Abstract
In the title compound {systematic name: methyl 1-[12-oxo-10-(3,4,5-trimethoxyphenyl)-4,6,13-trioxatetracyclo[7.7.0.03,7.011,15]hexadeca-1,3(7),8-trien-16-yl]-1H-1,2,3-triazole-4-carboxylate dichloromethane solvate}, C26H25N3O9·CH2Cl2, the tetrahydrofuran ring and the six-membered ring fused to it both display envelope conformations.
Related literature
For similar structures of 4β-N-substituted-4-desoxypodophyllotoxin and derivatives, see: Bilal et al. (2008 ▶); Yu & Chen (2008 ▶); Van Maanen et al. (1988 ▶). For a review of the structures of azides and triazides, see: Bräse et al. (2005 ▶). For additional background to 1,3-dipolar azide–alkyne cycloaddition reactions, see: Hainsworth et al. (1985 ▶); Huisgen (1963 ▶); Jacobsen et al. (1988 ▶); Lee (2004 ▶). 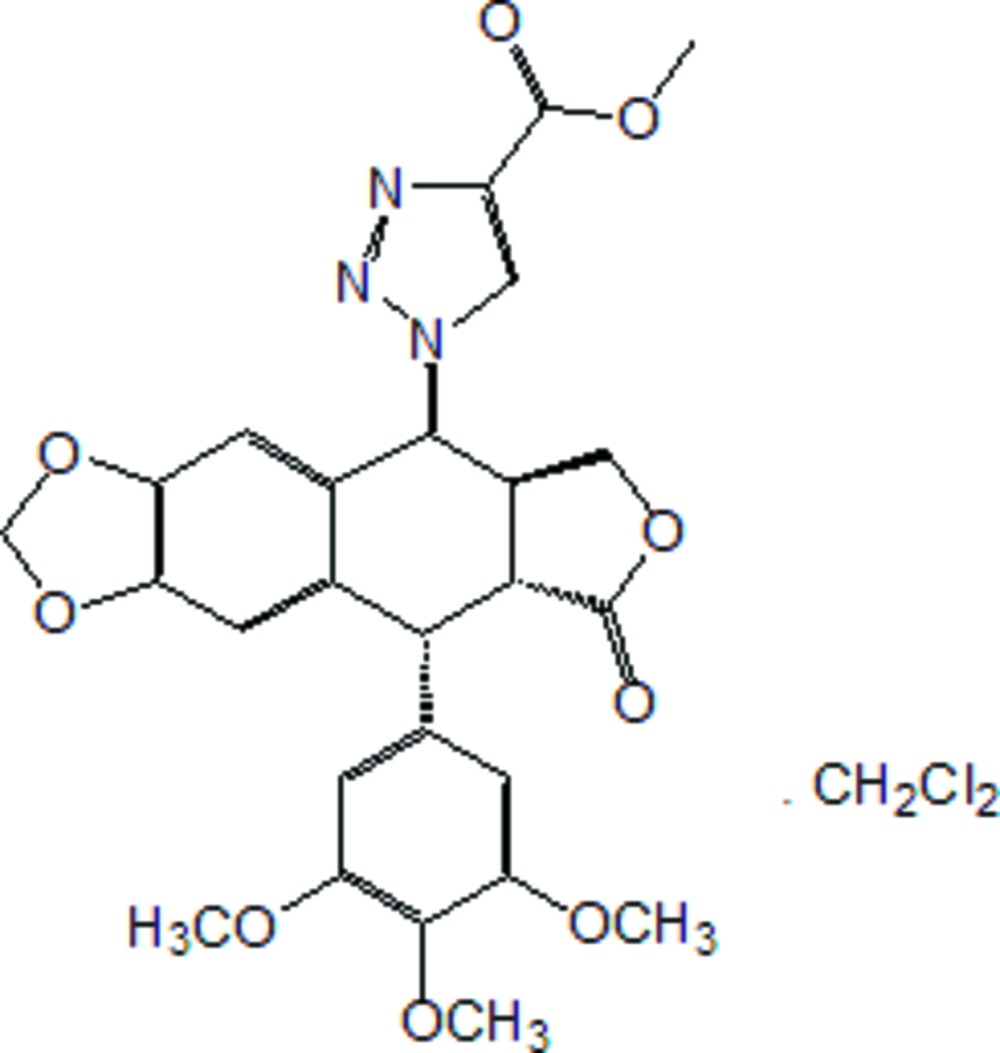
Experimental
Crystal data
C26H25N3O9·CH2Cl2
M r = 608.42
Orthorhombic,
a = 10.377 (2) Å
b = 12.639 (3) Å
c = 20.463 (4) Å
V = 2683.9 (10) Å3
Z = 4
Mo Kα radiation
μ = 0.30 mm−1
T = 113 K
0.28 × 0.24 × 0.12 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005 ▶) T min = 0.920, T max = 0.965
22515 measured reflections
6397 independent reflections
5768 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.079
S = 1.01
6397 reflections
374 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.34 e Å−3
Absolute structure: Flack (1983 ▶), 2800 Friedel pairs
Flack parameter: 0.04 (4)
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809050612/vm2011sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809050612/vm2011Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 30873363), the Great Program of the Science Foundation of Tianjin (09ZCKFNC01200) and the Program of the Science Foundation of Tianjin (08JCYBJC070000).
