Abstract
Estrogen induces a global change in the translation profile of Xenopus hepatocytes, replacing serum protein synthesis with production of the yolk protein precursor vitellogenin. This is accomplished by the coordinate destabilization of serum protein mRNAs and the transcriptional induction and subsequent stabilization of vitellogenin mRNA. Previous work identified an endonuclease activity whose appearance on polysomes correlated with the disappearance of serum protein mRNAs. This enzyme, polysomal ribonuclease 1 (PMR1), is a novel member of the peroxidase gene family. The current study examined the association of PMR1 with its mRNA targets on polysomes and mRNPs. The highest amount of polysome-bound PMR1 was observed prior to estrogen induction of mRNA decay. Its distribution on sucrose density gradients matched the absorbance profile of polysome-bound mRNA, suggesting that PMR1 forms a latent complex with mRNA. Following dissociation with EDTA the 62 kDa PMR1 sedimented with a larger complex of >670 kDa. Estrogen induces a 22-fold increase in unit enzymatic activity of polysome-bound PMR1, and a time-dependent loss of PMR1 from polysomes in a manner that mirrors the disappearance of albumin mRNA. These data suggest that the key step in the extensive estrogen-induced change in mRNA decay in Xenopus liver is activation of a latent mRNA endonuclease associated with its target mRNA.
INTRODUCTION
The turnover rate of any given mRNA represents the sum of numerous influences, including the presence of cis-acting stability or instability elements, bound proteins, the degree of ribosome loading and the integration of extracellular stimuli (reviewed in 1,2). Basic mechanisms of yeast mRNA decay have been well characterized and shown to involve shortening of the poly(A) tail, which triggers decapping and then 5′→3′ decay of the mRNA body by Xrn1p exoribonuclease (3). While there is ample evidence for deadenylation prior to mRNA decay for numerous vertebrate mRNAs (e.g. those containing AU-rich elements), data for decapping in vertebrate mRNA decay remains limited (4). An intimate link between mRNA decay and translation is underscored by the ‘closed loop’ model, in which the 3′ poly(A) tail is juxtaposed with the 5′ cap through the shared interaction of eIF4G with poly(A)-binding protein and eIF4E (5,6). While poly(A) shortening may disrupt this relationship, the key step initiating mRNA decay appears to be accessibility of the 5′ cap to Dcp1p (7,8). The vertebrate poly(A)-specific exonuclease PARN (9,10) is activated in vitro by a 5′ cap on a target mRNA (9,11), raising the possibility that the degree of eIF4E binding to the cap influences PARN activation, deadenylation and, ultimately, degradation of the mRNA body.
Vertebrate cells also make extensive use of endonucleases to catalyze mRNA decay, particularly in cases where mRNA turnover rates change rapidly in response to extracellular stimuli (12). On a theoretical level, endonuclease cleavage anywhere within the body of an mRNA will have functionally the same effect as deadenylation in disrupting the relationship between the 5′ cap and poly(A) tail. In keeping with this, many mRNAs targeted by endonuclease-mediated mRNA are not deadenylated prior to degradation (13). Polysomal RNase 1 (PMR1) is an mRNA endonuclease involved in the global regulation of mRNA decay in Xenopus liver. In this tissue, estrogen induces coordinate destabilization of the bulk of the serum protein mRNAs (14) and transcriptional induction and stabilization of the mRNA encoding the yolk protein precursor vitellogenin (15). The overall extent of this phenomenon is unique, resulting in reprogramming of the translation profile from the production of serum proteins to production of yolk proteins. PMR1 was identified as an estrogen-induced endoribonuclease activity that appeared on polysomes coincident with the activation of mRNA decay (16). Using generation of a unique degradation intermediate from albumin mRNA to assay activity we purified a 62/64 kDa endonuclease from polysomes of estrogen-treated frogs (17) and identified PMR1 as a novel member of the peroxidase gene family (18). Like its closest homolog myeloperoxidase, mature PMR1 is processed from a Mr = 80 000 precursor; however, it lacks both heme and N-linked carbohydrate.
Before PMR1 was purified and its cDNA cloned, sucrose density gradients were employed to characterize the association of enzymatic activity with liver polysomes from estrogen-treated frogs. Little ribonuclease activity was observed on polysomes isolated from control animals; however, a robust, albumin mRNA-selective activity was seen on polysomes from estrogen-treated frogs (16). Under the conditions used at the time this ribonuclease activity was found primarily on large polysome complexes bearing the 6.2 kb vitellogenin mRNA (19). EDTA treatment dissociated these complexes to 40S and 60S subunits as well as a population of 80S mRNA-bound monoribosomes, and PMR1 enzymatic activity was found either at the top of the gradient or in the 80S peak. Based on these results we hypothesized that PMR1 either joined the polysome complex after translation initiation or was present in a latent form with its target mRNAs as a component of the translating mRNP. Results presented here show that polysomes from control frogs contain PMR1 in a latent form that is released into a >670 kDa particle upon EDTA dissociation of ribosome subunits. Estrogen induces a 22-fold increase in PMR1 activity, which alters the polysome distribution in parallel with disappearance of its substrate mRNAs.
