Abstract
In the structure of the title complex, [Cu(C7H5O2)(OH)(C12H12N2)(C7H6O2)]·H2O, the CuII ion is pentacoordinated in a tetragonal-pyramidal geometry with one O atom of a hydroxide group, one O atom of a benzoate anion and two N atoms of a 4,4′-dimethyl-2,2′-bipyridine ligand occupying the basal plane, and one O atom of a benzoic acid molecule located at the apical site. The title complex was refined with a metal-coordinated OH group and a ‘free’ benzoic acid molecule, although it can be assumed that the proton is delocalized between the OH and the COOH group. The uncoordinated water molecule is equally disordered over two positions. The structure displays O—H⋯O hydrogen bonding.
Related literature
For selected 4,4′-dimethyl-2,2′-bipyridine copper complexes, see: Deschamps et al. (2002 ▶); Dong et al. (2006 ▶); Feng et al. (2007 ▶); Lin et al. (2008 ▶); Qian & Huang (2006 ▶); Willett et al. (2001 ▶).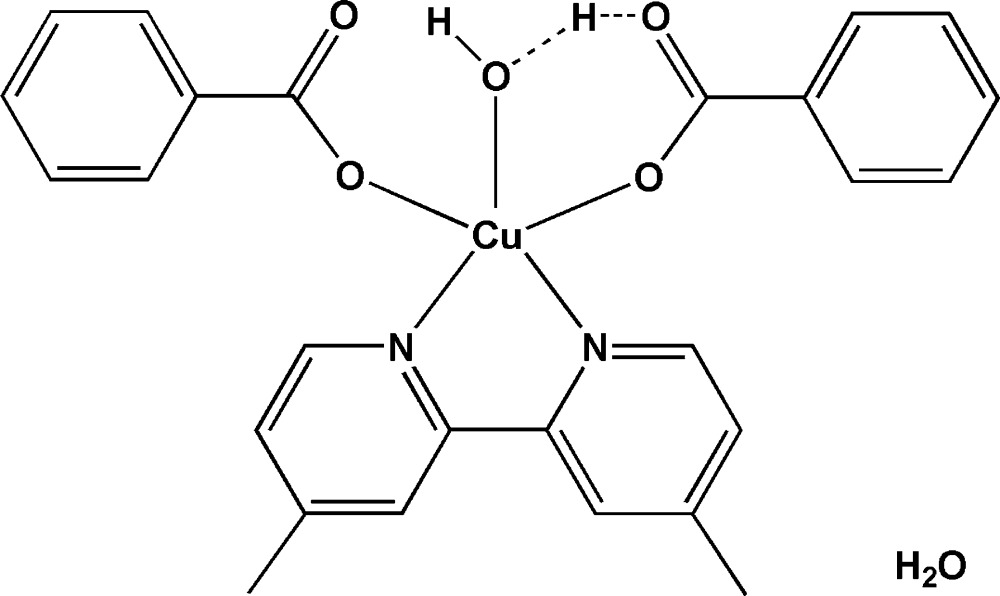
Experimental
Crystal data
[Cu(C7H5O2)(OH)(C12H12N2)(C7H6O2)]·H2O
M r = 526.03
Monoclinic,
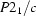
a = 11.3325 (15) Å
b = 17.155 (2) Å
c = 13.4007 (18) Å
β = 98.049 (3)°
V = 2579.5 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.89 mm−1
T = 296 K
0.28 × 0.26 × 0.25 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.789, T max = 0.808
13741 measured reflections
4538 independent reflections
3075 reflections with I > 2σ(I)
R int = 0.053
Refinement
R[F 2 > 2σ(F 2)] = 0.053
wR(F 2) = 0.146
S = 0.95
4538 reflections
329 parameters
H-atom parameters constrained
Δρmax = 0.53 e Å−3
Δρmin = −0.25 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809043189/zq2009sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809043189/zq2009Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2W—H2WB⋯O2i | 0.85 | 2.51 | 3.195 (8) | 138 |
| O2W—H2WA⋯O1W | 0.85 | 2.24 | 2.821 (10) | 125 |
| O1W—H1WB⋯O4 | 0.85 | 2.26 | 2.932 (6) | 136 |
| O1W—H1WA⋯O2W ii | 0.85 | 2.28 | 2.937 (10) | 134 |
| O5—H5A⋯O4 | 0.82 | 1.93 | 2.642 (4) | 145 |
| O2—H2⋯O5 | 0.82 | 1.86 | 2.636 (5) | 158 |
Symmetry codes: (i) 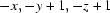 ; (ii)
; (ii) 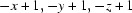 .