supplementary crystallographic information
Comment
Podophyllotoxin derivatives such as Etoposide and Teniposide are in clinical use as antineoplastic agents. NK611, as well as NPF, GL-311 and TOP53 are presently under clinical trials. These podophyllotoxin ligands block the catalytic activity of DNA-topoisomerase II by stabilizing a cleavable enzyme DNA complex in which the DNA is cleaved and covalently linked to the enzyme (Hainsworth,1985). Its high toxicity has limited its application as a drug in cancer chemotherapy. Hence, extensive structure modifications have been performed since the 1950's. Meanwhile, click chemistry with copper-catalyzed Huisgen 1,3-dipolar azide-alkyne cycloaddition (CuAAC) producing 1,2,3-triazoles has exhibited increasing importance in organic chemistry due to the chemoselective nature and mild reaction conditions (Bräse, 2005). Moreover, as a functional group the 1,2,3-triazole ring has found widespread occurrence in a variety of fields. These advantages have stimulated us to couple the podophyllotoxin parent nucleus with 1,2,3-triazole to give potential anticancer candidates. Herein we report the crystal structure of the title compound (Fig.1), which can be used as a candidate for anti-tumor molecules. The asymmetric unit consists of the organic molecule and one dichloromethane molecule, C26H25N3O9.CH2Cl2. The configuration of this derivative is the same as found for podophyllotoxin, which is compatible with the mild reaction conditions. The compound contains three planar moieties: plane A consisting of atoms C10 to C22, plane B consisting of atoms C1 to C6, and the triazole ring as plane C. The dihedral angles between the planes A/B, B/C and A/C are 83.82 (6)°, 32.06 (8)° and 86.54 (7)°, respectively.
Experimental
The title compound was synthesized from natural product podophyllotoxin by copper-catalyzed Huisgen 1, 3-dipolar azide-alkyne cycloaddition. Crystals of the title compound suitable for crystal structure analysis were obtained from a dichloromethane solution by slowly evaporating the solvent.
Refinement
All H atoms were located in difference Fourier maps and refined independently with isotropic displacement parameters.
Figures
Fig. 1.
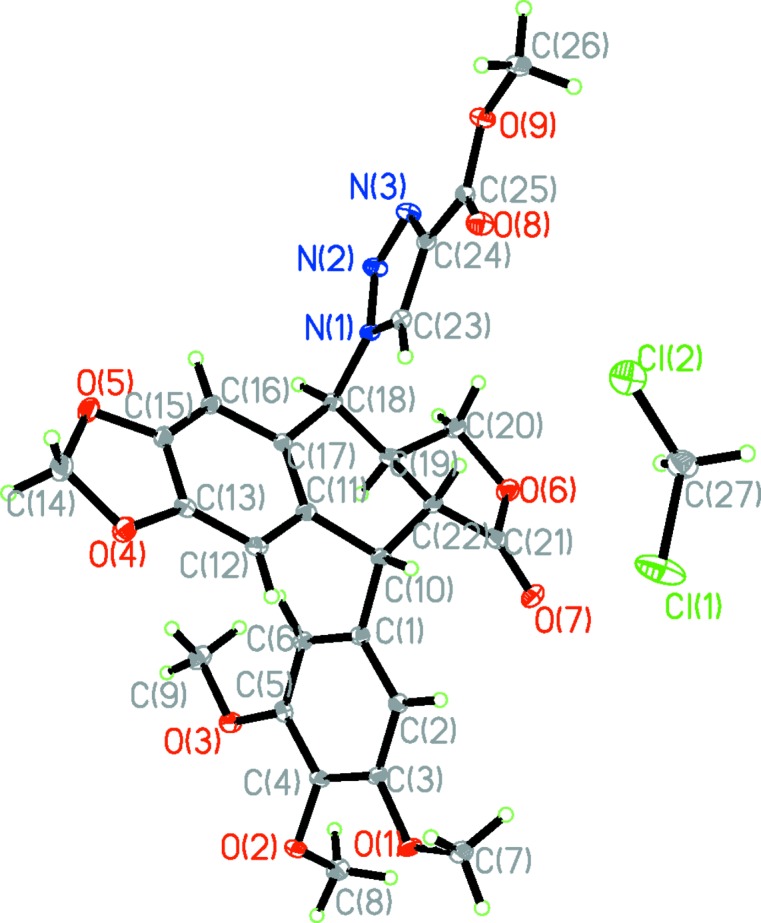
The molecular structure of (I), with atom labels and 50% probability displacement ellipsoids for non-H atoms.
