Abstract
Marine Synechococcus spp and marine Prochlorococcus spp are numerically dominant photoautotrophs in the open oceans and contributors to the global carbon cycle. Syn5 is a short-tailed cyanophage isolated from the Sargasso Sea on Synechococcus strain WH8109. Syn5 has been grown in WH8109 to high titer in the laboratory and purified and concentrated retaining infectivity. Genome sequencing and annotation of Syn5 revealed that the linear genome is 46,214bp with a 237bp terminal direct repeat. Sixty-one open reading frames (ORFs) were identified. Based on genomic organization and sequence similarity to known protein sequences within GenBank, Syn5 shares features with T7-like phages. The presence of a putative integrase suggests access to a temperate life-cycle. Assignment of eleven ORFs to structural proteins found within the phage virion was confirmed by mass-spectrometry and N-terminal sequencing. Eight of these identified structural proteins exhibited amino acid sequence similarity to enteric phage proteins. The remaining three virion proteins did not resemble any known phage sequences in GenBank as of August 2006. Cryoelectron micrographs of purified Syn5 virions revealed that the capsid has a single “horn”, a novel fibrous structure protruding from the opposing end of the capsid from the tail of the virion. The tail appendage displayed an apparent three-fold rather than six-fold symmetry. An 18Å-resolution icosahedral reconstruction of the capsid revealed a T=7 lattice, but with an unusual pattern of surface knobs. This phage/host system should allow detailed investigation of the physiology and biochemistry of phage propagation in marine photosynthetic bacteria.
Introduction
Marine cyanobacteria are numerically dominant photoautotrophs in the global oceans 1; 2. Two genera, Prochlorococcus and Synechococcus, contribute significantly to global photosynthesis 3; 4; 5; 6; 7. Marine viruses which infect these genera, termed “cyanophages”, have been isolated from various environments ranging from estuarine to open ocean. The infectious cyanophage virion concentrations in each sampled oceanic regime range from that of the host cells to an order of magnitude lower 8; 9; 10; 11. Due to their presence and prevalence within the upper surface layers, cyanophage have been implicated in the control of the cyanobacterial community structure and primary production, and therefore also in the flow of nutrients and energy throughout the marine ecosystems 12; 13; 14; 15. Establishment of laboratory phage/host systems for propagating and studying the interaction of cyanophages with their marine photosynthetic hosts would be valuable.
Despite their very different hosts and habitats, all cyanophage isolated to date share morphological similarity with the three families of dsDNA tailed phages established for phages of more common laboratory bacteria 8; 9; 10; 16: Myoviridae, long-contractile tailed phages; Siphoviridae, long-noncontractile tailed phages; and Podoviridae, short tailed phages (family designations refer to morphology only, as phage phylogenetic relationships are demonstrably more complicated). Cyanophage genomes show a marked similarity with the genomes of enteric phages 17; 18; 19 in terms of both gene similarity and the presence of similar cassettes found within mosaic genomes 20; 21. The presence of similar phage morphologies and genome sequences between cyanophages and other phages indicates that these phages may be descended from a common ancestor. Since the evolutionary emergence of cyanobacteria predates enteric bacteria by many hundreds of millions of years, the cyanophages may in fact more closely resemble the ancestral phage types.
While five cyanophage genomes have been sequenced and annotated to date, few biochemical studies have been performed to analyze their structural proteins. Preliminary investigations of the molecular biology of five cyanophage isolates was undertaken by Wilson et al10. These long, non-contractile tailed phages were concentrated from large volumes of crude lysate via polyethylene glycol (PEG) precipitation and centrifugation, and electrophoresed through SDS-gels. Banding patterns indicated that these phages contained structural proteins in the approximate number and proportion as the Siphoviridae. Phage S-PM2, a contractile-tailed phage, was concentrated in a similar fashion as the phages of Wilson et al10 and analyzed by SDS-gels. After the genome was sequenced, virion proteins were further examined via mass-spectrometry 19. Their results indicated that the SDS-banding pattern of S-PM2 and structural proteins are similar to the T4-like phages.
In the development of a laboratory system for biochemical work on cyanophage proteins, we chose a short-tailed phage which was expected to have a simpler virion protein composition than a long tailed phage, to aid the protein identification process. Syn5 was isolated by Waterbury and Valois 8 from the Sargasso Sea on Synechococcus strain WH8109 (a member of the Marine A cluster of Synechococcus, clade II). At least 15 other cyanophage isolates also propagate on WH1809, identifying it as one of the more phage sensitive strains in culture as tested by Waterbury and Valois (1993) and by Sullivan et al9. Syn5 did not propagate on at least 19 other Synechococcus and Prochlorococcus strains tested by Waterbury and Valois8 and by Sullivan et al 9. Most other Podoviridae tested by these two groups had similarly narrow host ranges. We have developed a system for large scale growth and purification of intact and infectious Syn5 phage particles. The resulting virions were sufficiently numerous and pure to characterize the structural proteins, to yield genomic DNA for sequencing, and to reconstruct a three-dimensional icosahedral model of the capsid using cryoelectron microscopy.
Results
Conditions for propagating cyanophage at high titer
Synechococcus WH8109 cultures for phage propagation were grown in SN medium at 26°C under a continuous irradiance of 50μEm−2s−1 and with constant aeration provided by a gas dispersion tube. While marine Synechococcus normally follows a diel cycle, under continuous irradiance and aeration these cultures achieved a maximum growth rate of approximately three doublings per day and a maximum cell density of 109 cells/ml 22. Lysis occurred approximately 10 hours after infection, and was characterized by a change in color of the culture from the initial dark red to bright yellow after ~eight hours and light green after ~ten hours. Accompanying this change in color was a profound loss in the turbidity of the culture. The maximum measured Syn5 burst size of 30 virions per cell was recorded when cultures of WH8109 were in log phase and at a concentration of approximately 1×108 cells/ml (data not shown). This burst size is about a fourth of the reported burst sizes for phages like T7 (120 phage/cell) 23 and phi-YeO-3 (132 phage/cell) 24.
Cryoelectron microscopy of intact Syn5 virions
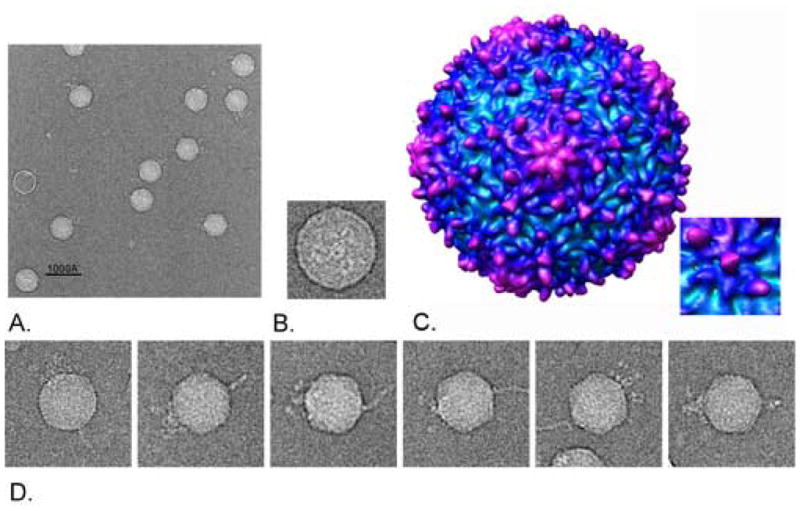
Electron micrographs of negatively stained virions (Figure 1) revealed the same short tailed icosahedral virion structure originally reported by Waterbury and Valois (1993). In an effort to obtain additional structural information, the particles were examined by cryoelectron microscopy (Figure 2). Individual images of the phage showed an isometric icosahedral capsid approximately 60nm in diameter, and a short knobby-appearing tail. Directly opposing the vertex of the capsid to which the tail was assembled was a novel protein structure, which we have named the “horn”. The horn is a slender elongated fibrous protrusion approximately 50nm long by 10nm wide at the base, tapering to 2–5nm wide at the tip. Every virion observed with an intact horn exhibited only a single horn, and only attached to the vertex directly opposite the tail (Figure 2A).. The horn was not visible in negatively-stained electron micrographs of Syn5, which may indicate that the horn structure was disrupted by the low pH or high ionic strength of the uranyl acetate stain.
Figure 1.