.
supplementary crystallographic information
Comment
As a contribution to structural characterization of 4,4'-dimethyl-2,2'-bipyridine copper complexes (Deschamps et al., 2002; Dong et al., 2006; Feng et al., 2007; Lin et al. 2008; Qian & Huang, 2006; Willett et al., 2001), we present here the crystal structure of the title complex, [CuLL'L''(OH)].H2O (L = benzoate, L' = benzoic acid, L'' = 4,4'-dimethyl-2,2'-bipyridine).
In the structure of the title complex, the short O2···O5 separation of 2.636 (5) Å clearly indicates a typical hydrogen bond. The corresponding hydrogen atom was located in the Fourier difference maps near O2 (hydroxido benzoic acid type complex A) although general chemical considerations would rather expect it on O5 (water benzoate type complex B). One can assume that the proton is delocalised somewhere in-between as presented in Scheme 1 but based on the X-ray data only, the title complex was finally refined with a metal-coordinated OH group and a "free" benzoic acid molecule.
In the complex, the Cu2+ ion is pentacoordinated, with two N atoms of 4,4'-dimethyl-2,2'-bipyridine, one O atom of a hydroxide group and one O atom of a benzoate anion in the basal plane and one O atom of a benzoic acid molecule completing the square-pyramidal geometry from the apical site (Fig. 1). The atoms N1, N2, O1 and O3 are nearly coplanar, and the Cu atom is displaced by 0.2309 (5) Å from this plane towards the apical O atom. The water solvent molecule is disordered over two positions in a 1:1 ratio.
With O—H···O hydrogen bonds (Table 1), an one-dimensional chain is formed as shown in Fig. 2.
Experimental
The title compound was synthesized hydrothermally in a Teflon-lined autoclave (25 ml) by heating a mixture of 4,4'-dimethyl-2,2'-bipyridine (0.2 mmol), benzoic acid (0.4 mmol) and CuSO4.5H2O (0.2 mmol) in water (10 ml) at 393 K for 3 d. Suitable crystals for an X-ray analysis were obtained.
Refinement
All H atoms were included in calculated positions, with C—H bond lengths fixed at 0.96 Å (methyl CH3), 0.93 Å (aryl group) and O—H = 0.85 Å and were refined in the riding-model approximation. Uiso(H) values were calculated at 1.5 Ueq(C) for methyl H atoms and 1.2 Ueq(C) for the other H atoms.
Figures
Fig. 1.
The molecular structure of the title compound with the atom-labelling scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are presented as small spheres of arbitrary radius.
Fig. 2.
Crystal packing of the title compound. Hydrogen-bond interactions are drawn with dashed lines.
Crystal data
| [Cu(C7H5O2)(OH)(C12H12N2)(C7H6O2)]·H2O | F(000) = 1092 |