Crystal data
| C26H25N3O9·CH2Cl2 | Dx = 1.506 Mg m−3 |
| Mr = 608.42 | Melting point = 212–213 K |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 9219 reflections |
| a = 10.377 (2) Å | θ = 1.9–27.9° |
| b = 12.639 (3) Å | µ = 0.30 mm−1 |
| c = 20.463 (4) Å | T = 113 K |
| V = 2683.9 (10) Å3 | Block, colorless |
| Z = 4 | 0.28 × 0.24 × 0.12 mm |
| F(000) = 1264 |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 6397 independent reflections |
| Radiation source: rotating anode | 5768 reflections with I > 2σ(I) |
| confocal | Rint = 0.036 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.9°, θmin = 1.9° |
| ω and φ scans | h = −13→13 |
| Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005) | k = −12→16 |
| Tmin = 0.920, Tmax = 0.965 | l = −26→26 |
| 22515 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H-atom parameters constrained |
| wR(F2) = 0.079 | w = 1/[σ2(Fo2) + (0.0476P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max = 0.001 |
| 6397 reflections | Δρmax = 0.18 e Å−3 |
| 374 parameters | Δρmin = −0.34 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 2800 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.04 (4) |
Special details
| Experimental. Both Cu+ and Cu2+ can be used as catalyst while combined with respective additives. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.81730 (11) | 0.46481 (8) | −0.07929 (6) | 0.0222 (2) | |
| O2 | 1.02230 (11) | 0.38471 (8) | −0.13992 (5) | 0.0200 (2) | |
| O3 | 1.07335 (11) | 0.17818 (9) | −0.13872 (6) | 0.0242 (3) | |
| O4 | 1.03010 (12) | 0.07454 (9) | 0.18541 (6) | 0.0279 (3) | |
| O5 | 1.04059 (12) | −0.10864 (9) | 0.17747 (6) | 0.0263 (3) | |
| O6 | 0.49041 (12) | −0.00212 (9) | −0.11823 (6) | 0.0273 (3) | |
| O7 | 0.50203 (13) | 0.17197 (9) | −0.09864 (6) | 0.0313 (3) | |
| O8 | 0.35597 (12) | −0.14406 (9) | 0.20717 (6) | 0.0254 (3) | |
| O9 | 0.30253 (11) | −0.31101 (8) | 0.17946 (6) | 0.0242 (3) | |
| N1 | 0.60318 (12) | −0.17930 (9) | 0.04644 (6) | 0.0167 (3) | |
| N2 | 0.56517 (13) | −0.28046 (10) | 0.03289 (7) | 0.0234 (3) | |
| N3 | 0.47699 (14) | −0.30643 (10) | 0.07549 (6) | 0.0226 (3) | |
| C1 | 0.78479 (14) | 0.18283 (11) | −0.03338 (7) | 0.0162 (3) | |
| C2 | 0.75602 (15) | 0.29059 (11) | −0.03655 (7) | 0.0176 (3) | |
| H2 | 0.6835 | 0.3180 | −0.0138 | 0.021* | |
| C3 | 0.83415 (15) | 0.35759 (11) | −0.07318 (8) | 0.0172 (3) | |
| C4 | 0.93996 (15) | 0.31698 (12) | −0.10722 (7) | 0.0165 (3) | |
| C5 | 0.96690 (15) | 0.20875 (12) | −0.10457 (7) | 0.0175 (3) | |
| C6 | 0.88965 (15) | 0.14181 (12) | −0.06738 (8) | 0.0178 (3) | |
| H6 | 0.9085 | 0.0683 | −0.0652 | 0.021* | |
| C7 | 0.72731 (16) | 0.51437 (13) | −0.03645 (8) | 0.0261 (4) | |
| H7A | 0.7549 | 0.5042 | 0.0089 | 0.039* | |
| H7B | 0.6419 | 0.4828 | −0.0427 | 0.039* | |
| H7C | 0.7232 | 0.5902 | −0.0462 | 0.039* | |
| C8 | 0.98547 (17) | 0.40051 (14) | −0.20675 (8) | 0.0245 (4) | |
| H8A | 0.9016 | 0.4361 | −0.2084 | 0.037* | |