MATERIALS AND METHODS
Preparation of liver extracts, mRNP and polysome fractions
Male Xenopus laevis were obtained from Xenopus One (Ann Arbor, MI) and maintained in plastic aquaria with a 12 h light/dark cycle. One milligram injections of estradiol were made in 90% propylene glycol/10% DMSO into the dorsal lymph sac. Detailed protocols for the preparation of post-nuclear extracts, mRNPs and polysomes from Xenopus liver have been published previously (19) and a compilation of these and other methods for studying PMR1 are presented elsewhere (20). Briefly, livers were perfused with 1× SSC, blotted dry and weighed. Diced tissue was homogenized in 2.5 vol of a buffer containing 40 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 7% sucrose, 2 mM dithiothreitol (DTT), 0.2 mM PMSF, 0.5 µg/ml leupeptin, 0.7 µg/ml pepstatin A, 2 µg/ml aprotinin, 50 mM NaF, 0.2 mM NaVO3. Post-nuclear extract, post-mitochondrial extract, P100 bearing membrane-bound polysomes and S100 were prepared by differential centrifugation (13,20). In the experiments in Figures 1 and 2 membrane-bound polysomes were applied to a linear 20–50% sucrose gradient in 30 mM Tris–HCl pH 7.4, 2 mM DTT, 5 mM EGTA, 5 mM MgCl2, in a clear 14 × 89 mm ultracentrifuge tube. This was centrifuged at 180 000 gmax at 4°C for 3.5 h in a Sorvall T-641 rotor. The absorbance at 254 nm was determined and aliquots of 0.5 ml fractions were stored at –80°C.
Figure 1.
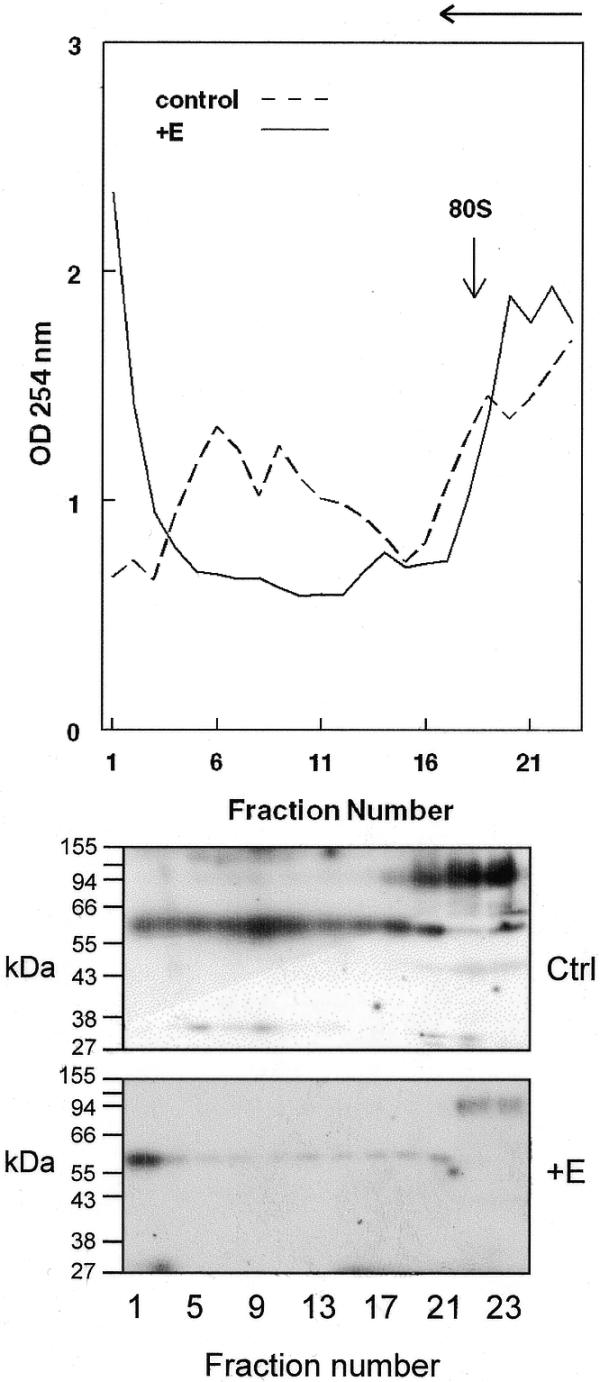
Distribution of PMR1 on membrane-bound polysomes. Liver membrane-bound polysomes from control or 24 h estrogen-treated frogs were separated on a 20–50% sucrose density gradient and 0.5 ml fractions were collected at 4°C. The absorbance at 254 nm was determined for each fraction and is plotted in the top panel. The direction of sedimentation is indicated by the arrow at the top of the diagram. In the bottom panel 30 µl of each odd numbered sample was analyzed by western blot using a polyclonal antibody to PMR1. The position of 80S monoribosome complexes is shown by an arrow.
Figure 2.