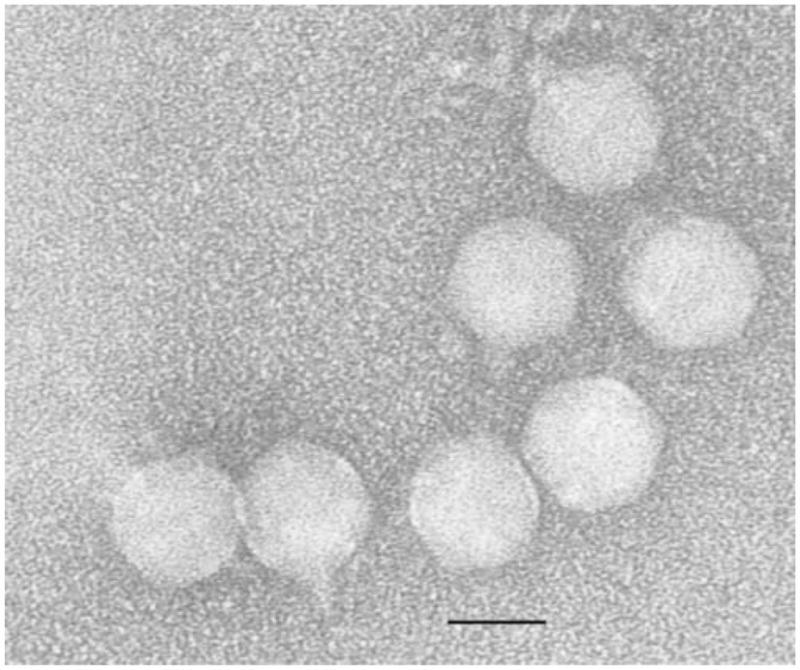
Electron Micrograph of Purified Syn5 Virions. Purified particles were placed on copper mesh grids and stained with 2% uranyl acetate. Particles exhibited the same morphology as originally observed by Waterbury and Valois; with an isometric icosahedral capsid approximately 60nm in diameter, and a short tail approximately 25nm in length.
Figure 2.
Cryoelectron micrographs of purified Syn5 particles and Icosahedral 18Å Reconstruction of the Capsid. Syn5 particles were flash-frozen and examined by cryoelectron microscopy. A) Syn5 virions. The extended horn is seen directly opposite the three-pronged tail on the isometric capsid. Scale bar = 100nm B) End view of the Syn5 particle. The tail tube is surrounded by tail proteins with threefold symmetry. C) 18Å Capsid Reconstruction with imposed icosahedral symmetry. The 18Å icosahedral reconstruction of the capsid is shown radially colored from cyan to purple with increasing radius. The map is in the T=7L form, but the hand of the capsid has yet to be resolved. D) Gallery of single Syn5 particles with horn structures.
When observed down the long axis, the phage tail exhibited three-fold symmetry (Figure 2B). An apparent tube is seen at the center and three extended structures, corresponding roughly to the positions expected for the tail structures seen in the side view in Figure 2D, surround the tube. Each of the three extended structures measure approximately 25nm by 5nm, and the apparent tube internal diameter is 6nm.
An 18Å-resolution icosahedral reconstruction of the Syn5 capsid demonstrated that the capsid is assembled with a triangulation number of 7, and appears to have significant depth variation in the proteins of the capsid surface (Figure 2C). While the pentameric and hexameric units characteristic of phage capsids are readily apparent, each hexamer appears to have a raised trio of protuberances (or “knobs”) along a single transect of the hexamer. The knobs fall in a straight line across the hexamer, with one in the center of the hexamer and two on opposite sides of the periphery of the hexamer. This arrangement is unusual in that to our knowledge there are no other examples of this arrangement of surface features among virus capsids of known structure. The positions of the knobs do not follow the quasi-equivalent positions of the capsid subunits. There are 180 knobs on the entire capsid, associated with the 60 hexamers. The central knob of each trio has a two-fold appearance, suggesting that it might be dimeric. The other two knobs show no obvious substructure at this resolution but appear to have volumes similar to the central knob. We speculate below on what proteins might make up these structures. There are no knobs associated with the pentamers, but there are five rather unusual curved ridges radiating out from each pentamer.
Thin-sections of Synechococcus infected with Syn5
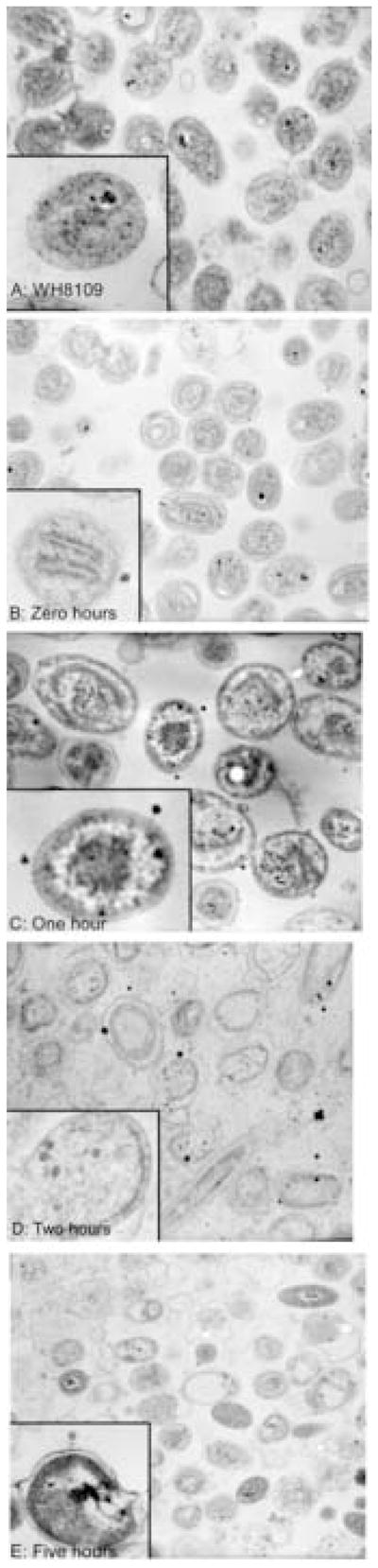
To observe the interaction of Syn5 virions with the host, ultra-thin sections of Syn5 infected cells fixed and embedded in resin were examined by transmission electron microscopy (TEM). Exponentially growing WH8109 cells were mixed with phage at a multiplicity of infection (moi) of 10 and incubated for various time periods at room temperature prior to fixation. Figure 3 chronicles a time course of Syn5 infection of WH8109. Figure 3A shows uninfected control cells. Cells were intact and unruptured; the thylakoid membranes were visible within many of the cells. Figure 3B shows the zero time point, in which phage and cells were mixed and immediately fixed with glutaraldehyde. The cells of the zero time point resembled the control cells, with intact visible cells.
Figure 3.
Thin-sections of Synechococcus WH8109 infected with Cyanophage Syn5. Time course of WH8109 cells infected with Syn5 in late exponential phase. A) Uninfected cells. B) Time Zero: phage were mixed with cells and immediately fixed with glutaraldehyde. C–E) Phage and cells were incubated for one, two, and five hours prior to fixation.
Fig. 3C shows infected cells one hour after infection. Numerous phage virions were adhered to the surface of the host cells, indicating that sufficient time had passed for adsorption to occur. There was no evidence of attachment to some particular site on the cells, or unusual mode of absorption. However, some of the virions appear to have elongated connections to the host cell surface, as if the tail structure was extensible. From the intensity of the staining, about 60% of the phage virions were empty of DNA while 40% still contained some DNA within their heads. The cellular structure was not systematically different from the uninfected or zero time cells. In particular, no virions were yet assembled within the host cells, an indication that one hour is still within the eclipse period of the phage’s life-cycle.
By two hours, Fig. 3D, 30% of the cells exhibited five or more assembled phage-like virions within the cell sections, distinguishable from the stain deposits noted below. Seventy percent of cells showed at least one internal phage virion. Some cells were visibly burst. By five hours, Fig. 3E, substantial numbers (~40%) of the cells within a field were ruptured and particles that appeared to be released virions were associated with some of these membranes. Given that complete lysis did not occur until approximately 10 hours after infection, the ruptured cells found at the later time points may be partially due to sensitivity of the phage-containing host cells to rupturing during the fixation or dehydration steps of the embedding process.
These thin-sections of WH8109 and Syn5 contained numerous small black deposits likely to be artifacts from the heavy metal staining step 25. These deposits could be distinguished from virions attached to the cells on the basis of size (in some cases), stain density, and shape. However, some of the intracellular dark spots could be carboxysomes. Nonetheless, their presence reduced the ability to monitor the development of new virions within the cells.