| H8B | 0.9794 | 0.3319 | −0.2288 | 0.037* | |
| H8C | 1.0502 | 0.4443 | −0.2287 | 0.037* | |
| C9 | 1.09771 (18) | 0.06681 (13) | −0.14314 (9) | 0.0279 (4) | |
| H9A | 1.0224 | 0.0314 | −0.1620 | 0.042* | |
| H9B | 1.1145 | 0.0384 | −0.0994 | 0.042* | |
| H9C | 1.1729 | 0.0545 | −0.1711 | 0.042* | |
| C10 | 0.70229 (15) | 0.11348 (11) | 0.01110 (7) | 0.0165 (3) | |
| H10 | 0.6540 | 0.1623 | 0.0407 | 0.020* | |
| C11 | 0.78712 (15) | 0.04464 (11) | 0.05453 (7) | 0.0161 (3) | |
| C12 | 0.86601 (15) | 0.09890 (12) | 0.09961 (8) | 0.0192 (3) | |
| H12 | 0.8637 | 0.1738 | 0.1029 | 0.023* | |
| C13 | 0.94594 (15) | 0.03996 (12) | 0.13838 (8) | 0.0188 (3) | |
| C14 | 1.09057 (17) | −0.01795 (14) | 0.21149 (9) | 0.0278 (4) | |
| H14A | 1.0721 | −0.0239 | 0.2588 | 0.033* | |
| H14B | 1.1851 | −0.0135 | 0.2056 | 0.033* | |
| C15 | 0.95192 (15) | −0.06919 (12) | 0.13392 (8) | 0.0188 (3) | |
| C16 | 0.87764 (15) | −0.12393 (12) | 0.09064 (8) | 0.0181 (3) | |
| H16 | 0.8828 | −0.1988 | 0.0875 | 0.022* | |
| C17 | 0.79280 (14) | −0.06537 (11) | 0.05070 (7) | 0.0156 (3) | |
| C18 | 0.70594 (15) | −0.13007 (11) | 0.00655 (7) | 0.0162 (3) | |
| H18 | 0.7587 | −0.1880 | −0.0133 | 0.019* | |
| C19 | 0.65391 (15) | −0.06140 (11) | −0.04854 (8) | 0.0174 (3) | |
| H19 | 0.7265 | −0.0457 | −0.0792 | 0.021* | |
| C20 | 0.53937 (17) | −0.09906 (12) | −0.08870 (8) | 0.0224 (3) | |
| H20A | 0.5667 | −0.1503 | −0.1226 | 0.027* | |
| H20B | 0.4734 | −0.1325 | −0.0605 | 0.027* | |
| C21 | 0.52869 (16) | 0.08367 (13) | −0.08212 (8) | 0.0224 (3) | |
| C22 | 0.60149 (15) | 0.04366 (11) | −0.02292 (7) | 0.0173 (3) | |
| H22 | 0.5353 | 0.0258 | 0.0108 | 0.021* | |
| C23 | 0.53786 (16) | −0.14106 (12) | 0.09778 (7) | 0.0172 (3) | |
| H23 | 0.5456 | −0.0731 | 0.1172 | 0.021* | |
| C24 | 0.45700 (15) | −0.22220 (12) | 0.11611 (8) | 0.0169 (3) | |
| C25 | 0.36761 (15) | −0.22017 (12) | 0.17193 (8) | 0.0192 (3) | |
| C26 | 0.21945 (17) | −0.31309 (14) | 0.23663 (8) | 0.0287 (4) | |
| H26A | 0.1500 | −0.2613 | 0.2312 | 0.043* | |
| H26B | 0.2700 | −0.2954 | 0.2756 | 0.043* | |
| H26C | 0.1824 | −0.3839 | 0.2416 | 0.043* | |
| Cl1 | 0.26269 (6) | 0.39288 (4) | 0.23482 (3) | 0.05616 (18) | |
| Cl2 | 0.21387 (5) | 0.18814 (4) | 0.29584 (2) | 0.03992 (13) | |
| C27 | 0.1834 (2) | 0.27027 (14) | 0.22719 (10) | 0.0343 (4) | |
| H27A | 0.2134 | 0.2342 | 0.1870 | 0.041* | |
| H27B | 0.0895 | 0.2823 | 0.2231 | 0.041* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0274 (6) | 0.0121 (5) | 0.0272 (6) | 0.0025 (4) | 0.0052 (5) | 0.0009 (4) |
| O2 | 0.0216 (6) | 0.0202 (6) | 0.0181 (5) | −0.0065 (4) | 0.0010 (5) | 0.0023 (4) |
| O3 | 0.0228 (6) | 0.0206 (6) | 0.0293 (6) | 0.0030 (5) | 0.0106 (5) | −0.0002 (5) |
| O4 | 0.0315 (7) | 0.0217 (6) | 0.0305 (7) | −0.0010 (5) | −0.0133 (6) | −0.0014 (5) |
| O5 | 0.0284 (6) | 0.0208 (6) | 0.0297 (7) | 0.0024 (5) | −0.0120 (5) | 0.0002 (5) |
| O6 | 0.0307 (7) | 0.0246 (6) | 0.0267 (6) | −0.0017 (5) | −0.0094 (5) | 0.0035 (5) |