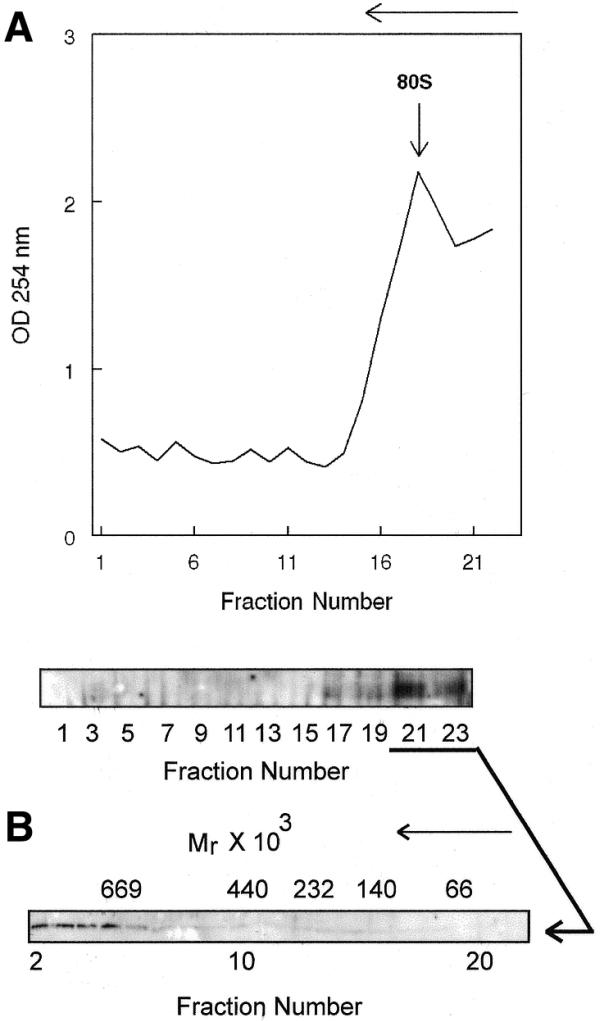
Glycerol gradient analysis of polysome-bound PMR1. (A) Membrane-bound polysomes prepared from control frogs were incubated for 45 min with gentle rocking at 4°C with 50 mM EDTA to dissociate ribosome-bound complexes, followed by fractionation on a 20–50% sucrose density gradient. The absorbance at 254 nm was determined for each fraction and is plotted in the top panel. In the bottom panel odd numbered samples were analyzed by western blot as in Figure 1 to identify PMR1-containing fractions. (B) Fractions 21–23 from the sucrose gradient in (A) were pooled, dialyzed and applied to a 10–40% glycerol gradient. Fractions obtained after centrifugation for 20 h at 83 000 gmax were analyzed by western blot as in (A). Molecular size markers consisting of a mixture of bovine serum albumin (Mr = 66 000), lactate dehydrogenase (Mr = 140 000), catalase (Mr = 232 000), ferritin (Mr = 440 000) and thyroglobulin (Mr = 669 000) were fractionated on a parallel gradient. Their sedimentation positions are indicated above the autoradiogram. The direction of sedimentation is indicated by an arrow above each figure.
In the experiments shown in Figures 3 and 4 polysomes and mRNPs were separated from post-mitochondrial extract on step gradients consisting of 2 ml of 10% sucrose and 2.5 ml of 35% sucrose prepared in 30 mM Tris–HCl, pH 7.4, 2 mM MgCl2, 2 mM DTT. The gradients were centrifuged at 147 000 gmax for 2 h at 4°C in a Beckman SW50.1 rotor. Polysomes were recovered from the pellet at the bottom of the tube and mRNP complexes were recovered at the interface between the 10 and 35% sucrose layers. The polysome-containing pellet was dissolved in the buffer used for the sucrose gradients, and both the polysome and mRNP fractions were dialyzed against 30 mM Tris–HCl, pH 7.4, 2 mM MgCl2, 2 mM DTT, 15% glycerol at 2°C and stored at –80°C. Polysome and mRNP fractions were quantified by Bradford assay for the amount of protein and for RNA by absorbance at 260 and 280 nm using Warburg–Christian analysis on a Beckman DU640 spectrophotometer.
Figure 3.
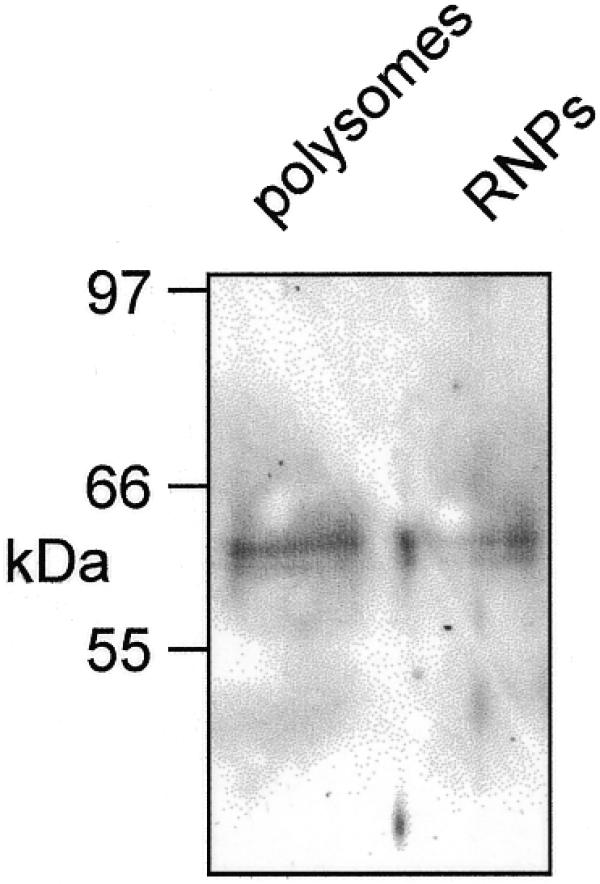
Recovery of PMR1 on oligo(dT)–cellulose. Post-nuclear liver extract prepared from 5 g liver from control frogs was fractionated on a discontinuous sucrose step gradient to prepare mRNPs and polysomes (20). These were bound to oligo(dT)–cellulose for 2 h at 4°C and the matrix was washed and eluted with water. The recovered material was dissolved in 20 µl of 10 mM Tris–HCl, pH 8.0, RNA was removed by digestion for 10 min at 37°C with 5 µg of RNase A and the sample was assayed by western blot for PMR1.
Figure 4.
