pro322, Tyr323, Asp354, Lys358, Asp373, Asn374, Arg375, and Pro376. The interactions between the adduct and the protein in the pocket are mainly hydrophobic. At the modified site, the damaged guanine is flipped out of the DNA. The simulation results suggest that the positions of the modified guanine and several of its neighboring bases are stabilized by stacking interactions with several amino acid residues (Figure 3A). Specifically, a stacking interaction between the modified guanine residue and Phe249 stabilizes the flipped-out base. It should be noted that there is an analogous stacking interaction between Phe249 and an undamaged cytosine in the crystal structure (26). Furthermore, Tyr96 stacks with the C5 which is on the 5′-side of the modified guanine, which retains a buckled base pair with G18; this stacking interaction was also present in the starting model, and remained stable throughout the simulations. In addition, on the outer strand, the base G16, which is the partner base of C7, is stacked with Tyr95 in the β-hairpin, and this interaction is also maintained throughout the simulation (Figure 3A).
Figure 3.
Model 1 of the adduct in the pocket of UvrB. The B[a]P-dG adduct is shown in stick with G6* in blue and B[a]P in magenta. Hydrogen atoms are deleted for clarity. (A) The stacking interactions that stabilize the binding of the DNA molecules to UvrB are shown. The residues involved in stacking interactions are shown in the stick model with color-coding by atom. (B) Accommodation of the B[a]P residue in the pocket behind the β-hairpin (the latter shown in cyan). The UvrB protein is shown in the surface model mode. The best representative structure is shown. A stereo view is given in Figure S7.
Possibilities for lesion translocation behind the UvrB β-hairpin
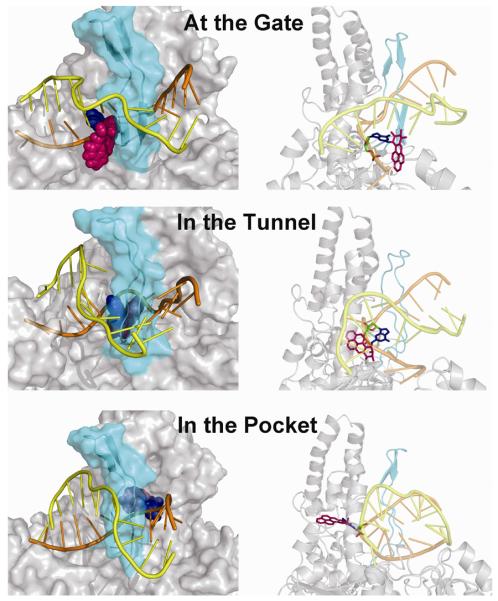
To investigate the possibility of translocation of the bulky lesion positioned on the inner DNA strand behind the β-hairpin of UvrB, we modeled the B[a]P adduct at three consecutive positions on this trajectory. These include the G6 position of the adduct with the adduct in the pocket as described above (Model 1), or the G5 position (Model 2) and the G4 position (Model 3) (Figure 1C). The hypothesis here is that the bulky adduct on the inner strand needs to translocate through the β-hairpin to achieve binding in the pocket as shown in Model 1 (see Movie in Supplementary Data). At the G4 position, the adduct is close to the β-hairpin where the aromatic rings are partly solvent exposed; however, the lesion does not enter the tunnel between the β-hairpin and the domain 1B of UvrB (Figure 4). At the G5 position, our models reveal that the adduct can reside behind the β-hairpin and become stably accommodated within the tunnel. It is not necessary for the β-hairpin to completely separate from domain 1B (Figure 4) in order to allow for the translocation of bulky adducts that are comparable in size to the (+)-cis-B[a]P-N2-dG adduct considered here. The upper part of the β-hairpin motif is still in contact with the UvrB domain 1B. However, the bottom of the β-hairpin changes conformation from a well organized β-sheet to a loop. This enables the root of the β-hairpin to move away from domain 1B, which enlarges the tunnel to allow the lesion to pass (Figure 4). When the adduct is at the G6 position in the pocket, the lesion may pass through the step that involves Model 2 in which it is in the tunnel (Figure 4).