| O7 | 0.0314 (7) | 0.0243 (6) | 0.0381 (7) | 0.0046 (5) | −0.0067 (6) | 0.0088 (6) |
| O8 | 0.0291 (7) | 0.0228 (6) | 0.0241 (6) | −0.0017 (5) | 0.0064 (5) | −0.0022 (5) |
| O9 | 0.0249 (6) | 0.0238 (6) | 0.0239 (6) | −0.0066 (5) | 0.0036 (5) | 0.0035 (5) |
| N1 | 0.0195 (6) | 0.0119 (6) | 0.0186 (6) | −0.0005 (5) | 0.0004 (5) | −0.0004 (5) |
| N2 | 0.0275 (8) | 0.0157 (7) | 0.0269 (8) | −0.0054 (5) | 0.0019 (6) | −0.0033 (6) |
| N3 | 0.0256 (8) | 0.0194 (7) | 0.0227 (7) | −0.0059 (6) | 0.0025 (6) | −0.0006 (6) |
| C1 | 0.0182 (7) | 0.0155 (7) | 0.0149 (7) | −0.0014 (6) | −0.0009 (6) | −0.0006 (6) |
| C2 | 0.0177 (8) | 0.0168 (8) | 0.0184 (8) | 0.0011 (6) | 0.0010 (6) | −0.0016 (6) |
| C3 | 0.0212 (8) | 0.0121 (7) | 0.0181 (8) | 0.0000 (6) | −0.0037 (6) | 0.0006 (6) |
| C4 | 0.0175 (8) | 0.0178 (7) | 0.0141 (7) | −0.0045 (6) | −0.0001 (6) | 0.0016 (6) |
| C5 | 0.0162 (8) | 0.0215 (8) | 0.0148 (7) | 0.0009 (6) | 0.0015 (6) | −0.0031 (6) |
| C6 | 0.0195 (8) | 0.0156 (7) | 0.0184 (8) | 0.0008 (6) | −0.0006 (6) | 0.0002 (6) |
| C7 | 0.0264 (9) | 0.0177 (8) | 0.0342 (10) | 0.0026 (6) | 0.0033 (8) | −0.0017 (7) |
| C8 | 0.0290 (9) | 0.0268 (9) | 0.0178 (8) | 0.0008 (7) | 0.0028 (7) | 0.0030 (7) |
| C9 | 0.0315 (9) | 0.0257 (9) | 0.0267 (9) | 0.0104 (7) | 0.0078 (8) | 0.0010 (7) |
| C10 | 0.0185 (8) | 0.0138 (7) | 0.0171 (8) | 0.0009 (6) | 0.0027 (6) | 0.0005 (6) |
| C11 | 0.0169 (8) | 0.0155 (7) | 0.0158 (7) | −0.0005 (6) | 0.0042 (6) | 0.0007 (5) |
| C12 | 0.0212 (8) | 0.0157 (8) | 0.0206 (8) | −0.0015 (6) | 0.0031 (6) | 0.0012 (6) |
| C13 | 0.0185 (8) | 0.0188 (8) | 0.0192 (8) | −0.0038 (6) | 0.0015 (6) | −0.0020 (6) |
| C14 | 0.0287 (9) | 0.0268 (9) | 0.0278 (9) | 0.0036 (7) | −0.0089 (7) | −0.0029 (7) |
| C15 | 0.0176 (8) | 0.0191 (8) | 0.0196 (8) | 0.0028 (6) | 0.0007 (6) | 0.0017 (6) |
| C16 | 0.0192 (8) | 0.0152 (8) | 0.0199 (8) | 0.0012 (6) | 0.0024 (6) | 0.0004 (6) |
| C17 | 0.0155 (7) | 0.0151 (7) | 0.0162 (7) | 0.0005 (5) | 0.0035 (6) | 0.0003 (5) |
| C18 | 0.0171 (8) | 0.0153 (7) | 0.0162 (7) | −0.0009 (6) | 0.0036 (6) | −0.0010 (6) |
| C19 | 0.0195 (8) | 0.0160 (8) | 0.0169 (8) | −0.0006 (6) | 0.0023 (6) | −0.0010 (6) |
| C20 | 0.0262 (9) | 0.0195 (8) | 0.0214 (8) | −0.0008 (7) | −0.0025 (7) | 0.0023 (6) |
| C21 | 0.0186 (8) | 0.0255 (9) | 0.0231 (8) | −0.0017 (6) | 0.0006 (7) | 0.0041 (7) |
| C22 | 0.0172 (7) | 0.0152 (8) | 0.0194 (8) | 0.0004 (6) | 0.0037 (6) | 0.0026 (6) |
| C23 | 0.0204 (8) | 0.0164 (7) | 0.0148 (8) | 0.0011 (6) | 0.0003 (6) | −0.0007 (6) |
| C24 | 0.0184 (8) | 0.0156 (7) | 0.0166 (8) | −0.0006 (6) | −0.0026 (6) | 0.0014 (6) |
| C25 | 0.0169 (8) | 0.0215 (8) | 0.0192 (8) | −0.0009 (6) | −0.0013 (7) | 0.0053 (6) |
| C26 | 0.0261 (9) | 0.0301 (9) | 0.0300 (9) | −0.0017 (8) | 0.0072 (8) | 0.0105 (8) |
| Cl1 | 0.0722 (4) | 0.0437 (3) | 0.0526 (3) | −0.0320 (3) | 0.0287 (3) | −0.0187 (3) |
| Cl2 | 0.0490 (3) | 0.0366 (3) | 0.0342 (3) | 0.0004 (2) | 0.0064 (2) | −0.0110 (2) |