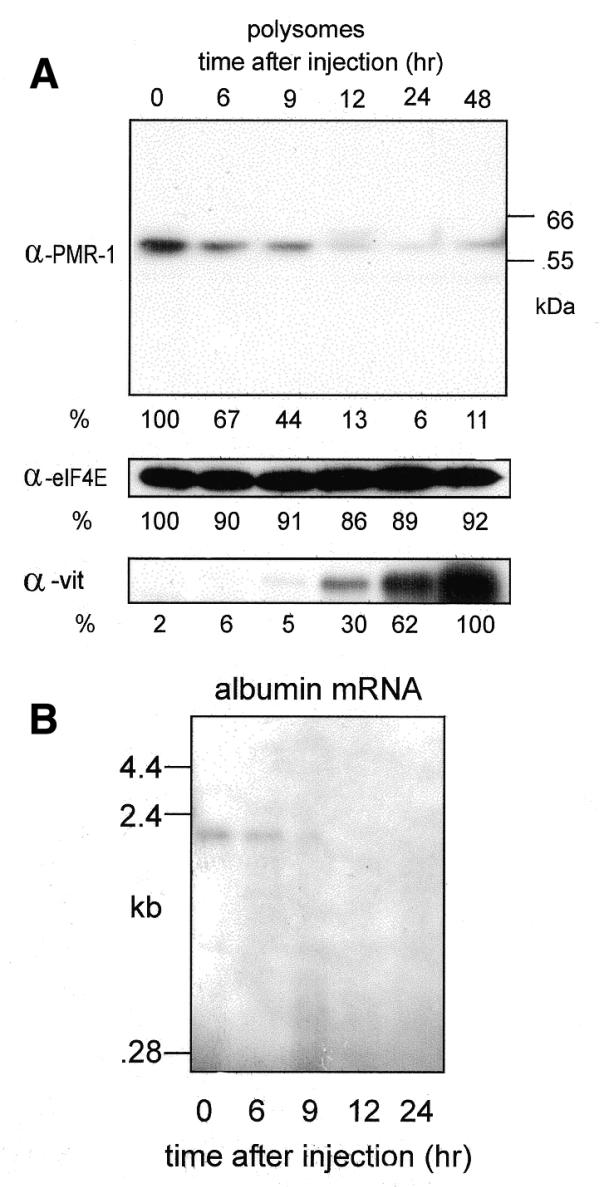
Estrogen-induced changes in polysome-bound PMR1. (A) Polysomes were prepared at the times indicated in the figure and normalized to 100 µg protein. These were separated by SDS–PAGE and analyzed by western blot using antibodies to PMR1, eIF4E and vitellogenin. The positions of the 55 and 66 kDa size markers are indicated in the PMR1 western. The relative signal intensity of each band was determined by scanning densitometry. For all but vitellogenin the signal at time 0 was arbitrarily set to 100%. For vitellogenin the maximal signal at 48 h was arbitrarily set to 100%. (B) Five micrograms of total RNA from each of the polysome fractions examined in (A) was analyzed by northern blot using an antisense probe for albumin mRNA.
Western blots
SDS–PAGE gels were electroblotted onto PVDF membrane (Immobilon P™, Millipore). The membrane was blocked with 5% non-fat dry milk in 20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20 (TBST) overnight followed by incubation with a rabbit polyclonal antibody to PMR1 (17), a rabbit polyclonal antibody to Xenopus eIF4E (kindly provided by Dr Simon Morley) or a sheep polyclonal antibody to Xenopus vitellogenin. Secondary HRP-coupled donkey anti-rabbit or anti-sheep antibodies were used for detection of immune complexes by enhanced chemiluminescence. Multiple exposures were obtained to ensure linearity, and signal intensities were quantified by scanning densitometry using a PDI 420oe scanning densitometer and Quantity One software.
Unit activity determination for mRNP and polysome-bound PMR1
mRNP and polysome fractions prepared from control and 24 h estrogen-treated frogs were adjusted to 100 µg protein, separated by SDS–PAGE and analyzed by western blot with a polyclonal antibody to PMR1. The relative amount of immunoreactive protein in each sample was determined by scanning densitometry of films obtained at several exposures to obtain an accurate determination of the amount of PMR1 in each sample. The samples were then normalized to equivalent amounts of immunoreactive PMR1 and the following amounts were added to individual reactions: time 0 polysomes, 0.39 µg; 24 h polysomes, 6.49 µg; time 0 mRNPs, 4.22 µg; 24 h mRNPs, 4.38 µg. Reactions were performed in 20 µl containing 250 fmol uniformly 32P-labeled 160 nt transcript derived from the portion of albumin mRNA bearing the mapped PMR1 cleavage sites (18,21). The reaction mixtures were separated on a denaturing 6% polyacrylamide–urea gel and unit activity was determined from the rate of disappearance of input transcript quantified by phosphorimager analysis.
RESULTS
PMR1 distributes with polysome-bound mRNA on sucrose density gradients
We previously showed that PMR1 is in the membrane-bound polysome fraction bearing both the serum protein mRNAs and vitellogenin mRNA (19). In the experiment in Figure 1 membrane-bound polysomes from control or estrogen-treated frogs were fractionated on a 20–50% sucrose gradient and the distribution of PMR1 immunoreactive material was compared with the profile of polysome-bound mRNA. The polysome profile changed with estrogen treatment in a manner consistent with the global estrogen-induced changes in protein synthesis. In control frogs the polysome-bound serum protein mRNAs distributed throughout the lower two-thirds of the gradient. These disappeared after estrogen treatment and were replaced by rapidly sedimenting polysomes bearing the 6.2 kb vitellogenin mRNA. These results confirm previous work on estrogen-induced changes in polysome distribution (22).
The distribution of PMR1 immunoreactivity is shown in the lower panel of Figure 1. Prior to estrogen treatment PMR1 distributed across the gradient in a manner that paralleled the distribution of polysome-bound mRNA. These results are consistent with the hypothesis that PMR1 is a component of the translating mRNP. An ∼120 kDa cross-reacting protein was observed in fractions at the top of the gradient, the amount of which was reduced following estrogen. The identity of this is unknown and is not likely to be the PMR1 precursor, which is 80 kDa (18). Following estrogen-induced destabilization of serum protein mRNAs the distribution of PMR1 shifted, so that it was now only observed in the rapidly sedimenting fractions bearing vitellogenin mRNA. As noted above, vitellogenin mRNA is stabilized in the presence of estrogen and we show elsewhere that, at least in vitro, the ability of PMR1 to cleave vitellogenin mRNA is inhibited by binding of the estrogen-induced multi-KH domain protein vigilin to the vitellogenin mRNA 3′-UTR (23) (see Discussion).