Figure 4.
Possible translocation mechanisms illustrated for the adduct in terms of Models 3, 2, and 1 (top to bottom, respectively). Left-hand panels: front view of the protein. The β-hairpin is shown in cyan and is semi-transparent so that the adduct behind the hairpin can be discerned. The adduct is in CPK model. The G6* is blue and B[a]P is magenta. The DNA inner strand is shown in orange and the outer strand in yellow. The right-hand panels show views of the DNA–UvrB complex looking into the tunnel from the left side. UvrB is in cartoon. The adduct is in stick model. Hydrogen atoms are deleted for clarity. The color code is the same as in the left-hand panels. These images suggest that the B[a]P lesion can be translocated past the β-hairpin; while the upper part of the β-hairpin remains in contact with domain 1B, the bottom part moves away in order to enlarge the tunnel. The best representative structures are shown. Stereo views are given in Figures S8 and S9.
Accommodation of the lesion on the outer strand: dynamic and solvent exposed
When the B[a]P adduct is on the outer strand in Model 4 the hydrophobic adduct fits nicely into a shallow cavity of the UvrB protein between the β-hairpin and domain 1A (Figures 5 and S10). The system is more dynamic than in the other models as shown by the RMSD plots of the damaged DNA (Figure S2). While there is substantial solvent exposure of the B[a]P aromatic rings, they appear to be minimizing this property by their neat fit into the UvrB cavity with further shielding by the flipped out base C17. Nonetheless the outer strand model has the greatest solvent exposure for the damaged nucleotide. We computed the solvent exposed surface area for the damaged nucleotide in the most representative structure in each of our models using Discovery Studio, and obtained the following values: Model 1 (pocket), 63 Å2, Model 2 (tunnel), 36 Å2, Model 3 (Gate), 151 Å2, Model 4 (outer strand), 269 Å2.
Figure 5.
Model 4 of the adduct positioned in the DNA outer strand in the UvrB complex. The UvrB protein is shown as a surface model. The β-hairpin is shown in cyan. The damaged DNA is indicated in cartoon mode. The DNA inner strand is shown in orange and the outer strand in yellow. The largely solvent-exposed B[a]P-dG adduct is shown in CPK model with G6* in blue and B[a]P in magenta. The best representative structure is shown. A stereo view is given in Figure S10.
Flipped out bases in our models
Our initial models contained flipped out bases, consistent with crystallographic, mutation, and fluorescence data (21,24-26,41). Specifically, in the pocket model (Model 1), the unmodified base G16 in the complementary strand is flipped out and stacked with Tyr95. The base C17, also in the complementary strand but opposite the lesion (Figure 1C), is also flipped out of the duplex in order to wrap the outer strand around the β- hairpin. The B[a]P-modified base G6* is flipped into the pocket and is stacked with Phe249. The C7 on the 3′ side of the damaged base G6* is also flipped out. In Model 2 and Model 3, the bases in the analogous positions (yellow colored bases in Figure 1B) are flipped out. In Model 4, the B[a]P-modified base G16 is flipped out and also stacks with Tyr95. However, the most representative structures following our MD simulations showed variable flipping patterns as summarized in Table 1 and shown in Figure S4. For example, in model 1, the C7 which is 3′ to the lesion rotated from its initial flipped out position, so that it was mainly stacked with T8 after simulation for 1 ns (Figure S5).
Table 1.
Flipping and stacking properties of the bases in representative structures of each model. a
| Model 1 (Pocket) |
Model 2 (Tunnel) |
Model 3 (Gate) |
Model 4 (Outer) |
|
|---|---|---|---|---|
| Base 6 | F/S | F/S | F/S | F/S |
| Base 7 | N | F | F | F |
| Base 16 | S | N | N | S |
| Base 17 | F | N | N | F |
F, flipped out bases; S, stacking interaction: base 6 stacks with Phe249, base 16 stacks with Tyr95; N designates no flipping or stacking with amino acid residues. Figure S4 shows these flipping and stacking properties for all the models.