| Mr = 526.03 | Dx = 1.355 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3905 reflections |
| a = 11.3325 (15) Å | θ = 2.8–24.2° |
| b = 17.155 (2) Å | µ = 0.89 mm−1 |
| c = 13.4007 (18) Å | T = 296 K |
| β = 98.049 (3)° | Block, colourless |
| V = 2579.5 (6) Å3 | 0.28 × 0.26 × 0.25 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 4538 independent reflections |
| Radiation source: fine-focus sealed tube | 3075 reflections with I > 2σ(I) |
| graphite | Rint = 0.053 |
| φ and ω scans | θmax = 25.0°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −13→13 |
| Tmin = 0.789, Tmax = 0.808 | k = −20→11 |
| 13741 measured reflections | l = −14→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.146 | H-atom parameters constrained |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0932P)2] where P = (Fo2 + 2Fc2)/3 |
| 4538 reflections | (Δ/σ)max < 0.001 |
| 329 parameters | Δρmax = 0.53 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cu1 | 0.09687 (4) | 0.20024 (2) | 0.53039 (3) | 0.05062 (19) | |
| N1 | 0.1716 (2) | 0.09925 (16) | 0.5797 (2) | 0.0480 (7) | |
| N2 | 0.0537 (2) | 0.13606 (16) | 0.40610 (19) | 0.0450 (7) | |
| O1 | −0.0650 (2) | 0.18703 (15) | 0.6049 (2) | 0.0656 (7) | |
| O2 | −0.1757 (3) | 0.2835 (2) | 0.5320 (3) | 0.0854 (9) | |
| H2 | −0.1116 | 0.2989 | 0.5178 | 0.128* | |
| O3 | 0.1942 (2) | 0.25605 (15) | 0.63954 (18) | 0.0631 (7) | |
| O4 | 0.1932 (3) | 0.37656 (17) | 0.5773 (2) | 0.0797 (9) | |
| C1 | −0.1583 (4) | 0.2267 (3) | 0.5918 (3) | 0.0622 (10) | |
| C2 | −0.2573 (3) | 0.2055 (2) | 0.6527 (3) | 0.0654 (11) | |
| C3 | −0.2304 (4) | 0.1691 (3) | 0.7438 (3) | 0.0709 (11) | |
| H3 | −0.1516 | 0.1562 | 0.7670 | 0.085* | |
| C4 | −0.3181 (5) | 0.1511 (3) | 0.8019 (4) | 0.0877 (14) | |
| H4 | −0.2984 | 0.1260 | 0.8635 | 0.105* | |
| C5 | −0.4335 (5) | 0.1702 (4) | 0.7687 (4) | 0.0992 (17) | |
| H5 | −0.4922 | 0.1588 | 0.8085 | 0.119* | |
| C6 | −0.4635 (4) | 0.2054 (4) | 0.6789 (4) | 0.1010 (19) | |
| H6 | −0.5429 | 0.2172 | 0.6567 | 0.121* | |
| C7 | −0.3759 (4) | 0.2245 (3) | 0.6186 (4) | 0.0898 (15) | |
| H7 | −0.3965 | 0.2494 | 0.5570 | 0.108* | |
| C8 | 0.2201 (3) | 0.3285 (2) | 0.6455 (3) | 0.0548 (9) | |
| C9 | 0.2891 (3) | 0.3544 (2) | 0.7448 (3) | 0.0543 (9) | |
| C10 | 0.3280 (3) | 0.4320 (2) | 0.7563 (3) | 0.0696 (11) | |
| H10 | 0.3137 | 0.4664 | 0.7023 | 0.083* | |
| C11 | 0.3871 (4) | 0.4575 (3) | 0.8471 (4) | 0.0825 (14) | |
| H11 | 0.4141 | 0.5087 | 0.8540 | 0.099* | |
| C12 | 0.4065 (4) | 0.4069 (3) | 0.9284 (4) | 0.0781 (13) | |
| H12 | 0.4451 | 0.4244 | 0.9901 | 0.094* | |