Identification of PMR1 in a >670 kDa complex
Earlier work showed that EDTA dissociation of polysomes released PMR1 in a form that sediments with mRNPs at the top of a sucrose gradient (19). If PMR1 forms a complex with translating mRNPs one would expect that the protein released by EDTA would sediment more rapidly on a glycerol gradient than free protein. In the experiment in Figure 2A polysomes from control frogs were treated with EDTA and fractionated on a 20–50% sucrose density gradient. The effectiveness of the EDTA treatment is evident from the change in the RNA absorbance profile. Western analysis of odd numbered fractions collected across the gradient showed that EDTA treatment released PMR1 in a form that sedimented at the top of the gradient. These fractions were then applied to a 10–40% glycerol gradient and the size distribution of PMR1-containing complexes was determined by western analysis of each of the fractions. The results in Figure 2B show that PMR1 sedimented in a >670 kDa complex. These data are consistent with the stable association of PMR1 with a large complex prior to activation of mRNA decay. To determine whether mRNA was present in the PMR1-containing complexes, mRNPs or EDTA-treated polysomes from control frogs were incubated with oligo(dT)–cellulose and recovered proteins were assayed by western blot. The results in Figure 3 show that both samples contain PMR1, confirming that the endonuclease is present in mRNA-containing complexes.
Effect of estrogen on the distribution of PMR1 on mRNPs and polysomes
Having shown that PMR1 is present on polysomes prior to estrogen treatment we examined the effect of estrogen on this association. In the experiments in Figures 4 and 5 frogs received estrogen at time 0 and step gradients were used to prepare mRNP and polysome complexes from livers obtained at times ranging from 6 to 48 h later (13). Samples from each time point were normalized to 100 µg protein and analyzed by western blot using a polyclonal antibody to PMR1. In the experiments in Figures 4 and 5 eIF4E was used as an internal control and in Figure 4 vitellogenin was used to document the effectiveness of estrogen stimulation.
Figure 5.
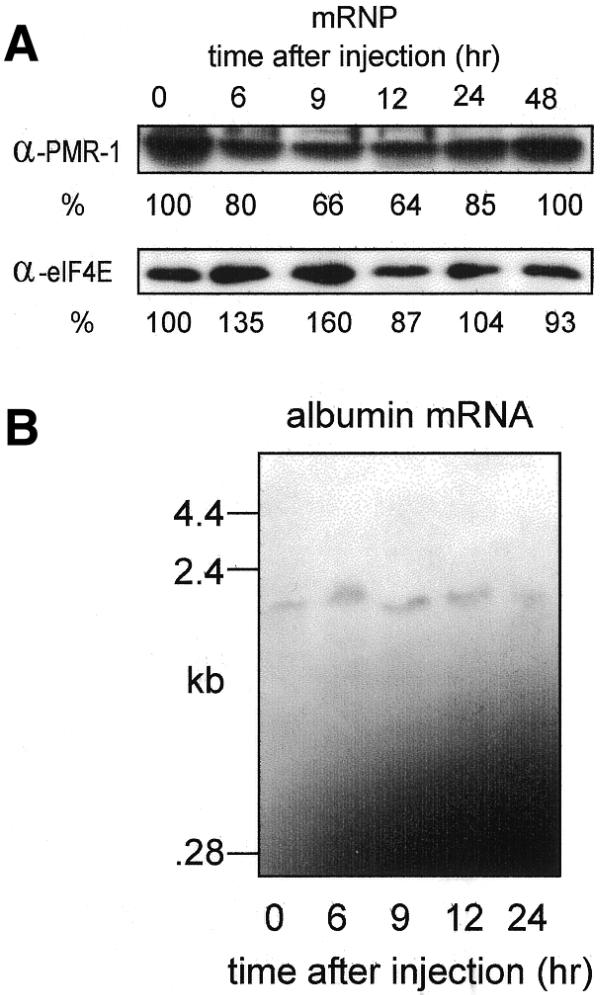
Estrogen-induced changes in mRNP-bound PMR1. (A) Samples of the mRNP fraction prepared from the same animals as in Figure 4 were normalized to 100 µg protein, separated by SDS–PAGE and analyzed by western blot as in Figure 4 using antibodies to PMR1 and eIF4E. The stronger PMR1 signal intensity seen here is a result of a longer exposure time and does not reflect the relative amount of PMR1 in these samples. mRNPs at time 0 contain 10% as much PMR1 as polysomes. (B) Five micrograms of total RNA from each of the polysome fractions examined in (A) was analyzed by northern blot using an antisense probe for albumin mRNA.
Polysome-bound PMR1 was highest prior to estrogen administration (Fig. 4A, top panel, time 0) and on a per microgram of protein (or per microgram of RNA) basis there is 10 times more PMR1 on polysomes than on mRNPs (data not shown). By 9 h, polysome-associated PMR1 dropped to 44% of control and by 12 h only 13% of the starting amount of polysomal PMR1 remained. Estrogen had little effect on the amount of polysome-bound eIF4E and induction of vitellogenin was clearly evident. The effect of estrogen on polysome-bound albumin mRNA present in the above fractions was determined by northern blot in Figure 4B. Albumin mRNA disappeared from these fractions with the same kinetics as seen for PMR1 and this disappearance matches previous results for the loss of albumin mRNA from total cytoplasmic RNA (24).