Genome sequence and gene identification
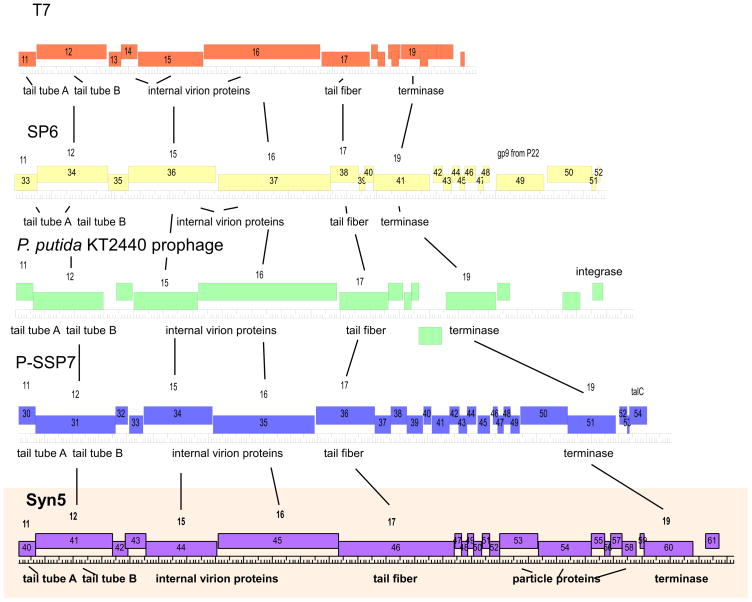
The Syn5 genome is 46,214 bp of double-stranded DNA, 55% G+C, including a 237 bp direct terminal repeat. Using the computer programs GeneMark, GLIMMER, and DNA Master, 61 open reading frames (ORFs) were identified as likely protein coding genes. The genes are all transcribed in the same direction (left to right as the map is drawn in Figure 4). The genes are tightly packed, with approximately 95% of the genome sequence devoted to protein coding genes. Translated Syn5 ORFs were compared to known protein sequences within the non-redundant (nr) database at http://www.ncbi.nlm.nih.gov/ using the BLASTp program 26. ORFs encoding sequences that matched known proteins with an e-value less than 0.001 are listed in Table 1 along with their putative function and the organism which contains the most similar sequence. Syn5 appeared to contain genes similar to those of the T7-like phages, as defined by Hausmann and Scholl et al 27; 28, including ten involved in DNA replication and eight encoding structural proteins. In terms of genomic organization, these recognizable T7-like genes demonstrated a high level of synteny between Syn5 and T7 (Figure 4). An additional three structural proteins were found in the virion by tandem mass-spectrometric analysis (see below).
Figure 4.
Map of the Syn5 Genome. The Syn5 genome is 46,214bp long with a 237bp terminal repeat. 61 genes are labeled. Genes identified with BLASTp as having similarity to known phage DNA replication genes are in blue, to structural proteins are in red, and to non-structural late genes in green. Unidentified genes are in magenta. Also labeled are the putative integrase (int), ribonucleotide reductase (nrt), thioredoxin (trd), and thymidylate synthase (thy) genes.
Table 1.
Identified ORFs within the Syn5 Genome. Each putative ORF within the Syn5 genome was compared to known protein sequences within the non-redundant database curated by NCBI using BLASTp. The most likely putative function of each predicted protein is listed, as well as the phage(s) and/or microbe(s) which contained the most closely related sequence(s). The phage or microbe with the most closely related sequence is listed first and that e-value is reported.
| ORF | Predicted Protein | Related Phage(s) or Microbes | E value |
|---|---|---|---|
| 1 | -- | P60 | 0.004 |
| 14 | -- | P60, Synechococcus elongatus | 4e-05 |
| 15 | RNA Polymerase | P60, P-SSP7, Vibriophage VP4, T7 | 5e-161 |
| 17 | Integrase | P-SSP7, Agrobacterium, Geobacillus, Burkholderia | 9e-26 |
| 21 | T7-like ssDNA binding protein | P-SSP7 | 9e-17 |
| 22 | Endonuclease | P-SSP7, Vibriophage VP4, Pseudomonas putida KT2440 | 9e-26 |
| 24 | Primase/Helicase | P60, P-SSP7, SIO1, phiYeO3-12, T3, Pseudomonas putida, T7 | 0.0 |
| 25 | Thioredoxin | P60 | 2e-6 |
| 27 | DNA polymerase | P60, Podovirus GOM, Podovirus SOG, Pseudomonas putida, Vibriophage VP4, T7 | 0.0 |
| 28 | -- | P-SSP7 | 4e-6 |
| 29 | Exonuclease | P60, P-SSP7, Pseudomonas putida, phiYeO3, T3, T7 | 1e-86 |
| 31 | -- | P60; Staphylococcus aureus phages 47, 96, 3A, phiN315, ROSA, 85, 53; Listeria phage 2389 | 3e-15 |
| 32 | -- | P60 | 6e-25 |
| 33 | Ribonucleotide reductase | P-SSP7 | 9e-162 |
| 34 | -- | P-SSP7 | 5e-62 |
| 35 | -- | P60, P-SSP7 | 2e-07 |
| 37 | Portal | P60, P-SSP7, Yersinia pestis phage phiA112, Pseudomonas putida, T7 | 0.0 |
| 38 | Scaffolding | P-SSP7, P60, Vibriophage VP4, T3, K1-5, T7 | 5e-30 |
| 39 | Capsid | P60, P-SSP7, Pseudomonas putida, T7 | 6e-74 |
| 40 | Tail tube A | P-SSP7, Pseudomonas putida, P60, T3, phiYeO3-12, T7 | 6e-26 |
| 41 | Tail tube B | P60, P-SSP7, Pseudomonas putida, Vibriophage VP4, T7 | 4e-140 |
| 42 | -- | P60, P-SSP7, epsilon15 | 2e-25 |
| 43 | Internal Virion Protein (gp 14) | P60 | 1e-10 |
| 44 | Internal Virion Protein (gp 15) | P60 | 3e-27 |
| 45 | Internal Virion Protein (gp 16) | P60, P-SSP7 | 5e-66 |
| 46 | Tail fiber | P-SSP7 (N-term 100aa) | 4e-19 |
| 47 | -- | P60 | 3e-13 |
| 49 | -- | P60, P-SSP7 | 2e-22 |
| 50 | -- | P60 | 3e-15 |
| 51 | HNH endonuclease | P60, P-SSM2, P-SSP7 | 8e-33 |
| 53 | Large protein involved in aggregation | Microbulbifer degradans | 0.001 |
| 54 | Outer membrane protein, putative RTX toxin | Burkholderia cepacia R18194, Shewanella oneidensis MR-1 | 3e-06 |
| 58 | Fiber | P-SSM4, P60 | 0.054, 0.21 |
| 60 | Terminase | P-SSP7, P60, Pseudomonas putida, T3, phiYeO3-12, T7 | 0.0 |
| 61 | Thymidylate synthase | Prochlorococcus marinus strain 9313, P60, S-PM2, P-SSM2, P-SSM4, Mycobacterium phage L5 | 4e-51 |
Other genes identified in the Syn5 genome include those encoding ribonucleotide reductase, thymidylate synthase, and thioredoxin. These enzymes are all thought to be used during phage DNA replication to scavenge host nucleotides. The Syn5 genes were similar to genes found in the cyanophages P-SSP7, P60, and S-PM218; 19. The phage-encoded thioredoxin in Syn5 may have a dual role in phage DNA replication, both as a co-factor of ribonucleotide reductase, and as a DNA polymerase (DNAP) accessory protein to increase the binding affinity of the DNAP to template DNA, similarly to the roles of thioredoxin in T7.
Holins are proteins that insert into the cell membrane and allow the endolysin access to the bacterial cell walls during phage infection, and regulation of their conformation determines the timing of lysis. The putative holin gene in the Syn5 genome, although lacking similarity to known holin sequences, has the organization of the lambda S holin. In lambda, two forms of the S holin (beginning at codon 1 and codon 3) are expressed in a two to one ratio; with the longer form acting as an inhibitor of the shorter. The expression of the two forms is controlled by a stem-loop located directly over the ribosome binding site of the gene 29. The translation of Syn5 gene 47 exhibited all of these holin-like characteristics: a transmembrane domain at the N-terminus of the protein, positively charged residues immediately preceding the transmembrane domain, and a putative regulatory loop prior to the gene. Finally, the putative holin has a similar location in the genome of Syn5 as the T7 holin does in its genome 30.
Despite the identification of a putative holin gene in the Syn5 genome, no corresponding endolysin is apparent in the genome sequence. Currently, only two cyanophage genomes, S-PM2 and P60, contain genes with recognizable endolysin sequences. In Syn5, there are multiple candidate genes of unknown function located immediately downstream from the putative holin which could encode an endolysin, but the sequences of those genes are not similar to anything in the NCBI database.