| C13 | 0.3686 (4) | 0.3309 (3) | 0.9178 (3) | 0.0744 (12) | |
| H13 | 0.3821 | 0.2968 | 0.9721 | 0.089* | |
| C14 | 0.3098 (3) | 0.3052 (2) | 0.8252 (3) | 0.0616 (10) | |
| H14 | 0.2843 | 0.2537 | 0.8182 | 0.074* | |
| C15 | −0.0037 (3) | 0.1604 (2) | 0.3175 (2) | 0.0522 (9) | |
| H15 | −0.0264 | 0.2125 | 0.3107 | 0.063* | |
| C16 | −0.0304 (3) | 0.1116 (2) | 0.2362 (3) | 0.0562 (9) | |
| H16 | −0.0706 | 0.1309 | 0.1760 | 0.067* | |
| C17 | 0.0021 (3) | 0.0342 (2) | 0.2435 (2) | 0.0545 (9) | |
| C18 | −0.0228 (4) | −0.0190 (3) | 0.1528 (3) | 0.0783 (13) | |
| H18A | −0.0952 | −0.0030 | 0.1120 | 0.117* | |
| H18B | −0.0310 | −0.0717 | 0.1751 | 0.117* | |
| H18C | 0.0420 | −0.0161 | 0.1139 | 0.117* | |
| C19 | 0.0616 (3) | 0.0085 (2) | 0.3355 (2) | 0.0499 (8) | |
| H19 | 0.0845 | −0.0434 | 0.3437 | 0.060* | |
| C20 | 0.0867 (3) | 0.06038 (19) | 0.4148 (2) | 0.0423 (8) | |
| C21 | 0.1553 (3) | 0.03960 (19) | 0.5139 (2) | 0.0431 (8) | |
| C22 | 0.1997 (3) | −0.0338 (2) | 0.5383 (2) | 0.0480 (8) | |
| H22 | 0.1856 | −0.0741 | 0.4916 | 0.058* | |
| C23 | 0.2658 (3) | −0.0484 (2) | 0.6329 (3) | 0.0509 (9) | |
| C24 | 0.3132 (4) | −0.1282 (2) | 0.6618 (3) | 0.0672 (11) | |
| H24A | 0.3985 | −0.1262 | 0.6768 | 0.101* | |
| H24B | 0.2909 | −0.1637 | 0.6070 | 0.101* | |
| H24C | 0.2805 | −0.1459 | 0.7202 | 0.101* | |
| C25 | 0.2833 (3) | 0.0141 (2) | 0.6973 (3) | 0.0624 (10) | |
| H25 | 0.3281 | 0.0076 | 0.7605 | 0.075* | |
| C26 | 0.2360 (3) | 0.0853 (2) | 0.6704 (3) | 0.0589 (10) | |
| H26 | 0.2486 | 0.1260 | 0.7165 | 0.071* | |
| O5 | 0.0289 (3) | 0.29651 (13) | 0.46085 (19) | 0.0677 (8) | |
| H5A | 0.0560 | 0.3350 | 0.4924 | 0.102* | |
| O1W | 0.3806 (6) | 0.4206 (5) | 0.4579 (5) | 0.116 (3) | 0.50 |
| H1WA | 0.4507 | 0.4049 | 0.4531 | 0.139* | 0.50 |
| H1WB | 0.3631 | 0.3985 | 0.5107 | 0.139* | 0.50 |
| O2W | 0.3699 (7) | 0.5847 (4) | 0.4468 (5) | 0.112 (2) | 0.50 |
| H2WA | 0.3466 | 0.5477 | 0.4814 | 0.135* | 0.50 |
| H2WB | 0.3060 | 0.6076 | 0.4216 | 0.135* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0706 (3) | 0.0357 (3) | 0.0434 (3) | −0.0046 (2) | 0.0005 (2) | −0.00101 (19) |
| N1 | 0.0625 (17) | 0.0394 (15) | 0.0420 (15) | −0.0075 (14) | 0.0068 (13) | −0.0005 (13) |
| N2 | 0.0549 (16) | 0.0372 (15) | 0.0427 (15) | −0.0046 (14) | 0.0066 (13) | 0.0021 (13) |
| O1 | 0.0714 (18) | 0.0529 (16) | 0.0737 (18) | 0.0049 (14) | 0.0140 (14) | −0.0017 (13) |
| O2 | 0.086 (2) | 0.089 (2) | 0.076 (2) | 0.004 (2) | −0.0082 (17) | 0.0052 (18) |
| O3 | 0.0863 (18) | 0.0440 (16) | 0.0535 (15) | −0.0054 (14) | −0.0096 (13) | −0.0086 (12) |