The experiment in Figure 5 examined the effect of estrogen on PMR1 and albumin mRNA present in mRNP complexes isolated from the same frogs as used for Figure 4. In contrast to the results observed with polysomes, estrogen had little effect on the amount of PMR1 present on mRNP complexes (Fig. 5A). The northern blot in Figure 5B shows that estrogen also had little effect on the amount of albumin mRNA present on mRNP complexes. As with PMR1, this fraction contains <10% of the cytoplasmic albumin mRNA.
Estrogen-induced changes in the unit activity of PMR1
Estrogen has no effect on the total amount of cellular PMR1 (18). Since PMR1 was originally identified as an estrogen-induced RNase activity on polysomes (16), the disappearance of immunoreactive material in Figure 4A could be explained by selective activation of polysome-bound enzyme followed by degradation of substrate mRNA. To test this, unit activity determinations were performed for PMR1 on polysomes and mRNP complexes isolated at time 0 and 24 h after estrogen. The samples used for these experiments were normalized to equal amounts of immunoreactive protein by scanning densitometry of western blots. The autoradiogram of this experiment is shown in the upper panel of Figure 6 and the quantified results are shown underneath. Polysome-bound PMR1 had 40 U activity at time 0; however, 24 h after estrogen the same amount of immunoreactive protein had 893 U, a 22-fold increase. Estrogen had little effect on the enzymatic activity of mRNP-associated PMR1. The selective degradation of albumin versus ferritin RNA confirmed PMR1 as the induced polysome-associated RNase activity rather than a non-specific endo- or exonuclease. These results are consistent with the estrogen-induced appearance of PMR1 activity on polysomes (16) and provide an explanation for the parallel disappearance of PMR1 immunoreactive material and albumin mRNA from polysomes following estrogen. PMR1 does not reciprocally accumulate in mRNP complexes and we have not examined further the fate of enzyme released from polysomes.
Figure 6.
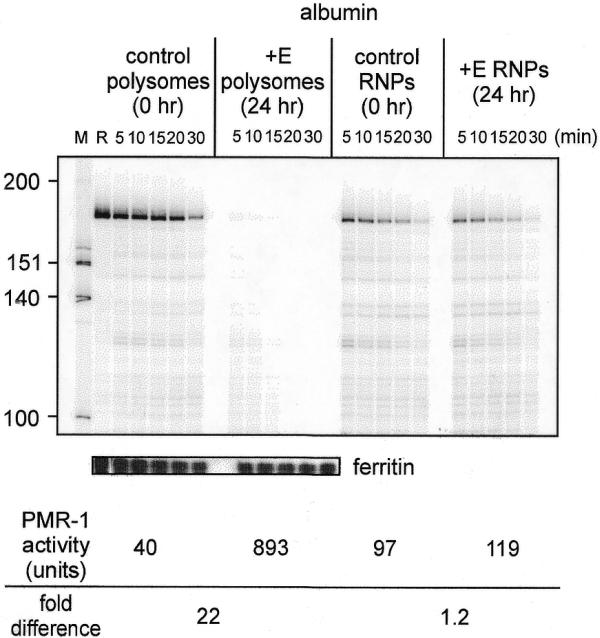
Estrogen-induced increase in PMR1 unit activity. Polysomes and mRNP complexes isolated from control and 24 h estrogen-treated frogs were normalized by scanning densitometry to equal amounts of immunoreactive PMR1. These were incubated with a uniformly 32P-labeled 160 nt albumin transcript (18) or a full-length 32P-labeled ferritin transcript for the indicated times and separated on a denaturing polyacrylamide–urea gel. The input transcript is denoted R in lane 2. The units of PMR1 enzymatic activity were determined by phosphorimager analysis of data obtained from the resulting albumin RNA decay curves. One unit of PMR1 activity equals the amount of enzyme that completely degrades 7 fmol albumin transcript substrate in 30 min at 22°C (17).
DISCUSSION
Estrogen activates a global reorganization of translation in Xenopus liver involving coordinate destabilization of the bulk of the serum protein mRNAs and transcriptional induction and stabilization of the mRNA for the yolk protein precursor vitellogenin. In contrast to a deadenylation-linked process that mediates the degradation of many unstable mRNAs, a substantial body of data indicates that estrogen activates a pathway in which the mRNA endonuclease PMR1 is the principal effector of global mRNA decay. Our previous work using activity assays on sucrose gradient fractions indicated that PMR1 was either brought to polysomes as part of an mRNP complex with its substrate mRNA or that it was recruited to polysomes after translation initiation (19). The current study employed antibody reagents developed in the course of purifying PMR1 and cloning its cDNA to directly evaluate the relationship between activation of mRNA decay by estrogen and association of PMR1 with polysomes.
Prior to the activation of mRNA decay PMR1 distributed across sucrose gradient fractions in a manner that generally reflected the RNA absorbance pattern. This pattern suggested that a latent PMR1 is associated either directly or indirectly with its substrate mRNAs. Support for this came from the glycerol gradient analysis in Figure 2, showing that EDTA released PMR1 from polysomes in a >670 kDa complex rather than as free protein. Co-localization of a latent endonuclease with its substrate mRNA solves the problem of targeting enzyme to substrate, at least for the initial stage of mRNA decay. Association of an endonuclease activity with the β-globin mRNP complex was proposed by Brawerman and co-workers in 1990 (25) and more recently a DMSO-inducible endonuclease activity was identified on both nuclear and cytoplasmic mRNPs (26).