With the exception of P60, all of the other sequenced cyanophage contain at least one gene which encodes a photosynthetic protein, generally psbA, 31; 32. However, Syn5 contains no identifiable photosynthesis genes, consistent with the lack of psbA and psbB observed in the genomes of Synechococcus podoviridae by Sullivan et al 33
Transcription
Three E. coli RNAP promoters are located within the first 850bp of the T7 genome, and these are recognized and transcribed by E. coli RNAP, facilitating the entrance of the rest of the genome to the cell 30. Given the high level of synteny between Syn5 and T7, it seems plausible that the mechanism of infection of their respective host cells is likewise conserved, indicating that there may be Synechococcus RNAP promoters located within the first ~4000 bp of the Syn5 genome, prior to the phage-encoded RNAP. Cyanobacterial sigma70-like promoters exhibit high levels of similarity to the sigma70 promoters of E. coli 34. Therefore, it was possible to screen for these types of promoters using programs designed to search for these sites in enteric sequences, such as BPROM (www.softberry.com). Indeed, a number of early genes appeared to have sigma70-like promoter sequences, indicating that they may be transcribed by the host’s RNAP (Figure 4).
The later T7 genes are transcribed by the phage-encoded RNAP and therefore are located behind one of the phage’s 17 T7-RNAP promoters 30. These promoter sites are highly conserved ~30bp stretches culminating in ribosome binding sites and are located directly before genes such as the capsid protein, the scaffolding protein, the ssDNA binding protein, as well as directly after the RNAP gene 30. By aligning the analogous sequences in Syn5, it is possible to identify an approximately 10 bp sequence: 5′-CCTTAATTAACT-3′. This sequence, or a similar one, appears at several other sites within the middle/late part of the Syn5 genome (Figure 4). The earliest copies of the putative promoter sequences appear before the T7-like ssDNA binding protein and within the DNA primase/helicase gene. Other putative promoter sites include regions immediately preceding several unidentified ORFs located between the DNA replication genes and the structural proteins, as well as in the middle of the tailfiber gene. There is no obvious protein that could be expressed from this last promoter, so it is unclear what its function may be. However, both SP6 and T7 30; 35 also have promoter-like sequences of unknown function located within the middle of functional genes.
Syn5 has three apparent rho-independent terminators. As in T7, one of them is located after the major capsid subunit gene (Figure 4). The other two are near the two ends of the genome but somewhat differently placed than the corresponding T7 terminators.
Translation
The majority of the Syn5 genes begin with the start codon AUG, with only 5% of the 61 genes of Syn5 predicted to begin with an unusual start codon (GUG or UUG). This was similar to many other phages, including other short-tailed cyanophages, where start codons other than AUG are quite rare 36; and in contrast to at least one myocyanophage, S-PM2, where 15% of predicted genes used an alternate start codon 19.
Many cyanobacterial and chloroplast genes do not exhibit recognizable Shine-Dalgarno (S-D) ribosome binding sites, despite the presence of sequence complementary to the consensus S-D sequence at the 3′ end of cyanobacterial 16S ribosomal RNA 37. To examine the quality of the potential S-D sites within the Syn5 genome, the nucleotide sequence was examined with program DNA Master. S-D sequences upstream of bacteriophage T7 genes generally scored 300 or better. A score of 450 represents an exact match to the S-D consensus and 9 represents the lowest possible score. Random sequence scores between 50 and 100. In Syn5, only two genes (genes 50 and 54) exhibited S-D scores of ≥300; the rest scored between 280–80; with an average of 168. The phage genes with the most similarity to non-cyanophage genes had some of the lowest scoring S-D sequences, including portal (180), DNAP (150), and RNAP (128).
The lack of identifiable promoters and strong S-D sequences along with the occurrence of genes with overlapping start and stop codons may be an indication of translational coupling. This phenomenon, in which the translation of the downstream gene is dependent on the translation of the preceding genes, has been identified in T4 and suggested to occur in S-PM2. Thirty-five of the Syn5 ORFs could be translationally coupled by these criteria.
Proteomic analysis of virion proteins
Protein composition and stoichiometry of purified virions was examined using SDS-polyacrylamide gel electrophoresis and Coomassie Brilliant Blue staining. Phage virions were boiled in SDS gel buffer to fully denature all phage proteins prior to electrophoresis. The samples were electrophoresed through a 10% polyacrylamide gel containing SDS. Syn5 exhibited 12 distinct polypeptide chains, ranging in molecular weight from ~140kDa to ~15kDa (Figure 5). When Syn5 particles were electrophoresed through a 4–20% polyacrylamide SDS gel, no additional bands were visible (data not shown). With the sequence of the Syn5 genome, it became possible to identify the gene that encodes each protein using mass-spectrometry and N-terminal sequencing of proteins extracted from an SDS-gel. Mass-spectrometric analysis was performed with a linear trap quadrupole mass-spectrometer (LTQ), which yielded internal amino acid sequence from each of the detected peptides as well as total molecular weight of each peptide.
Figure 5.
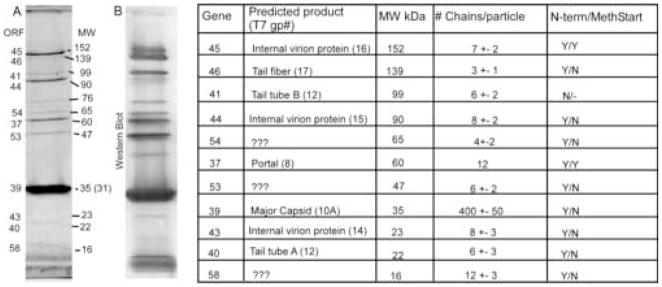
SDS-polyacrylamide gel and Western Blot of Syn5 virion proteins. Purified Syn5 particles were electrophoresed through 10% polyacrylamide containing SDS. A) Polyacrylamide gel stained with Coomassie Blue. 12 protein bands are visible and are marked with: the encoding gene number as determined by tandem mass-spectrometry, the approximate molecular weights in kDa as determined by relative mobility, polypeptide chain copy number rounded to the nearest whole number followed by the equivalent protein’s copy number in T7 in parentheses, confirmation of mass-spectrometric identification by N-terminal sequencing (Y for yes, N for no) and presence of N-terminal methionine on the protein (Y for yes, N for no). B) Western blot. After electrophoresis, proteins were blotted to PVDF membrane and probed with polyclonal antiSyn5 rabbit sera. The bands were then visualized using the ECF anti-rabbit kit (Amersham)
Mass-spectrometric analysis was performed on each of the protein bands labeled in Figure 5A. Based on stringent conditions, as defined by Sequest Search parameters, it was possible to match eleven of the twelve visible protein bands in Figure 5A to a corresponding gene in the Syn5 genome. An internal positive control lay in the fact that the protein identified by sequence comparison as the capsid protein, which is present in the highest copy number in most phage virions of any structural protein, was confirmed by mass-spectrometric analysis as the most intense band visible on the SDS-gel. It was also possible to assign the portal protein (encoded by gene 37) to the 60kDa band as well as the proteins encoded by genes 54, 53, 40, 43, and 58 to the remaining bands visible in Figure 5A, with one exception. For the 72 kDa band in Figure 5A, no peptides were detected that were not already assigned to one of the other bands on the gel. It is possible that this protein band represents a host protein which co-purified with the phage virions, as the library of sequences to which peptide spectra were compared was limited to sequences from a six-frame translation of the Syn5 genome. It was not possible to include the WH8109 genome in the library as it has not yet been sequenced.
N-terminal sequencing of the visible protein bands produced by electrophoresis of Syn5 virions through an SDS-gel was performed to confirm that the N-terminal sequence of a particular band matched the predicted N-terminal sequence encoded by the assigned gene. Eleven of the twelve virion proteins were visible after transferring them to a PVDF membrane, and these were subjected to multiple rounds of Edman degradation. The resulting amino acids were analyzed using HPLC and a reverse phase column to determine the N-terminal amino acid sequence.
Most of these sequences confirmed the gene assignments made through mass-spectrometry. Gene/protein assignments of 9 of the 11 Syn5 bands were confirmed by N-terminal sequencing. One band was not visible after transfer (encoded by gene 43), and one band (encoded by gene 41) had high background signals making amino acid detection impossible. Also noted in Figure 5A is whether or not the N-terminal methionine of each protein was recovered during degradation.