| C27 | 0.0350 (11) | 0.0262 (9) | 0.0417 (11) | −0.0044 (8) | 0.0050 (9) | −0.0084 (8) |
Geometric parameters (Å, °)
| O1—C3 | 1.3722 (18) | C9—H9A | 0.9800 |
| O1—C7 | 1.4258 (19) | C9—H9B | 0.9800 |
| O2—C4 | 1.3823 (17) | C9—H9C | 0.9800 |
| O2—C8 | 1.4339 (19) | C10—C11 | 1.524 (2) |
| O3—C5 | 1.3631 (19) | C10—C22 | 1.535 (2) |
| O3—C9 | 1.433 (2) | C10—H10 | 1.0000 |
| O4—C13 | 1.3710 (19) | C11—C17 | 1.394 (2) |
| O4—C14 | 1.430 (2) | C11—C12 | 1.411 (2) |
| O5—C15 | 1.3747 (19) | C12—C13 | 1.368 (2) |
| O5—C14 | 1.438 (2) | C12—H12 | 0.9500 |
| O6—C21 | 1.371 (2) | C13—C15 | 1.384 (2) |
| O6—C20 | 1.4575 (19) | C14—H14A | 0.9900 |
| O7—C21 | 1.1985 (19) | C14—H14B | 0.9900 |
| O8—C25 | 1.2084 (19) | C15—C16 | 1.363 (2) |
| O9—C25 | 1.3409 (19) | C16—C17 | 1.411 (2) |
| O9—C26 | 1.454 (2) | C16—H16 | 0.9500 |
| N1—C23 | 1.340 (2) | C17—C18 | 1.516 (2) |
| N1—N2 | 1.3664 (18) | C18—C19 | 1.522 (2) |
| N1—C18 | 1.4800 (19) | C18—H18 | 1.0000 |
| N2—N3 | 1.3057 (19) | C19—C20 | 1.521 (2) |
| N3—C24 | 1.367 (2) | C19—C22 | 1.528 (2) |
| C1—C6 | 1.392 (2) | C19—H19 | 1.0000 |
| C1—C2 | 1.396 (2) | C20—H20A | 0.9900 |
| C1—C10 | 1.526 (2) | C20—H20B | 0.9900 |
| C2—C3 | 1.392 (2) | C21—C22 | 1.515 (2) |
| C2—H2 | 0.9500 | C22—H22 | 1.0000 |
| C3—C4 | 1.398 (2) | C23—C24 | 1.377 (2) |
| C4—C5 | 1.397 (2) | C23—H23 | 0.9500 |
| C5—C6 | 1.392 (2) | C24—C25 | 1.472 (2) |
| C6—H6 | 0.9500 | C26—H26A | 0.9800 |
| C7—H7A | 0.9800 | C26—H26B | 0.9800 |
| C7—H7B | 0.9800 | C26—H26C | 0.9800 |
| C7—H7C | 0.9800 | Cl1—C27 | 1.7613 (19) |
| C8—H8A | 0.9800 | Cl2—C27 | 1.775 (2) |
| C8—H8B | 0.9800 | C27—H27A | 0.9900 |
| C8—H8C | 0.9800 | C27—H27B | 0.9900 |
| C3—O1—C7 | 117.47 (12) | O4—C14—O5 | 108.23 (13) |
| C4—O2—C8 | 112.53 (12) | O4—C14—H14A | 110.1 |
| C5—O3—C9 | 116.98 (12) | O5—C14—H14A | 110.1 |
| C13—O4—C14 | 106.31 (12) | O4—C14—H14B | 110.1 |
| C15—O5—C14 | 105.43 (12) | O5—C14—H14B | 110.1 |
| C21—O6—C20 | 109.90 (12) | H14A—C14—H14B | 108.4 |
| C25—O9—C26 | 114.00 (12) | C16—C15—O5 | 128.01 (14) |
| C23—N1—N2 | 110.51 (12) | C16—C15—C13 | 121.57 (15) |
| C23—N1—C18 | 130.16 (12) | O5—C15—C13 | 110.42 (14) |
| N2—N1—C18 | 119.31 (12) | C15—C16—C17 | 117.59 (14) |
| N3—N2—N1 | 107.58 (12) | C15—C16—H16 | 121.2 |
| N2—N3—C24 | 108.46 (12) | C17—C16—H16 | 121.2 |
| C6—C1—C2 | 120.50 (14) | C11—C17—C16 | 121.13 (14) |
| C6—C1—C10 | 121.51 (13) | C11—C17—C18 | 123.14 (14) |
| C2—C1—C10 | 117.91 (13) | C16—C17—C18 | 115.68 (13) |
| C3—C2—C1 | 119.62 (14) | N1—C18—C17 | 109.06 (12) |
| C3—C2—H2 | 120.2 | N1—C18—C19 | 113.12 (13) |
| C1—C2—H2 | 120.2 | C17—C18—C19 | 110.17 (12) |
| O1—C3—C2 | 125.16 (14) | N1—C18—H18 | 108.1 |
| O1—C3—C4 | 114.67 (14) | C17—C18—H18 | 108.1 |
| C2—C3—C4 | 120.17 (14) | C19—C18—H18 | 108.1 |
| O2—C4—C5 | 120.10 (14) | C20—C19—C18 | 119.93 (12) |
| O2—C4—C3 | 119.95 (13) | C20—C19—C22 | 100.32 (12) |
| C5—C4—C3 | 119.81 (13) | C18—C19—C22 | 111.58 (13) |