The finding that estrogen induces a significant increase in the unit activity of polysome-bound PMR1 provides a logical explanation for the parallel disappearance of albumin mRNA and PMR1 (Fig. 4) from polysomes as a function of time after estrogen administration. Without its substrate and associated mRNA-binding proteins PMR1 would no longer be held to polysomes. However, PMR1 does not reciprocally accumulate in mRNP complexes and we have yet to determine the fate of the released protein, except for the small portion that associates with vitellogenin mRNA (see below). No change was observed in the amount or unit activity of mRNP-bound PMR1; however, this population represents <10% of total PMR1 on a unit protein basis. PMR1 is not induced by estrogen (18) and the destabilization of albumin mRNA is blocked by anti-estrogens (27) but not by inhibitors of translation (28), suggesting that a non-genomic mechanism transduces the signal from the estrogen receptor to polysome-bound PMR1. Also, an inhibitor of PMR1 activity was observed in S100 at the same time we identified PMR1 (16). Based on these results and our current findings we propose the model shown in Figure 7, in which PMR1 and albumin mRNA are co-localized on a polysome-bound mRNP and PMR1 is maintained in a latent form by a bound inhibitor. In this model a signal activated by estrogen binding to the estrogen receptor targets the inhibitor (denoted IPA, inhibitor of PMRI activity) in a manner analogous to the regulation of translation initiation by phosphorylation of 4E-BPs (29), resulting in dissociation of the PMR1–inhibitor complex and activation of mRNA decay. The activated enzyme then cleaves its polysome-bound substrate and is released in the process. Various aspects of this model are currently under investigation in our laboratory.
Figure 7.
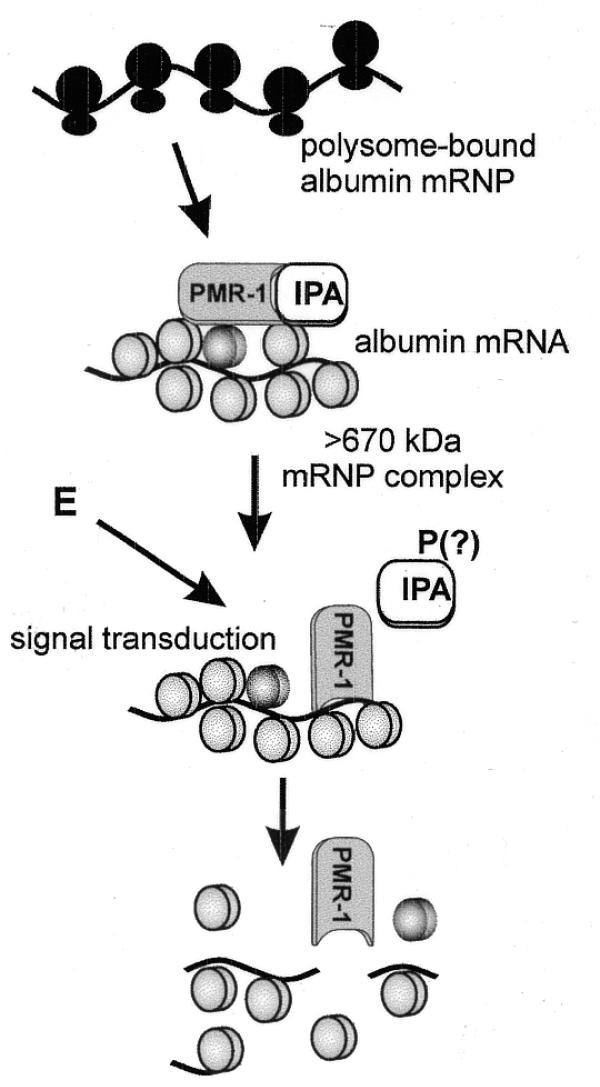
A model for activation of mRNA decay by PMR1. In this model PMR1 present on polysomes as part of a large RNP complex is maintained in a latent form by a bound inhibitory protein IPA (inhibitor of PMRI activity). Estrogen (E) activates a signal transduction pathway resulting in post-translational modification of IPA (indicated here as phosphorylation, but that is only speculation), causing it to dissociate from PMR1 and activate cleavage of albumin mRNA within the complex.
Finally, the results in Figure 1 and our previous work (19) indicate that a fraction of the enzymatically active PMR1 sediments with large vitellogenin mRNA-containing polysomes. The vitellogenin mRNA 3′-UTR has two consensus PMR1 cleavage sites (21), yet vitellogenin mRNA is stabilized by estrogen (30). This raises the question why is vitellogenin mRNA not degraded like the serum protein mRNAs by the activated endonuclease? Shapiro and co-workers have linked the stabilization of vitellogenin mRNA to estrogen induction of the muli-KH domain RNA-binding protein vigilin (31,32). Vigilin binds over the region of the vitellogenin mRNA 3′-UTR bearing the PMR1 cleavage sites (31) and we have recently demonstrated that this binding inhibits in vitro cleavage by PMR1 (23). Selective stabilization of vitellogenin mRNA versus albumin mRNA results from the significantly higher affinity of vigilin for the vitellogenin mRNA 3′-UTR than for albumin mRNA and these findings may be generally applicable to the regulated decay of other mRNAs that are degraded through endonuclease-mediated pathways.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Simon Morley for providing the antibody to Xenopus eIF4E and Kathleen Boris-Lawrie for her helpful comments on this paper. This work was supported by NIH grant GM38277 to D.R.S. K.S.C. was supported by a Presidential Fellowship from The Ohio State University.
References
- 1.Ross J. (1996) Control of messenger RNA stability in higher eukaryotes. Trends. Genet., 12, 171–175. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson A. and Peltz,S.W. (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem., 65, 693–739. [DOI] [PubMed] [Google Scholar]
- 3.Tharun S. and Parker,R. (1997) Mechanisms of mRNA turnover in eukaryotic cells. In Harford,J. and Morris,D.R. (eds), mRNA Metabolism and Post-transcriptional Gene Regulation. Wiley, New York, NY, pp. 181–200.