| O4 | 0.113 (2) | 0.0570 (17) | 0.0635 (17) | −0.0284 (17) | −0.0080 (16) | 0.0101 (15) |
| C1 | 0.076 (3) | 0.052 (2) | 0.053 (2) | −0.003 (2) | −0.011 (2) | −0.018 (2) |
| C2 | 0.059 (2) | 0.065 (3) | 0.070 (3) | 0.000 (2) | 0.002 (2) | −0.036 (2) |
| C3 | 0.073 (3) | 0.063 (3) | 0.077 (3) | −0.003 (2) | 0.010 (2) | −0.013 (2) |
| C4 | 0.098 (4) | 0.079 (3) | 0.090 (3) | −0.009 (3) | 0.028 (3) | −0.021 (3) |
| C5 | 0.086 (4) | 0.121 (5) | 0.095 (4) | −0.012 (3) | 0.028 (3) | −0.031 (4) |
| C6 | 0.062 (3) | 0.143 (6) | 0.096 (4) | 0.006 (3) | 0.005 (3) | −0.041 (4) |
| C7 | 0.072 (3) | 0.118 (4) | 0.075 (3) | 0.007 (3) | −0.006 (2) | −0.028 (3) |
| C8 | 0.062 (2) | 0.048 (2) | 0.055 (2) | −0.008 (2) | 0.0096 (18) | −0.0103 (19) |
| C9 | 0.052 (2) | 0.051 (2) | 0.060 (2) | −0.0014 (18) | 0.0075 (17) | −0.0180 (19) |
| C10 | 0.071 (3) | 0.056 (2) | 0.080 (3) | −0.007 (2) | 0.005 (2) | −0.017 (2) |
| C11 | 0.069 (3) | 0.068 (3) | 0.106 (4) | −0.011 (2) | −0.001 (3) | −0.041 (3) |
| C12 | 0.060 (3) | 0.094 (4) | 0.077 (3) | 0.002 (3) | −0.003 (2) | −0.042 (3) |
| C13 | 0.065 (3) | 0.092 (4) | 0.064 (3) | 0.012 (3) | 0.000 (2) | −0.012 (3) |
| C14 | 0.060 (2) | 0.059 (3) | 0.065 (2) | 0.005 (2) | 0.0040 (19) | −0.012 (2) |
| C15 | 0.065 (2) | 0.043 (2) | 0.047 (2) | 0.0012 (18) | 0.0028 (17) | 0.0039 (17) |
| C16 | 0.066 (2) | 0.056 (2) | 0.043 (2) | 0.000 (2) | −0.0053 (17) | 0.0024 (17) |
| C17 | 0.062 (2) | 0.053 (2) | 0.047 (2) | −0.0061 (19) | 0.0019 (16) | −0.0065 (18) |
| C18 | 0.106 (3) | 0.070 (3) | 0.054 (2) | 0.006 (3) | −0.006 (2) | −0.017 (2) |
| C19 | 0.058 (2) | 0.0419 (19) | 0.049 (2) | 0.0008 (17) | 0.0061 (16) | 0.0009 (16) |
| C20 | 0.0477 (18) | 0.0387 (18) | 0.0416 (18) | −0.0057 (16) | 0.0102 (14) | −0.0001 (15) |
| C21 | 0.0487 (18) | 0.0388 (18) | 0.0427 (18) | −0.0083 (16) | 0.0096 (14) | 0.0025 (15) |
| C22 | 0.057 (2) | 0.0390 (19) | 0.0483 (19) | −0.0057 (17) | 0.0074 (15) | 0.0050 (16) |
| C23 | 0.055 (2) | 0.046 (2) | 0.052 (2) | −0.0039 (18) | 0.0106 (16) | 0.0110 (17) |
| C24 | 0.076 (3) | 0.057 (2) | 0.068 (3) | 0.011 (2) | 0.009 (2) | 0.019 (2) |
| C25 | 0.078 (3) | 0.060 (3) | 0.044 (2) | −0.009 (2) | −0.0075 (18) | 0.0094 (19) |
| C26 | 0.084 (3) | 0.048 (2) | 0.042 (2) | −0.009 (2) | −0.0007 (18) | 0.0015 (17) |
| O5 | 0.107 (2) | 0.0367 (14) | 0.0542 (15) | −0.0051 (15) | −0.0079 (14) | 0.0008 (12) |
| O1W | 0.115 (5) | 0.130 (7) | 0.119 (6) | 0.009 (5) | 0.069 (5) | 0.051 (5) |
| O2W | 0.152 (6) | 0.083 (5) | 0.101 (5) | −0.008 (5) | 0.013 (5) | 0.017 (4) |
Geometric parameters (Å, °)