Polypeptide stoichiometry in phage virions
Phage virions are made out of many discreet polypeptide chains which are generally present at the same stoichiometry in each individual particle. Through a series of dilutions of Syn5 virions on SDS-gels stained with Coomassie Blue, we determined the rough copy number of each protein per Syn5 virion. The intensity of each protein band was determined using ImageQuant (Molecular Dynamics) software. Each band intensity, taken to be proportional to the total mass of protein, was normalized to the predicted molecular weight of the peptide chain. Determination the copy number of each chain was achieved by comparing the normalized intensity of each peptide chain to the normalized intensity of the portal band. The portal band was assumed to be present in a copy number of 12 chains per virion, a conserved number within all studied dsDNA tailed phages. By using this factor, it was possible to assign a copy number to each of the other peptides (Figure 5A). Copy numbers were rounded to the nearest whole number, as was the standard error. This method assumes that each of the proteins binds equivalent amounts of Coomassie Blue; however, this may not actually be true, and therefore the copy numbers could be significantly different than is determined by this calculation.
Each detectable Syn5 protein was similar in copy number to its corresponding protein in T7, with the exception of the tail fiber. T7 tailfibers are trimers, and each virion contains six homotrimers, for a total of 18 tail fiber peptide chains per virion. In contrast, the Syn5 virion appeared to contain only 3 tail fiber polypeptide chains, which may indicate that the Syn5 virion only has one tail fiber, or has a non-trimeric tail fiber; alternatively, the Syn5 tail fiber may not be homologous with the T7 tail fiber, or not sensitive to staining with Coomassie Brilliant blue or silver nitrate. Further information about the apparently 3-fold symmetric tail structure of Syn5 may help resolve these questions.
Immunogenicity of Virion Proteins
To prepare polyclonal antibodies, NZ white rabbits were immunized with purified Syn5 virions. A Western blot was then performed, using purified Syn5 virions. Virion proteins were electrophoresed through a 10% polyacrylamide gel containing SDS, electroblotted to PVDF membrane, probed with anti-Syn5 polyclonal rabbit sera and secondary mouse anti-rabbit antibodies, and visualized using ECF substrate.
The resulting visible bands are shown in Figure 5B. Most of the major structural proteins were detectable, including those encoded by genes 45, 46, 41, 54, 37, 53, 39 and 40. Also visible were the discreet bands that were determined by mass-spectrometry to be conglomerates of other structural proteins, including the bands at 76kDa, 38kDa, and 28kDa. Not detected were bands encoded by genes 43, 44, and 58. Proteins encoded by genes 43 and 44 may not have been detected as they are predicted to be internal virion proteins, and therefore may not have been well exposed to the immune system of the rabbit during antibody production. Gene 45 is also predicted to encode an internal virion protein, however, the corresponding protein found in T7 (gp 16) is the first protein that comes out of the virion after adhesion to a host cell to form the extensible tail that crosses the membrane of the host cell and facilitates DNA injection 38. If the protein encoded by gene 45 acts in a similar manner as gp16 of T7, and cellular membranes were present within the preparation, free virions might bind to these membranes and begin the process of infection. The protein encoded by gene 45 would then travel out of the virion. Therefore, this protein could have been exposed to the immune system of the rabbit and have antibodies created against it. The protein encoded by gene 58 has unknown function, but may also be located internally in the virion as it was not recognized by the rabbit antibodies.
The intensity of most of the bands of the Western blot varied in rough proportion to the intensity of the protein bands seen on the SDS-gel. Phage tail fibers are known to be highly antigenic and may stimulate antibody production at higher rates than other phage proteins 39. The Syn5 tail fiber band (encoded by gene 46) was more intense on the Western blot than it appeared on the SDS-gel, as were bands produced by Tail Tube B, and the proteins encoded by genes 53 and 54. This may indicate that these two unknown proteins are also highly antigenic and may play a role in host cell recognition.
Discussion
Since the identification of the first cyanophage and the discovery of their prevalence within the surface waters of the ocean, researchers have primarily focused on the ecology of these phages. We report here the development of a robust phage/host laboratory system, for investigating the biochemistry and physiology of phage infection of marine photosynthetic cyanobacteria.
Our methods of Synechococcus growth and phage purification resulted in reliable production of significant quantities of highly concentrated cyanophage virions (1012 virions/ml) that remained intact and infectious despite exposure to CsCl and various detergents. Using these samples, it was possible to initiate more synchronous infections for following growth of the phage with the host cell. The dependence of burst size on the growth rate of the host cells and more specifically, on the rates of DNA and protein synthesis, remain to be determined 40; 41; 42. In the field, under nutrient limited conditions where Synechococcus cells are growing more slowly on a diel cycle, the Syn5 burst size may be substantially smaller while the latent and eclipse periods may be longer than the calculated laboratory numbers. Observations supporting this have been made in the field by Steward et al. (1996) 43
Electron micrographs show that Syn5 virions have an icosahedral head approximately 60nm in diameter and a short conical tail approximately 10nm in diameter and 20nm long; closely resembling T7-like podoviridae. The cryoelectron micrographs of Syn5 virions also revealed a fibrous structure, which we have termed the horn, extending from a capsid vertex opposite the tail. It is possible that the horn of Syn5 may be an additional host recognition structure comprised of the highly antigenic proteins encoded by genes 53 and/or 54.
The thin-sections of Syn5 interacting with its host, WH8109, demonstrate that these virions bind to the outer surface of the host cells via their tails within one hour. While bound, the phage virions appear to have a longer tail than unbound virions, which could indicate the presence of an internal T7-like extensible tail. During T7 infection, three proteins found inside the capsid of the free phage virions form a bridge from the capsid through the outer membrane of the host cell, facilitating the initial entry the phage DNA to the cell 44. Similar proteins may exist within Syn5 and aid infection of WH8109 in the same manner.
The genome sequence of Syn 5 shared similarity with the genomes of other T7-like phages, both in terms of organization of the genome as well as similarity of genes at the amino acid sequence level (Figure 6). There were no similarities to the marine phages of the Corticoviridae type, such as PM2, described by Bamford and coworkers. In T7, the structural genes are located towards the end of the genome, as are the Syn5 genes. In T7-like phages, not only the location but the specific order of major structural genes appears to be conserved: portal, followed by scaffold, major/minor capsid, tail tube A, tail tube B, several internal virion proteins, and lastly, the tail fiber. The similarity of this genomic region within these phages is also apparent in the relative lengths of these genes as well. Given high level of conservation exhibited in gene location, order, and length within the T7 structural arm, the plasticity at the extreme right end of the genome becomes unmistakable. T7 appears to be the most streamlined of these phages with the fewest total genes in this area, while the genomes of SP6, Pseudomonas putida KT2440 prophage, P-SSP7, and Syn5 demonstrate that this type of phage is capable of supporting an additional module of genes that is several kb in length without apparent detriment. The gene content of the additional module is not conserved with regard to sequence, length, or number of genes in the five examples shown here; suggesting that this area of the T7-like genome may be able to support a wide variety of inserts which might provide a competitive advantage to the phage in a particular ecological niche.
Figure 6.
Structural arms of short-tailed phage genomes. Comparison of structural arms of five T7-like phages. Genes are represented by rectangles, and labeled both according to T7 nomenclature (above) and the number at which they occur when counting genes from the left end of the genome to the right end of the genome (inside). Each full length hatch mark on the ruler = 1kb. The synteny and conservation of length of the T7-like genes at the right end of the genome is readily apparent, as is the plasticity at the extreme right end genomes with regards to the inclusion of an additional gene module. Figure generated by DNA Master.
Cyanophage Syn5 virion proteins were directly analyzed through SDS-polyacrylamide gel electrophoresis, Western blotting, mass-spectrometry, and N-terminal sequencing. Syn5 virions exhibited 12 distinct bands when electrophoresed through a SDS- polyacrylamide gel, ranging from approximately 154 kDa to 15 kDa. Using tandem mass-spectrometry and N-terminal sequencing, it was possible to assign 11 cyanophage structural proteins to predicted genes within the Syn5 genome, including 8 proteins similar to the T7 structural proteins: portal, major capsid, tail tube A and B, internal virion proteins, and tail fiber (Figure 5B). The three Syn5 proteins without similarity to T7 proteins corresponded to large genes located at the right end of the genome, beyond the tail fiber. Two of these genes, 53 and 54, encode large proteins, 47 and 65 kDa respectively. The third gene, 58, encodes a 16 kDa protein, the smallest protein detected in the virion via SDS-gel electrophoresis. The larger two proteins were both similar to known fibrous sequences: the protein encoded by 53 is similar to a large protein involved in aggregation in Microbulbifer degradans, and the protein encoded by 54 is similar to the RTX protein and outermembrane proteins of Shewanella and Burkholderia. Both Syn5 proteins were highly antigenic, in a similar manner as the putative tail fiber, as shown by the abundance of antibody recognizing them when examined by Western blotting.