| O3—C5—C6 | 125.09 (14) | C20—C19—H19 | 108.1 |
| O3—C5—C4 | 114.81 (13) | C18—C19—H19 | 108.1 |
| C6—C5—C4 | 120.07 (14) | C22—C19—H19 | 108.1 |
| C1—C6—C5 | 119.82 (14) | O6—C20—C19 | 103.49 (12) |
| C1—C6—H6 | 120.1 | O6—C20—H20A | 111.1 |
| C5—C6—H6 | 120.1 | C19—C20—H20A | 111.1 |
| O1—C7—H7A | 109.5 | O6—C20—H20B | 111.1 |
| O1—C7—H7B | 109.5 | C19—C20—H20B | 111.1 |
| H7A—C7—H7B | 109.5 | H20A—C20—H20B | 109.0 |
| O1—C7—H7C | 109.5 | O7—C21—O6 | 121.17 (15) |
| H7A—C7—H7C | 109.5 | O7—C21—C22 | 130.67 (15) |
| H7B—C7—H7C | 109.5 | O6—C21—C22 | 108.15 (13) |
| O2—C8—H8A | 109.5 | C21—C22—C19 | 101.14 (13) |
| O2—C8—H8B | 109.5 | C21—C22—C10 | 120.70 (12) |
| H8A—C8—H8B | 109.5 | C19—C22—C10 | 114.36 (13) |
| O2—C8—H8C | 109.5 | C21—C22—H22 | 106.6 |
| H8A—C8—H8C | 109.5 | C19—C22—H22 | 106.6 |
| H8B—C8—H8C | 109.5 | C10—C22—H22 | 106.6 |
| O3—C9—H9A | 109.5 | N1—C23—C24 | 104.66 (13) |
| O3—C9—H9B | 109.5 | N1—C23—H23 | 127.7 |
| H9A—C9—H9B | 109.5 | C24—C23—H23 | 127.7 |
| O3—C9—H9C | 109.5 | N3—C24—C23 | 108.78 (14) |
| H9A—C9—H9C | 109.5 | N3—C24—C25 | 125.55 (14) |
| H9B—C9—H9C | 109.5 | C23—C24—C25 | 125.63 (14) |
| C11—C10—C1 | 110.59 (12) | O8—C25—O9 | 124.24 (15) |
| C11—C10—C22 | 109.26 (12) | O8—C25—C24 | 122.69 (14) |
| C1—C10—C22 | 116.22 (12) | O9—C25—C24 | 113.07 (14) |
| C11—C10—H10 | 106.8 | O9—C26—H26A | 109.5 |
| C1—C10—H10 | 106.8 | O9—C26—H26B | 109.5 |
| C22—C10—H10 | 106.8 | H26A—C26—H26B | 109.5 |
| C17—C11—C12 | 119.80 (14) | O9—C26—H26C | 109.5 |
| C17—C11—C10 | 124.14 (14) | H26A—C26—H26C | 109.5 |
| C12—C11—C10 | 116.03 (12) | H26B—C26—H26C | 109.5 |
| C13—C12—C11 | 117.78 (14) | Cl1—C27—Cl2 | 111.18 (11) |
| C13—C12—H12 | 121.1 | Cl1—C27—H27A | 109.4 |
| C11—C12—H12 | 121.1 | Cl2—C27—H27A | 109.4 |
| C12—C13—O4 | 128.28 (14) | Cl1—C27—H27B | 109.4 |
| C12—C13—C15 | 122.11 (15) | Cl2—C27—H27B | 109.4 |
| O4—C13—C15 | 109.60 (14) | H27A—C27—H27B | 108.0 |
| C23—N1—N2—N3 | −0.34 (17) | C13—C15—C16—C17 | −0.7 (2) |
| C18—N1—N2—N3 | 178.32 (12) | C12—C11—C17—C16 | −0.9 (2) |
| N1—N2—N3—C24 | 0.55 (17) | C10—C11—C17—C16 | 177.31 (13) |
| C6—C1—C2—C3 | −1.0 (2) | C12—C11—C17—C18 | 176.30 (13) |
| C10—C1—C2—C3 | 175.82 (14) | C10—C11—C17—C18 | −5.5 (2) |
| C7—O1—C3—C2 | 11.4 (2) | C15—C16—C17—C11 | 1.2 (2) |
| C7—O1—C3—C4 | −168.31 (14) | C15—C16—C17—C18 | −176.13 (14) |
| C1—C2—C3—O1 | −178.91 (14) | C23—N1—C18—C17 | 37.7 (2) |
| C1—C2—C3—C4 | 0.7 (2) | N2—N1—C18—C17 | −140.67 (13) |
| C8—O2—C4—C5 | 92.48 (17) | C23—N1—C18—C19 | −85.29 (18) |
| C8—O2—C4—C3 | −91.68 (17) | N2—N1—C18—C19 | 96.36 (15) |
| O1—C3—C4—O2 | 4.1 (2) | C11—C17—C18—N1 | −103.10 (16) |
| C2—C3—C4—O2 | −175.64 (14) | C16—C17—C18—N1 | 74.20 (16) |
| O1—C3—C4—C5 | 179.91 (13) | C11—C17—C18—C19 | 21.6 (2) |
| C2—C3—C4—C5 | 0.2 (2) | C16—C17—C18—C19 | −161.08 (13) |
| C9—O3—C5—C6 | 8.2 (2) | N1—C18—C19—C20 | −41.98 (18) |
| C9—O3—C5—C4 | −173.77 (14) | C17—C18—C19—C20 | −164.34 (13) |
| O2—C4—C5—O3 | −3.2 (2) | N1—C18—C19—C22 | 74.75 (15) |