- 4.Couttet P., Fromont-Racine,M., Steel,D.M., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarun S.Z.,Jr, Wells,S.E., Deardorff,J.A. and Sachs,A.B. (1997) Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA, 94, 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz D.C. and Parker,R. (2000) mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol., 20, 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tharun S., He,W., Mayes,A.E., Lennertz,P., Beggs,J.D. and Parker,R. (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- 9.Koerner C.G. and Wahle,E. (1997) Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem., 272, 10448–10456. [DOI] [PubMed] [Google Scholar]
- 10.Dehlin E., Wormington,M., Körner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M., Fritz,D.T., Ford,L.P. and Wilusz,J. (2000) Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell, 5, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoenberg D.R. and Chernokalskaya,E. (1997) Ribonucleases involved in eukaryotic mRNA turnover. In Harford,J. and Morris,D.R. (eds), mRNA Metabolism and Post-transcriptional Gene Regulation. Wiley, New York, NY, pp. 217–240.
- 13.Schoenberg D.R. and Cunningham,K.S. (1999) Characterization of mRNA endonucleases. Methods, 17, 60–73. [DOI] [PubMed] [Google Scholar]
- 14.Pastori R.L., Moskaitis,J.E., Buzek,S.W. and Schoenberg,D.R. (1991) Coordinate estrogen-regulated instability of serum protein-coding messenger RNAs in Xenopus laevis.Mol. Endocrinol., 5, 461–468. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen D.A. and Shapiro,D.J. (1990) Insights into hormonal control of messenger RNA stability. Mol. Endocrinol., 4, 953–957. [DOI] [PubMed] [Google Scholar]
- 16.Pastori R.L., Moskaitis,J.E. and Schoenberg,D.R. (1991) Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry, 30, 10490–10498. [DOI] [PubMed] [Google Scholar]
- 17.Dompenciel R.E., Garnepudi,V.R. and Schoenberg,D.R. (1995) Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J. Biol. Chem., 270, 6108–6118. [DOI] [PubMed] [Google Scholar]
- 18.Chernokalskaya E., DuBell,A.N., Cunningham,K.S., Hanson,M.N., Dompenciel,R.E. and Schoenberg,D.R. (1998) A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA, 4, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastori R.L. and Schoenberg,D.R. (1993) The nuclease that selectively degrades albumin messenger RNA in vitro associates with Xenopus liver polysomes through the 80S ribosome complex. Arch. Biochem. Biophys ., 305, 313–319. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham K.S., Hanson,M.N. and Schoenberg,D.R. (2001) Polysomal ribonuclease I. In Nicholson,A.W. (ed.), Methods in Enzymology: Ribonucleases. Academic Press, New York, NY, in press. [DOI] [PubMed]
- 21.Chernokalskaya E., Dompenciel,R.E. and Schoenberg,D.R. (1997) Cleavage properties of a polysomal ribonuclease involved in the estrogen-regulated destabilization of albumin mRNA. Nucleic Acids Res., 25, 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berridge M.V., Farmer,S.R., Green,C.D., Henshaw,E.C. and Tata,J.R. (1976) Characterization of polysomes from Xenopus liver synthesizing vitellogenin and translation of vitellogenin and albumin messenger RNA’s in vitro. Eur. J. Biochem., 62, 161–171. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham K.S., Dodson,R.E., Nagel,M.A., Shapiro,D.J. and Schoenberg,D.R. (2000) Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-UTR by the mRNA endonuclease PMR1. Proc. Natl Acad. Sci. USA, 97, 12498–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenberg D.R., Moskaitis,J.E., Smith,J.H.,Jr and Pastori,R.L. (1989) Extranuclear estrogen-regulated destabilization of Xenopus laevis serum albumin mRNA. Mol. Endocrinol., 3, 805–814. [DOI] [PubMed] [Google Scholar]
- 25.Bandyopadhyay R., Coutts,M., Krowczynska,A. and Brawerman,G. (1990) Nuclease activity associated with mammalian mRNA in its native state: possible basis for selectivity in mRNA decay. Mol. Cell. Biol., 10, 2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantinova I.M., Kulichkova,V.A., Evteeva,I.N., Mittenberg,A.G., Volkova,I.V., Ermolaeva,J.B. and Gause,L.N. (1999) The specific endoribonuclease activity of small nuclear and cytoplasmic alpha-RNPs. FEBS Lett., 462, 407–410. [DOI] [PubMed] [Google Scholar]
- 27.Riegel A.T., Aitken,S.C., Martin,M.B. and Schoenberg,D.R. (1987) Posttranscriptional regulation of albumin gene expression in Xenopus liver: evidence for an estrogen receptor-dependent mechanism. Mol. Endocrinol., 1, 160–167. [DOI] [PubMed] [Google Scholar]
- 28.Moskaitis J.E., Buzek,S.W., Pastori,R.L. and Schoenberg,D.R. (1991) The estrogen-regulated destabilization of Xenopus albumin mRNA is independent of translation. Biochem. Biophys. Res. Commun., 174, 825–830. [DOI] [PubMed] [Google Scholar]
- 29.Gingras A.C., Gygi,S.P., Raught,B., Polakiewicz,R.D., Abraham,R.T., Hoekstra,M.F., Aebersold,R. and Sonenberg,N. (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev., 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brock M.L. and Shapiro,D.J. (1983) Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell, 34, 207–214. [DOI] [PubMed] [Google Scholar]
- 31.Dodson R.E. and Shapiro,D.J. (1994) An estrogen-inducible protein binds specifically to a sequence in the 3′ untranslated region of estrogen-stabilized vitellogenin mRNA. Mol. Cell. Biol., 14, 3130–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodson R.E. and Shapiro,D.J. (1997) Vigilin, an ubiquitous protein with 14 KH domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region binding protein. J. Biol. Chem., 272, 12249–12252. [DOI] [PubMed] [Google Scholar]