The cryoEM structure of the Syn5 capsid shows that it has the triangulation number, T=7, expected for a phage that packages this size of genome, but it also shows some unexpected features in the form of the surface knobs described above. Features of this general appearance on the surface of capsids are often indicative of “decoration proteins”, phage-coded proteins that add to and stabilize the capsid during capsid maturation. However, we do not believe the Syn5 knobs are decoration proteins, for two reasons. First, our analysis of the protein composition of virions (Figure 5) shows no candidates for proteins present in the large number of copies per virion needed to account for the knobs in the structure. Second, the positions of the knobs mean that a decoration protein would need to bind to two non-equivalent positions on the capsid: at the center of the hexamer and at two positions at the periphery of the hexamer, but not at the other four symmetry related positions at the periphery of the hexamer.
We suggest instead that the knobs are made of portions of the major capsid subunits themselves. Although there is no high resolution structure for the Syn5 capsid protein or its T7 homologue, there are strong suggestions that all capsid proteins of tailed phages, as well as Herpesviruses, share the same basic polypeptide fold 45, which is known at high resolution for the capsid of phage HK97 46; 47. In HK97 the N-terminal arm of the capsid subunit is disordered in the procapsid but in the mature capsid it wraps tightly around the adjacent subunit, presumably serving to knit the structure together 47; 48. We speculate that the N-terminal arms of the Syn5 subunit serve the same stabilizing function but do so by associating in pairs on the outside of the capsid and forming the knobs. The pairing scheme implied by this model—two pairs of subunits from opposite sides of the hexamer forming the two peripheral protuberances and the remaining two subunits forming the central protuberance by joining across the center of the hexamer—produces a hexamer with two-fold rather than six-fold symmetry. This feature of the hexamers of the mature Syn5 capsid may derive from the strong two-fold character of the hexamers observed in all known T=7 procapsids, including procapsids of phage T7 and presumably Syn5. The pentamers, with an odd number of subunits, do not have any associated knobs. Possibly the N-terminal arms of the pentamer subunits form the five radial curved ridges that surround the central part of the pentamers.
The presence of the horn on Syn5 raises interesting questions with respect to assembly of the virion. In the assembly pathway of many well-studied tailed phages, assembly of the icosahedral capsid is thought to proceed by the rapid addition of capsid subunits to an initial nucleus of proteins which contains the dodecameric portal ring. As the formation of the nucleus – onto which the tail proteins later assemble – occurs at a much slower rate than that of capsid subunit addition, the mature virion is unlikely to have multiple tails. The portal containing complex extends into the interior of the capsid, as was demonstrated in an asymmetric cryoEM reconstruction of T7-like Salmonella phage epsilon15 49, but it does not extend as far as the opposite vertex, at least in the mature virion. Thus for the known tailed phages, all the minor capsid components are located at a single vertex. A similar statement can probably be made for the capsids of other dsDNA viruses with a unique capsid vertex, including bacteriophages of the Tectiviridae such as PRD1 50, members of the Corticoviridae such as marine PM2 51, and members of the Phycodnaviridae found abundantly in the marine environment, such as PBCV1 52. None of these examples provide clear indications of how the vertex opposite the tail might be identified for addition of the horn because it appears that the presence of a distinct structure at this position is unique to Syn5 among studied viruses. We speculate that the prohead assembly scaffold could identify the horn vertex if it assembled with cylindrical symmetry under the influence of the portal complex.
The study of the cyanophage structural proteins, specifically the host recognition proteins, is the first step towards understanding cyanophage/host interactions and the rates at which these interactions occur in the oceans. Elucidation of these rates is critical towards determination of the amount of lateral gene transfer between seemingly unrelated host organisms. With an estimated ≤0.0002% of phage genomic sequences sampled globally, and the presence of genes of unknown function within the majority of phage genomes, phages represent one of the largest reservoirs of unknown genomic sequences on Earth 53. As there are numerous examples of prophages conferring new abilities on infected host cells with detrimental effects on human health 54; 55, the potential exists for a novel phage/host combination with such detrimental effects to arise from promiscuous gene transfer. The increases of E. coli and other sewage bacteria in the coastal areas of the oceans from human activities highlights the questions of how much phage mediated transfer exists between these bacteria and the marine cyanobacterial genera (which produce a significant portion of our oxygen); and what effect, if any, this gene transfer may have. Further analysis of the cyanophages may yield insight to these areas.
Materials and Methods
Cell growth
Synechococcus strain WH8109 (herein after referred to as WH8109) was grown in SN media 56 at 26°C under continuous light at an irradiance of 50μEm−2s−1 supplied by cool white fluorescent light (40W bulbs). WH8109 was grown in a culture vessel with a glass fritted aerator, constructed from a 2L rectangular polycarbonate bottle (Nalgene) and the three-holed polypropelene cap (Nalgene). Silicon tubing (Cole-Parmer) was attached to an aquarium pump, filtered through a 0.4μ bacterial air vent (VWR) and attached to the air input of the culture vessel. Opposite the air input, within the culture vessel, was attached a glass–fritted aeration tube (VWR) such that the aeration tube ended just above the bottom of the vessel. The air output was kept sterile via a piece of silicon tubing twisted into a Pasteur loop. The final port in the cap was kept closed except during sampling, during which it provided easy access to the culture within the vessel. Cells counts were performed by filtering culture samples onto a 0.4μ polycarbonate filter and quantitatively analyzing the filter using epifluorescence microscopy. Ten microscope fields were counted per filter, with between 30 and 100 cells per field, and the average was used to determine cell concentration 2.
Phage Purification and Concentration
Phage were concentrated and purified using a modified version of polyethelene glycol (PEG) precipitation 57. WH8109 was grown to a density of 1×108 cells/ml at 26°C in SN media at an irradiance of 50μE m−2s−1. Syn 5 was added at a moi of 0.1. The culture was incubated until lysis, approximately 10 hours after infection (noted by a change in turbidity and color). The crude lysate was spun at 8,000 g for 15 minutes at 4°C to pellet cellular debris. The supernatant was then filtered through a Whatmann glass fiber filter and a 0.4μm polycarbonate filter to further remove cellular debris, specifically cell membranes, which may bind free phage and prevent their recovery. After the incubation, the lysate was moved to a 4°C room, and NaCl was added to a final concentration of 0.5M and stirred to dissolution. PEG 8000 was added to a final concentration of 10%w/v, mixed to dissolution, and incubated with stirring for two hours at 4°C. Phage were then pelleted by centrifugation at 9,000g for 30 minutes. The supernatant was poured off, and the pellet was resuspended by gentle stirring at 4°C in an appropriate amount of SN (10–15ml per liter of crude lysate) supplemented with 50mM Tris-HCl, pH 8.0 and 100mM MgCl2.
Several non-ionic detergents and a range of salt concentrations were tested in conjunction with PEG precipitation to determine the effect on cyanophage yield. Variations in salt concentrations were tested to determine if the addition of 0.5M NaCl was optimal when working with phage in a seawater-based medium. Maximum phage yields were recovered when 0.5M NaCl was added to the phage solution prior to PEG precipitation 22. Initially, detergents were added prior to phage precipitation to minimize interactions between cyanophage and any remaining cellular membranes and to allow more cyanophage to be recovered from the lysate; however none of the detergents tested (Triton X-100, Brij 58, Elugent, Tween-20, and NP40) appeared to have a significant effect on cyanophage yield 22. Cyanophage concentration and purification for all remaining experiments was therefore undertaken by the standard protocol for PEG precipitation of phage developed by Yamamoto et al 57.
The suspension was layered onto a step-gradient made of 20% sucrose (w/v), ρ=1.4 CsCl, and ρ=1.6 CsCl. Each gradient layer was made with SN. The gradients were ultracentrifuged at 40,000 rpm for 1.5 hours in a SW50.1 rotor or 28,000 rpm for 4 hours in a SW28 rotor. An opalescent phage band was visible at the interface between the ρ=1.4 CsCl band and ρ=1.6 CsCl steps. The phage band was collected and dialyzed for 30 minutes against 1M NaCl, 100mM MgCl2, 50mM Tris-HCl, pH 8.0, and then against 100mM NaCl, 100mM MgCl2, 50 mM Tris-HCl pH 8.0 in a Slide-A-Lyzer dialysis cassette (Pierce).