| C3—C4—C5—O3 | −179.03 (14) | C17—C18—C19—C22 | −47.60 (17) |
| O2—C4—C5—C6 | 174.94 (14) | C21—O6—C20—C19 | −22.57 (16) |
| C3—C4—C5—C6 | −0.9 (2) | C18—C19—C20—O6 | 160.39 (13) |
| C2—C1—C6—C5 | 0.3 (2) | C22—C19—C20—O6 | 37.97 (14) |
| C10—C1—C6—C5 | −176.39 (14) | C20—O6—C21—O7 | 177.91 (16) |
| O3—C5—C6—C1 | 178.54 (14) | C20—O6—C21—C22 | −3.11 (17) |
| C4—C5—C6—C1 | 0.6 (2) | O7—C21—C22—C19 | −153.98 (19) |
| C6—C1—C10—C11 | 45.76 (19) | O6—C21—C22—C19 | 27.17 (16) |
| C2—C1—C10—C11 | −131.05 (14) | O7—C21—C22—C10 | −26.7 (3) |
| C6—C1—C10—C22 | −79.53 (18) | O6—C21—C22—C10 | 154.47 (14) |
| C2—C1—C10—C22 | 103.66 (16) | C20—C19—C22—C21 | −38.84 (14) |
| C1—C10—C11—C17 | −114.25 (16) | C18—C19—C22—C21 | −166.96 (12) |
| C22—C10—C11—C17 | 14.9 (2) | C20—C19—C22—C10 | −170.17 (13) |
| C1—C10—C11—C12 | 63.98 (17) | C18—C19—C22—C10 | 61.71 (17) |
| C22—C10—C11—C12 | −166.88 (13) | C11—C10—C22—C21 | −163.16 (13) |
| C17—C11—C12—C13 | −0.1 (2) | C1—C10—C22—C21 | −37.2 (2) |
| C10—C11—C12—C13 | −178.42 (14) | C11—C10—C22—C19 | −42.11 (17) |
| C11—C12—C13—O4 | −179.74 (15) | C1—C10—C22—C19 | 83.85 (16) |
| C11—C12—C13—C15 | 0.7 (2) | N2—N1—C23—C24 | −0.01 (17) |
| C14—O4—C13—C12 | −179.90 (17) | C18—N1—C23—C24 | −178.48 (15) |
| C14—O4—C13—C15 | −0.29 (18) | N2—N3—C24—C23 | −0.56 (18) |
| C13—O4—C14—O5 | 0.88 (18) | N2—N3—C24—C25 | −178.25 (14) |
| C15—O5—C14—O4 | −1.12 (18) | N1—C23—C24—N3 | 0.34 (17) |
| C14—O5—C15—C16 | −179.58 (17) | N1—C23—C24—C25 | 178.03 (14) |
| C14—O5—C15—C13 | 0.96 (18) | C26—O9—C25—O8 | −3.0 (2) |
| C12—C13—C15—C16 | −0.3 (3) | C26—O9—C25—C24 | 176.46 (13) |
| O4—C13—C15—C16 | −179.94 (14) | N3—C24—C25—O8 | 178.28 (16) |
| C12—C13—C15—O5 | 179.20 (14) | C23—C24—C25—O8 | 1.0 (3) |
| O4—C13—C15—O5 | −0.43 (19) | N3—C24—C25—O9 | −1.2 (2) |
| O5—C15—C16—C17 | 179.92 (15) | C23—C24—C25—O9 | −178.50 (15) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: VM2011).
References
- Bilal, A. & Bhat, P. (2008). Eur. J. Med. Chem. 43, 2067–2072. [DOI] [PubMed]
- Bräse, S., Gil, C. & Knepper, K. (2005). Angew. Chem. Int. Ed. 44, 5188–5240. [DOI] [PubMed]
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Hainsworth, J. D., Williams, S. D., Einhorn, L. H. & Birch, R. (1985). J. Clin. Oncol. 3, 666–671. [DOI] [PubMed]
- Huisgen, R. (1963). Angew. Chem. Int. Ed. 2, 565–598.
- Jacobsen, E. N., Marko, I. & Sharpless, K. B. (1988). J. Am. Chem. Soc. 110, 1968–?.
- Lee, K. H. (2004). J. Nat. Prod. 273–283 [DOI] [PubMed]
- Rigaku/MSC (2005). CrystalClear. Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Van Maanen, J. M. S., De Vries, J. & Pinedo, H. M. (1988). J. Natl Cancer Inst. 80, 1526–1533. [DOI] [PubMed]
- Yu, P. F. & Chen, H. (2008). Chem. Pharm. Bull. 56, 831–834. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809050612/vm2011sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809050612/vm2011Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report