DISCUSSION
Inner and outer strand models: the (+)-cis-B[a]P-N2-dG adduct can be accommodated in UvrB on the inner strand and can be housed in a pocket behind the β-hairpin
Since a crystal structure of a UvrB / DNA complex is available which reveals that the DNA duplex is partially separated by the β-hairpin protruding into the double helix, the question of whether the damaged base is located on the inner or the outer DNA strand, relative to the hairpin, has remained a matter of debate (26). In a repair study, DNA photolyase bound to a pyrimidine photodimer was shown to stimulate incision by the UvrABC repair system (29). DNA photolyases catalyze the cleavage of the cyclobutane ring, using blue or near-UV light as the energy source (51,52). A crystal structure of the photolyase / DNA complex (28) showed that the damaged pyrimidines were flipped out into the photolyase pocket. These observations suggest that in this special case, the UvrAB complex recognizes DNA photolyase bound to the pyrimidine dimer, which is apparently located on the outer strand of the UvrB-DNA complex to avoid steric interference of the two repair systems (26). Our Model 4 shows that if the damaged base is on the outer strand, the lesion as well as its 3′-flanking neighbor base would both be exposed to the solvent (Figures 5 and S10), although the B[a]P ring system is accommodated in a cavity between the β-hairpin and domain 1A. This model seems to be in disagreement with fluorescence studies (21) which suggested that the base flanking a cholesterol residue-modified lesion on the 3′-side is flipped into a hydrophobic cavity of the UvrB protein. However, in our Model 4 C17 is solvent exposed (Figure 5). In addition, for efficient UvrC incision to occur, a base on the unmodified strand must be flipped out and interact with Tyr95 (21,22). It was suggested that this base is the 5′-nearest neighbor to the partner base in the complementary strand opposite the lesion. However, G6 in Model 4 is flipped out but stacked with Phe249 (Figure S4). These considerations suggest that the damaged base is positioned on the inner strand.
The crystal structures of the UvrB/DNA complex provide further insights into the location of the damaged base. The structure used as our modeling template suggests the existence of a binding pocket behind the β-hairpin which could be a location where the modified base can be accommodated (26). However, the structure of UvrB bound to a fluorescein-adducted DNA duplex suggested a “steric gate” model of the UvrB protein for this much larger-size adduct (26,27). In this model there is a narrow tunnel between the β-hairpin and the UvrB domain 1B (also shown in our Model 3, Figure 4) which accommodates the undamaged DNA single strand; however, a large lesion such as fluorescein with its long and flexible linkage to the modified base cannot pass through the tunnel. Hence, translocation is blocked and the binding of UvrC and the subsequent incisions would be triggered by this blocked translocation. The “steric gate” model can only be supported if the adduct is located on the inner strand, but is stopped before entering the tunnel behind the β-hairpin.
It is possible that the location of the damaged base in the DNA – UvrB complex, i.e., before UvrC is recruited, depends on the nature of the lesion and the specific recognition mechanism. For example, while the hydrophobic pocket behind the β-hairpin appears to be a plausible site for accommodating the hydrophobic (+)-cis-B[a]P-N2-dG adduct, a hydrophilic lesion likely would not enter this pocket; one could speculate that it might remain in the tunnel if it were small enough to enter it. In the case of UvrABC mediated repair of cyclobutane pyrimidine dimer bound by photolyase, the cyclobutane pyrimidine ring would have to be rotated outward or could possibly be located on the outer strand, to accommodate bound DNA photolyase. However this would require UvrB to engage the non-damaged strand behind the hairpin. Perhaps, if the stoichiometry of UvrA and UvrB is two molecules of each as reported by Goosen and co-workers (10,53), one of the UvrB's could engage either strand once a helical perturbation is first sensed by UvrA. However, in the case of bulky adducts, such as cholesterol, fluorescein, menthol, and benzo[a]pyrene-derived adducts, which require that UvrC be utilized for incising more than 10 base pairs around the damaged site, the positioning of hydrophobic adducts on the inner strand appears favored , as suggested by the crystal structures(26,34,35), fluorescence experiments(21,22), and our models. Current knowledge of the repair mechanisms (7) and our models, suggest that UvrB acts as a gauge to accurately recognize the location of the damaged base; the UvrC then binds to the UvrB/DNA complex to specifically incise the damaged strand at defined sites on the 3′ and 5′ sides of the damaged base. The UvrC endonuclease does not necessarily need to interact directly with the adduct in order to perform the incisions. A recent crystal structure of the C-terminal half of UvrC revealed a likely binding mode of this enzyme with DNA, via a positively charged patch on the enzyme's surface (54). The full UvrB/UvrC interaction mechanism will undoubtedly become clearer as crystal structures of such complexes become available.