Ultramicrotomy of phage infected cells
Synechococcus cells were grown to a density of 7×105 cells/ml, and Syn5 virions were added at a moi of 10. 25ml of infected culture were pelleted by centrifugation (10min at 8000g) and the supernatant was poured off. The pellet was fixed in a 1% glutaraldehyde solution buffered with 0.1M sodium cacodylate buffer on ice in the dark for 30 min. The pellet was then gently washed three times in buffer for at least an hour per wash and fixed again with 1% osmium tetroxide in 0.1M sodium cacodylate buffer. After three more washes in buffer with an hour per wash, the pellet was dehydrated with increasing concentrations of ethanol, 5 min at each of 50%, 75%, 90%, and finally 3 times at 100%. After dehydration, the pellet was infused first with 100% propylene oxide for one hour and then propylene oxide mixed 1:1 with Spurr’s Low viscosity resin (prepared according to manufacturer’s instructions) for one hour while being constantly rotated for even infusion. Finally, the pellet was infiltrated overnight in pure resin on a rotator, and then transferred to a 2ml BEEM capsule in fresh resin. The capsule was baked at 60°C for 24 hours. The hardened resin was trimmed with razor blades and glass knives to expose the sample, and then 70 nm ultra-thin sections were cut using a diamond knife (Diatome) and a microtome. The sections were transferred onto 200 or 300 uncoated mesh copper grids that had been briefly rinsed in 100% ethanol and distilled water. The sections on grids were then stained with 1% uranyl acetate for 15 minutes, washed three times in double-distilled water (ten quick immersions per wash), stained in 1% lead citrate for 4 minutes, and washed a final three times (ten quick immersions per wash) in double-distilled water. The sections were then examined with an electron microscope.
Cryoelectron microscopy
A Vitrobot (http://www.vitrobot.com) was used to flash-freeze a 3 μl aliquot of purified Syn5 virions onto a copper Quantifoil R2/2 grid. The sample was loaded onto a Gatan 626 cryoholder and imaged in a JEOL 2010F cryomicroscope operated at 200KV and at liquid nitrogen temperature. Using JAMES software 58, 362 images were collected on a Gatan 4K CCD, with a dose of 10–15 e/A2 and a defocus of 1–5 um at 40,000X magnification (CCD magnification of 55,360X). The virions were automatically selected with the program “ethane” 59 followed by manual screening using “boxer” from EMAN60; 61. The CTF parameters were determined using an automated routine (Yang, unpublished). ~5000 virions were refined and reconstructed with icosahedral symmetry imposed using SAVR62, with ~2,600 virions in the final map of 18Å resolution by 0.5 Fourier shell correlation. The map was visualized with Chimera (UCSF).
DNA sequencing
The Syn5 genome was sequenced at the Pittsburgh Bacteriophage Institute at the University of Pittsburgh (Pittsburgh, PA). Methods for DNA sequencing and contig assembly have been reported elsewhere 20; 35. Briefly, 10μg genomic phage DNA was sheared hydrodynamically and repaired with T4 ligase to produce blunt ends. 1 to 3kb fragments of blunt-ended DNA were ligated into the EcoRV site of the pBluescript II KS+vector. Ligated plasmids were transformed into E. coli XL1-Blue cells. Plasmids were recovered from clones via Eppendorf PerfectPrep Plasmid 96 Vac Direct Bind System purification kit (Eppendorf, Hamburg Germany). Inserts were sequenced from both ends using the Applied Biosystems BigDye v3.0 dye terminator chemistry and universal sequencing primers. Labeled reaction mixtures were separated and analyzed using an ABI Prism 3730 DNA analyzer with a 48 capillary array. Underrepresented areas of the genome were covered and assembly was achieved by design of oligonucleotide primers and whole genome template in further sequencing reactions. Approximately eight-fold coverage was obtained.
Sequence Assembly and Analysis
Sequence chromatograms were assembled and analyzed using the Phred, Phrap and Consed sequence analysis software 63. The Syn5 genome terminal repeat was verified by primer extension. The completed genome sequence was assembled in the proper orientation from the Consed assembly. Open reading frames were identified using GeneMark 64, Glimmer 65; 66, and DNA Master (J. G. Lawrence) (http://cobamide2.bio.pitt.edu) software and visual inspection. Translated ORFs were compared with known protein sequences using BLASTp 26. Sigma-70 promoter sequences were identified using the BPROM program; rho-independent terminators were identified using FindTerm; potential hairpins were identified with BestPal, all found at http://www.softberry.com. Transmembrane helix regions were identified with TMHMM67.
Phage protein analysis
CsCl-purified Syn5 virions were mixed with SDS-buffer, boiled for 3 minutes, and loaded onto a 10% polyacrylamide gel containing SDS. The samples were electrophoresed at 20mA constant current until the dye line ran off the end of the gel. The gels were stained with Coomassie blue. 12 protein bands were visible and their approximate molecular weights were determined by comparison to a protein standard (Broad Range Protein Marker, New England Biolabs).
The Syn5 structural protein bands were then excised with a razor blade and individually packaged in microcentrifuge tubes for tandem mass-spectrometric analysis. A slice of gel was included as a negative control and to provide background correction. The gel bands were digested with trypsin as according to the MIT Biopolymers trypsin digest protocol (proprietary). 2μL of the extracted sample was diluted to 50 μL with 0.1% acetic acid. 15 μL of this dilution was run through a reverse-phase capillary column using high-pressure liquid chromatography (HPLC) with an Agilent Model 1100 Nanoflow HPLC and into a linear trap quadrupole (LTQ) mass-spectrometer (Thermo Electron Corporation, FL), where the peptides were ionized with 35kV. Detected peptides, as represented by well-resolved peaks with a high relative abundance at a specific mass/charge, were fragmented. Spectra of the resulting B- and Y- ions produced by peptide fragmentation were recorded every 0.2ms. Resulting spectra were compared to a database produced from the six-frame translation of the entire nucleotide sequence of the Syn5 genome and assigned a putative peptide sequence. Quality spectra indicative of true peptide matches were determined by visual inspection, and the scores (XC, Delta Cn, Sp, RSp) computed by the Sequest Database Search Software 68.
N-terminal sequencing of structural proteins
Phage virions were electrophoresed through an SDS-polyacrylamide gel. Protein bands were transferred from the gel to PVDF membrane (Millipore) via a Criterion plate-electrode transfer apparatus (BioRad) in transfer buffer (10% methanol, 25mM Tris pH 8.3, 192mM glycine) at a constant voltage of 100V for 1 hour at 4°C. The membrane was stained with Coomassie Blue to visualize the transferred protein bands. Visible bands were subjected to multiple cycles of Edman degradation. Amino acids recovered from each band after each cycle were identified using High Pressure Liquid Chromatography (HPLC) with a reverse phase column in conjunction with amino acid standards. Amino acid assignment was based on quantity of each amino acid detected after each round compared to an amino acid standard; the highest quantity above background resulted in residue assignment.
Western blot analysis of Syn5 proteins
Purified Syn5 virions were prepared. Immune sera were produced at Covance Research Products, Inc. by the following protocol: 150 μl of a 1010 phage/ml solution was brought up to 1 ml volume with phosphate buffered saline (PBS), split into 0.5ml volumes, and injected into two New Zealand White Rabbits. Two boosts of antigen at the same concentration were performed at two week intervals. After six weeks, serum was collected from each rabbit, and another antigen boost was injected. After eight weeks, more serum was collected and a final antigen boost was performed. Final bleeds were performed ten weeks and twelve weeks after initial inoculation.
Purified Syn5 virions were electrophoresed at 20mA through a 10% polyacrylamide gel containing SDS. The gel was immersed in transfer buffer (20% methanol, 25mM Tris, 192mM glycine) for 15 min, and then electroblotted to PVDF membrane (Millipore) in transfer buffer using a Criterion electroplate transfer apparatus (BioRad) at 100V for 1hr at 4°C. The membrane was then stored in wash buffer (PBS + 0.1% Tween20, pH 7.5) overnight. The membrane was probed using an ECF anti-Rabbit antibody kit (Amersham) according to manufacturer’s instructions. Briefly, the membrane was placed in blocking solution (Blocking Agent (Amersham)(5%w/v) in wash buffer) for 1 hour at 4°C, then washed 3X for 15min, then 2X for 5min in wash buffer at 4°C. The membrane was then probed with polyclonal Syn5 rabbit sera diluted 1:10 in wash buffer for 1 hr at 4°C, and washed as above. Finally, the membrane was probed with 2° anti-rabbit antibodies (Amersham) diluted 1:10000 in wash buffer for 1hr at 4°C and washed as above. The membrane was then incubated at room temperature for 5 min with ECF substrate (Amersham) and scanned using a FluroImager 595 (Molecular Dynamics).
Acknowledgments
We thank John Waterbury and Freddie Valois for their kind donation of Syn5 and WH8109; and Ian Molineux, Matt Sullivan, Debbie Lindell, Penny Chisholm, Craig Peebles, Jeffrey Lawrence, and the members of King, Hendrix, Hatfull, and Chiu labs for helpful discussions. WHP was supported by NSF grant EIA0225609 to JK. PRW and JK received funding from NIEHS grant 1-P50-ES012742 and NSF grant OCE-0430724. Work in Pittsburgh was supported by NIH grant GM51975 to RWH, GFH and JGL. Work in Houston was supported by the Robert Welch Foundation and NIH grant P41 RR02250 to WC. Tryptic digest/mass spectrometry analysis was performed by Alla Leshinsky and Richard Cook, and N-terminal sequencing was performed by Heather Amoroso, all of the MIT Biopolymers Core Facility.
Footnotes
Nucleotide Sequence Accession Number: The Syn5 genome sequence has been deposited in GenBank and has an accession number of EF372997.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterbury JB, Watson SW, Valois FW, Franks DG. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 3.Goericke R. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Research Part II. 1993;40:2283–2294. [Google Scholar]
- 4.Li WKW. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnology and Oceanography. 1994;39:169–175. [Google Scholar]
- 5.Liu H, Nolla HA, Campbell L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquatic Microbial Ecology. 1997;12:39–47. [Google Scholar]
- 6.Liu H, Campbell L, Landry MR, Nolla HA, Brown SL, Constantinou J. Prochlorococcus and Synechococcus growth rates and contributions in the Arabian Sea during the 1995 Southwest and Northeast monsoons. Deep Sea Research Part II. 1998;45:2327–2352. [Google Scholar]
- 7.Veldhuis MJW, Kraay GW, VanBleijswijk JDL, Baars MA. Seasonal and spatial variability in phytoplankton biomass, productivity and growth in the northwestern Indian Ocean: The southwest and northeast monsoon, 1992–1993. Deep-Sea Research Part I-Oceanographic Research Papers. 1997;44:425–449. [Google Scholar]
- 8.Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophage abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–51. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 10.Wilson WH, Joint IR, Carr NG, Mann NH. Isolation and Molecular Characterization of 5 Marine Cyanophages Propagated on Synechococcus Sp Strain Wh7803. Applied and Environmental Microbiology. 1993;59:3736–3743. doi: 10.1128/aem.59.11.3736-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suttle CA, Chan AM. Dynamics and distribution of cyanophages and their effects on marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–3174. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergh O, Borsheim KY, Bratbak G, Heldal M. High Abundance of Viruses Found in Aquatic Environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 13.Proctor LM, Fuhrman JA. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. [Google Scholar]
- 14.Suttle CA, Chan AM, Cottrell MT. Infection of Phytoplankton by Viruses and Reduction of Primary Productivity. Nature. 1990;347:467–469. [Google Scholar]
- 15.Bratbak G, Heldal M, Thingstad TF, Riemann B, Haslund OH. Incorporation of viruses into the budget of microbial C-transfer: A first approach. Marine Ecology Progress Series. 1992;83:273–280. [Google Scholar]
- 16.Suttle CA, Chan AM. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Marine Ecology Progress Series. 1993;92:99–109. [Google Scholar]
- 17.Chen F, Lu JR. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Applied and Environmental Microbiology. 2002;68:2589–2594. doi: 10.1128/AEM.68.5.2589-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann NH, Clokie MR, Millard A, Cook A, Wilson WH, Wheatley PJ, Letarov A, Krisch HM. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol. 2005;187:3188–200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Barbarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. Origins of highly mosiac mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope WH. PhD. Massachusetts Institute of Technology and Woods Hole Oceanographic Institution; 2005. Genes and Structural Proteins of the Phage Syn5 of the Marine Cyanobacteria, Synechococcus. [Google Scholar]
- 23.Wang WF, Margolin W, Molineux IJ. Increased synthesis of an Escherichia coli membrane protein suppresses F exclusion of bacteriophage T7. J Mol Biol. 1999;292:501–12. doi: 10.1006/jmbi.1999.3088. [DOI] [PubMed] [Google Scholar]
- 24.Pajunen M, Kiljunen S, Skurnik M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol. 2000;182:5114–20. doi: 10.1128/jb.182.18.5114-5120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozzola JJ, Russell LD. Electron Microscopy. Jones and Bartlett Publishers; Sudbury, MA: 1992. [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Hausmann R. The T7 Group. In: Calendar R, editor. The Bacteriophage. I Plenum; New York, NY: 1988. [Google Scholar]
- 28.Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J Mol Biol. 2004;335:1151–71. doi: 10.1016/j.jmb.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 29.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 30.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 31.Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci U S A. 2004;101:11013–8. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. Plos Biology. 2006;4:1344–1357. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis S, Martin J. The transcription apparatus and the regulation of transcription. In: Bryant D, editor. The molecular biology of cyanobacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1994. [Google Scholar]
- 35.Dobbins AT, George M, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. Journal of Bacteriology. 2004;186:1933–1944. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiology and Molecular Biology Reviews. 2003;67:86. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fargo DC, Zhang M, Gillham NW, Boynton JE. Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Molecular and General Genetics. 1998;257:271–282. doi: 10.1007/s004380050648. [DOI] [PubMed] [Google Scholar]
- 38.Struthers-Schlinke JS, Robins WP, Kemp P, Molineux IJ. The internal head protein Gp16 controls DNA ejection from the bacteriophage T7 virion. J Mol Biol. 2000;301:35–45. doi: 10.1006/jmbi.2000.3940. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RS, Lielausis I. Serological studies with mutants of phage T4D defective in genes determining tail fiber structure. Genetics. 1965;52:1187–1200. doi: 10.1093/genetics/52.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohannan B, Lenski R. Linking genetic change to community evolution:insights from studies of bacteria and bacteriophage. Ecol Letters. 2000;3:362–377. [Google Scholar]
- 41.Rabinovitch A, Fishov I, Hadas H, Einav M, Zaritsky A. Bacteriophage T4 development in Escherichia coli is growth rate dependent. J Theor Biol. 2002;216:1–4. doi: 10.1006/jtbi.2002.2543. [DOI] [PubMed] [Google Scholar]
- 42.Hadas H, Einav M, Fishov I, Zaritsky A. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology. 1997;143 ( Pt 1):179–85. doi: 10.1099/00221287-143-1-179. [DOI] [PubMed] [Google Scholar]
- 43.Steward GF, Smith DC, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Sea. Marine Ecology Progress Series. 1996;131:287–300. [Google Scholar]
- 44.Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–65. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 45.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. Journal of virology. 2005;79:14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wikoff WR, Duda RL, Hendrix RW, Johnson JE. Crystallographic analysis of the dsDNA bacteriophage HK97 mature empty capsid. Acta crystallographica. 1999;55:763–71. doi: 10.1107/s0907444998017661. [DOI] [PubMed] [Google Scholar]
- 47.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. Journal of molecular biology. 2003;334:885–99. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 48.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–8. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 49.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–6. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gowen B, Bamford JKH, Bamford DH, Fuller SD. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. Journal of Virology. 2003;77:7863–7871. doi: 10.1128/JVI.77.14.7863-7871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huiskonen JT, Kivela HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nature Structural & Molecular Biology. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 52.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Advances in Virus Research. 2006;66:293. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohwer F. Global phage diversity. Cell. 2003;113:141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 54.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 55.Boyd EF, Davis BM, Musser JM. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 2002;9:137–144. doi: 10.1016/s0966-842x(01)01960-6. [DOI] [PubMed] [Google Scholar]
- 56.Waterbury JB, Willey JM. Isolation and growth of marine planktonic cyanobacteria. Methods in Enzymology. 1988;167:100–105. [Google Scholar]
- 57.Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of Polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- 58.Booth CR, Jiang W, Baker ML, Zhou ZH, Ludtke SJ, Chiu W. A 9 angstroms single particle reconstruction from CCD captured images on a 200 kV electron cryomicroscope. Journal of structural biology. 2004;147:116–27. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. Journal of structural biology. 2000;131:126–34. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 60.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 61.Ludtke SJ, Jakana J, Song JL, Chuang DT, Chiu W. A 11.5 A single particle reconstruction of GroEL using EMAN. Journal of molecular biology. 2001;314:253–62. doi: 10.1006/jmbi.2001.5133. [DOI] [PubMed] [Google Scholar]
- 62.Jiang W, Li Z, Zhang Z, Booth CR, Baker ML, Chiu W. Semiautomated icosahedral particle reconstruction at sub-nanometer resolution. Journal of structural biology. 2001;136:214–25. doi: 10.1006/jsbi.2002.4439. [DOI] [PubMed] [Google Scholar]
- 63.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Research. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 64.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Research. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Research. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 68.MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal Chem. 2002;74:5593–9. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]