DNA-protein crosslinks are also subject to repair by the UvrABC system if the protein is less than ~12-14 kDa in size and also may be repaired by human NER (55). In our models, such unusually bulky lesions would need to be situated either at the steric gate (Model 3) or on the outer strand (Model 4).
β-hairpin “breathing” may facilitate lesion translocation behind it
One important feature of the UvrB protein is its ability to partially separate the damaged DNA duplex into its separate strands and locate the lesion by short translocations of the damaged DNA strand through the β-hairpin motif (26,27,35). To investigate translocation, we built 3 models to simulate the trajectory of the adducted DNA translocating through the β-hairpin in three consecutive steps. Model 3 shows the adduct positioned at the gate of the tunnel between the β-hairpin and domain 1B. Model 2 shows the adducted DNA translocated one base further in the 3′-direction of the damaged strand, with the B[a]P-modified base positioned in a location between the β-hairpin and domain 1B. Model 1, the pocket model, shows the DNA translocated one additional base step and the adduct has passed through the tunnel.
Model 2 reveals that the β-hairpin does not need to be completely separated from domain 1B in order to allow the damaged DNA with an adduct the size of the (+)-cis-B[a]P-N2-dG to translocate through the narrow tunnel. The bottom part of the β-hairpin changes it's conformation to accommodate the bulky adduct, but the top is still in strong contact with domain 1B. On the other hand, a padlock model has been proposed (7,15), which suggests that the β-hairpin can separate from domain 1B to insert itself between the double strands of the damaged DNA, so that one strand is clamped between the β-hairpin and domain 1B. This separation would be required at the time of initial binding and separation of the damaged DNA duplex, at least for the case of closed circular DNA. The padlock model does not require translocation through the hairpin, although it may still occur after one DNA strand is clamped by the β-hairpin. The flexibility of the β-hairpin, seen in our simulations, suggests that the extent of hairpin breathing could vary, depending on the lesion shape and size, and that the hairpin could open as in the padlock model by separating from domain 1B if necessary. In the most extreme case, the lesion may be completely blocked at the tunnel entrance as in the “steric gate” model (27). A recent fluorescence study suggests the possibility that the nature of the lesion determines how far the damaged strand can translocate behind the β-hairpin (22). A cholesterol lesion linked directly to the DNA backbone is suggested to be translocated further behind the β-hairpin as compared to a menthol lesion which is directly linked to a base.
Hindered lesion translocation in UvrB may provide insights into the observations of Zou et al. (19), which show that in the case of the (+)-cis-B[a]P-N2-dG lesion, 3′ incision occurs at the fifth, sixth and seventh phosphates 3′ to the lesion, with the extent of incisions decreasing in the order of fifth > sixth > seventh phosphates. This was also observed for DNA photoproducts in different sequence contexts (17). These three different incision modes, producing fragments of increasing length, might be explained by our three models: the pocket model, the tunnel model, and the gate model; together, these models might explain incisions at the fifth, sixth, and seventh phosphates on the 3′-side of the lesion, respectively. If the UvrC dual incision is facilitated by translocation stalling (7), the non-uniform probabilities of 3′-incisions at the phosphodiester sites could result: the relative incision rate at the 7th phosphate would be lowest when the lesion reaches the gate suggesting that the adduct is stalled at this position for the shortest time. A further translocation would place the lesion within the tunnel where it may reside for a longer time, thus increasing the probability of incisions at the sixth phosphate. Finally, the lesion is stalled in the pocket thus leading to the highest cleavage probability at the fifth phosphate. This hypothesis suggests a dynamic aspect of the dual incision mechanism that may depend on the size, shape, and conformation of the lesion (19).
Flip-out of bases and β-hairpin intrusion into the double-helix
The 2-AP studies by Malta et al (21) showed that several bases around the damaged site including the base immediately adjacent to the lesion on the 3′ side, as well as its partner base on the complementary strand are flipped out of the DNA duplex. Using two different lesions, namely a cholesterol lesion linked on the DNA backbone and a menthol modified thymine, a more recent fluorescence study suggested that the specific position of the flipped base on the 3′-side of the lesion is dependent on the type of lesion (18). However, flipping of the nucleotide on the undamaged strand was shown to be independent of lesion type, and both flipped bases are protected from solvent (22). Fluorescence measurements and experiments with mutants show that the Tyr95 and Phe249 stack with the flipped-out bases on the damaged and partner strands (21,24,25,41). Our modeling studies are consistent with the possibility that the type of lesion determines which specific base on the 3′-side of the lesion is flipped, since, as detailed in the Results section, flipping patterns differ in the different models (Table 1).
A flip-out mechanism appears to be common to different DNA repair pathways and is well established in the base excision repair (BER) pathway (56,57). In the NER pathway, a helix-opening mechanism (58) that is likely to be accompanied by base flipping (59), seems important for lesion recognition and subsequent incisions. The crystal structure of the yeast Rad4 NER protein, the orthologue of the human NER recognition factor XPC, shows that the partners of the cyclobutane pyrimidine photodimer (CPD) damage are flipped out (36). The extrusion of a base out of the duplex originates from local duplex distortion/destabilization by the lesion which facilitates intrusion of a β-hairpin between the strands at the lesion site. Thus the prokaryotic (26) and eukaryotic (36) NER lesion recognition mechanisms share common structural features such as base flipping and β-hairpin intrusion between the strands of the DNA duplex at the lesion site.
CONCLUSION
We have utilized molecular modeling and molecular dynamics simulations evaluating all available experimental data to explore lesion housing and lesion translocation in UvrB. Our results suggest that the (+)-cis-B[a]P-N2-dG lesion is located on the DNA strand behind the UvrB β-hairpin. It might pass through a tunnel formed by the β-hairpin and domain 1B and be housed in a pocket behind the β-hairpin. The undamaged DNA strand is on the outside of the tunnel. Overall, our models suggest that lesion size and shape would govern possibilities for traversing the tunnel or remaining jammed in front of it prior to excision by UvrC, possibly explaining differently sized excision products. Additionally, our results suggest unifying insights concerning the functioning of the prokaryotic UvrB and the yeast Rad4 ortholog of the human XPC protein (36). Specifically both utilize the β-hairpin motif to intrude between the two strands of the damaged double helix and base flipping, manifesting common recognition features involving local thermodynamic destabilization with base extrusion (60). Interestingly, the UV DDB1-DDB2 damage recognition complex also utilizes hairpin intrusion and lesion extrusion for damage recognition (61).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, National Cancer Institute Grants CA28038 to S.B. and CA099194 to N.E.G, and the NIEHS, NIH, Intramural Research Program to B.V.H.. Partial support for computational infrastructure and systems management was also provided by Grant CA75449 to S.B. Computational support was provided in part by the National Science Foundation Partnerships for Advanced Computational Infrastructure. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations
- ROS
reactive oxygen species
- NER
nucleotide excision repair
- (+)-anti-B[a]PDE
(+)-7r,8t-dihydrodiol-t9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- (+)-cis-B[a]P-N2-dG
10R-(+)-cis-anti-B[a]P-N2-dG
- SD
steepest descent
- CG
conjugate gradient
- RMSD
root-mean-square deviations
- BER
base excision repair
- CPD
cyclobutane pyrimidine photodimer
Footnotes
Supporting Information Available
The molecular dynamics protocol and a translocation movie are provided. Figure S1 shows the superimposition of the ATP binding site when modeling the ATP from 1D9Z into 2FDC, and the superimposition of protein residues 420-460 of 2FDC to residues 418-458 of 2D7D to model a missing loop from 2D7D to 2FDC. Figure S2 shows plots of the all-atom RMSD of the current relative to the starting structure as a function of time for the whole UvrB/DNA complex, and for just the damaged and undamaged DNA strands. Figure S3 shows plots of the torsion angles χ, α′ and β′ for all complex models in the selected time frames for analysis. Figure S4 shows the flipping and stacking properties of the bases in representative structures of each model. Figure S5 shows the C7 base in the flipped-out and flipped-in position of Model 1. Figures S6-S10 show stereo views of Figures 2-5. Table S1 gives glycosidic torsion angle χ values in the initial models for MD simulations. Table S2 gives AMBER atom type, connection type, and partial charge assignments for the (+)-cis-B[a]P-N2-dG adduct. Table S3 gives AMBER atom type, connection type, and partial charge assignments for ATP. Table S4 gives AMBER atom type, connection type, and partial charge assignments for Mg2+. Table S5 gives box sizes and number of waters and counterions in the MD simulation initial models. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 4.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 5.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 6.Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol. 2006 doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 7.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. How relevant is the Escherichia coli UvrABC model for excision repair in eukaryotes? J Cell Sci. 1991;100(Pt 4):687–691. doi: 10.1242/jcs.100.4.687. [DOI] [PubMed] [Google Scholar]
- 9.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci U S A. 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhoeven EE, Wyman C, Moolenaar GF, Goosen N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. Embo J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DellaVecchia MJ, Croteau DL, Skorvaga M, Dezhurov SV, Lavrik OI, Van Houten B. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J Biol Chem. 2004;279:45245–45256. doi: 10.1074/jbc.M408659200. [DOI] [PubMed] [Google Scholar]
- 12.Pakotiprapha D, Inuzuka Y, Bowman BR, Moolenaar GF, Goosen N, Jeruzalmi D, Verdine GL. Crystal Structure of Bacillus stearothermophilus UvrA Provides Insight into ATP-Modulated Dimerization, UvrB Interaction, and DNA Binding. Mol Cell. 2007 doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Lu M, Tang MS, Van Houten B, Ross JB, Weinfeld M, Le XC. DNA wrapping is required for DNA damage recognition in the Escherichia coli DNA nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0902281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage sensing complex in bacterial nucleotide excision repair. J Biol Chem. 2009 doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theis K, Skorvaga M, Machius M, Nakagawa N, Van Houten B, Kisker C. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat Res. 2000;460:277–300. doi: 10.1016/s0921-8777(00)00032-x. [DOI] [PubMed] [Google Scholar]
- 16.Van Houten B, Illenye S, Qu Y, Farrell N. Homodinuclear (Pt,Pt) and Heterodinuclear (Ru,Pt) Metal-Compounds as DNA-Protein Cross-Linking Agents - Potential Suicide DNA Lesions. Biochemistry. 1993;32:11794–11801. doi: 10.1021/bi00095a007. [DOI] [PubMed] [Google Scholar]
- 17.Myles GM, Van Houten B, Sancar A. Utilization of DNA Photolyase, Pyrimidine Dimer Endonucleases, and Alkali Hydrolysis in the Analysis of Aberrant Abc Excinuclease Incisions Adjacent to Uv-Induced DNA Photoproducts. Nucleic Acids Research. 1987;15:1227–1243. doi: 10.1093/nar/15.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Houten B, Gamper H, Holbrook SR, Hearst JE, Sancar A. Action Mechanism of Abc Excision Nuclease on a DNA Substrate Containing a Psoralen Cross-Link at a Defined Position. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 20.Sibghat U, Sancar A, Hearst JE. The repair patch of E. coli (A)BC excinuclease. Nucleic Acids Res. 1990;18:5051–5053. doi: 10.1093/nar/18.17.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair. J Biol Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 22.Malta E, Verhagen CP, Moolenaar GF, Filippov DV, van der Marel GA, Goosen N. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair (Amst) 2008 doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Moolenaar GF, Schut M, Goosen N. Binding of the UvrB dimer to non-damaged and damaged DNA: residues Y92 and Y93 influence the stability of both subunits. DNA Repair (Amst) 2005;4:699–713. doi: 10.1016/j.dnarep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Moolenaar GF, Hoglund L, Goosen N. Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. Embo J. 2001;20:6140–6149. doi: 10.1093/emboj/20.21.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou Y, Ma H, Minko IG, Shell SM, Yang Z, Qu Y, Xu Y, Geacintov NE, Lloyd RS. DNA damage recognition of mutated forms of UvrB proteins in nucleotide excision repair. Biochemistry. 2004;43:4196–4205. doi: 10.1021/bi035992a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 27.Waters TR, Eryilmaz J, Geddes S, Barrett TE. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 2006;580:6423–6427. doi: 10.1016/j.febslet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 28.Mees A, Klar T, Gnau P, Hennecke U, Eker AP, Carell T, Essen LO. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 29.Sancar A, Franklin KA, Sancar GB. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc Natl Acad Sci U S A. 1984;81:7397–7401. doi: 10.1073/pnas.81.23.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 31.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 32.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo(a)pyrene diol epoxides. Nature. 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 33.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–7843. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eryilmaz J, Ceschini S, Ryan J, Geddes S, Waters TR, Barrett TE. Structural insights into the cryptic DNA-dependent ATPase activity of UvrB. J Mol Biol. 2006;357:62–72. doi: 10.1016/j.jmb.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 35.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. Embo J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007 doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 37.Moolenaar GF, Franken KL, Dijkstra DM, Thomas-Oates JE, Visse R, van de Putte P, Goosen N. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J Biol Chem. 1995;270:30508–30515. doi: 10.1074/jbc.270.51.30508. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- 39.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 40.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorvaga M, DellaVecchia MJ, Croteau DL, Theis K, Truglio JJ, Mandavilli BS, Kisker C, Van Houten B. Identification of residues within UvrB that are important for efficient DNA binding and damage processing. J Biol Chem. 2004;279:51574–51580. doi: 10.1074/jbc.M409266200. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. AMBER 8. University of California; San Francisco: 2004. [Google Scholar]
- 43.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 44.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 45.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 46.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 47.Simmerling C, Kollman P. MOIL-View: A program for visualization of structure and dynamics of biomolecules. Abstracts of Papers of the American Chemical Society. 1996;211:92. Comp. [Google Scholar]
- 48.Xie XM, Geacintov NE, Broyde S. Origins of conformational differences between cis and trans DNA adducts derived from enantiomeric anti- benzo[a]pyrene diol epoxides. Chem Res Toxicol. 1999;12:597–609. doi: 10.1021/tx990021a. [DOI] [PubMed] [Google Scholar]
- 49.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, et al. AMBER 7. University of California; San Francisco: 2002. [Google Scholar]
- 50.DeLano WL. 2002:0.99. on World Wide Web http://www.pymol.org.
- 51.Carell T, Burgdorf LT, Kundu LM, Cichon M. The mechanism of action of DNA photolyases. Curr Opin Chem Biol. 2001;5:491–498. doi: 10.1016/s1367-5931(00)00239-8. [DOI] [PubMed] [Google Scholar]
- 52.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 53.Malta E, Moolenaar GF, Goosen N. Dynamics of the UvrABC nucleotide excision repair proteins analyzed by fluorescence resonance energy transfer. Biochemistry. 2007;46:9080–9088. doi: 10.1021/bi7002235. [DOI] [PubMed] [Google Scholar]
- 54.Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 56.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 57.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. Embo J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 60.Scharer OD. A molecular basis for damage recognition in eukaryotic nucleotide excision repair. Chembiochem. 2008;9:21–23. doi: 10.1002/cbic.200700619. [DOI] [PubMed] [Google Scholar]
- 61.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.