| Cu1—O3 | 1.955 (2) | C12—C13 | 1.373 (6) |
| Cu1—O5 | 1.997 (2) | C12—H12 | 0.9300 |
| Cu1—N2 | 1.999 (3) | C13—C14 | 1.396 (5) |
| Cu1—N1 | 1.999 (3) | C13—H13 | 0.9300 |
| Cu1—O1 | 2.219 (3) | C14—H14 | 0.9300 |
| N1—C21 | 1.346 (4) | C15—C16 | 1.374 (5) |
| N1—C26 | 1.348 (4) | C15—H15 | 0.9300 |
| N2—C15 | 1.338 (4) | C16—C17 | 1.378 (5) |
| N2—C20 | 1.352 (4) | C16—H16 | 0.9300 |
| O1—C1 | 1.249 (5) | C17—C19 | 1.391 (4) |
| O2—C1 | 1.258 (5) | C17—C18 | 1.514 (5) |
| O2—H2 | 0.8200 | C18—H18A | 0.9600 |
| O3—C8 | 1.276 (5) | C18—H18B | 0.9600 |
| O4—C8 | 1.237 (5) | C18—H18C | 0.9600 |
| C1—C2 | 1.521 (6) | C19—C20 | 1.385 (4) |
| C2—C3 | 1.367 (6) | C19—H19 | 0.9300 |
| C2—C7 | 1.396 (6) | C20—C21 | 1.484 (4) |
| C3—C4 | 1.381 (6) | C21—C22 | 1.378 (5) |
| C3—H3 | 0.9300 | C22—C23 | 1.402 (5) |
| C4—C5 | 1.361 (7) | C22—H22 | 0.9300 |
| C4—H4 | 0.9300 | C23—C25 | 1.373 (5) |
| C5—C6 | 1.347 (8) | C23—C24 | 1.502 (5) |
| C5—H5 | 0.9300 | C24—H24A | 0.9600 |
| C6—C7 | 1.404 (7) | C24—H24B | 0.9600 |
| C6—H6 | 0.9300 | C24—H24C | 0.9600 |
| C7—H7 | 0.9300 | C25—C26 | 1.363 (5) |
| C8—C9 | 1.514 (5) | C25—H25 | 0.9300 |
| C9—C14 | 1.362 (5) | C26—H26 | 0.9300 |
| C9—C10 | 1.404 (5) | O5—H5A | 0.8200 |
| C10—C11 | 1.375 (6) | O1W—H1WA | 0.8500 |
| C10—H10 | 0.9300 | O1W—H1WB | 0.8500 |
| C11—C12 | 1.386 (7) | O2W—H2WA | 0.8500 |
| C11—H11 | 0.9300 | O2W—H2WB | 0.8500 |
| O3—Cu1—O5 | 94.85 (10) | C13—C12—H12 | 120.0 |
| O3—Cu1—N2 | 159.99 (11) | C11—C12—H12 | 120.0 |
| O5—Cu1—N2 | 91.90 (10) | C12—C13—C14 | 119.7 (5) |
| O3—Cu1—N1 | 90.50 (10) | C12—C13—H13 | 120.2 |
| O5—Cu1—N1 | 171.29 (11) | C14—C13—H13 | 120.2 |
| N2—Cu1—N1 | 80.85 (11) | C9—C14—C13 | 121.0 (4) |
| O3—Cu1—O1 | 97.39 (11) | C9—C14—H14 | 119.5 |
| O5—Cu1—O1 | 90.31 (12) | C13—C14—H14 | 119.5 |
| N2—Cu1—O1 | 101.40 (10) | N2—C15—C16 | 122.6 (3) |
| N1—Cu1—O1 | 95.82 (10) | N2—C15—H15 | 118.7 |
| C21—N1—C26 | 117.6 (3) | C16—C15—H15 | 118.7 |
| C21—N1—Cu1 | 115.4 (2) | C15—C16—C17 | 120.3 (3) |
| C26—N1—Cu1 | 127.0 (2) | C15—C16—H16 | 119.8 |
| C15—N2—C20 | 118.1 (3) | C17—C16—H16 | 119.8 |
| C15—N2—Cu1 | 126.8 (2) | C16—C17—C19 | 117.3 (3) |
| C20—N2—Cu1 | 115.1 (2) | C16—C17—C18 | 120.4 (3) |
| C1—O1—Cu1 | 128.1 (3) | C19—C17—C18 | 122.2 (4) |
| C1—O2—H2 | 109.5 | C17—C18—H18A | 109.5 |
| C8—O3—Cu1 | 128.6 (2) | C17—C18—H18B | 109.5 |
| O1—C1—O2 | 124.4 (4) | H18A—C18—H18B | 109.5 |
| O1—C1—C2 | 117.8 (4) | C17—C18—H18C | 109.5 |
| O2—C1—C2 | 117.7 (4) | H18A—C18—H18C | 109.5 |
| C3—C2—C7 | 118.9 (4) | H18B—C18—H18C | 109.5 |
| C3—C2—C1 | 120.0 (4) | C20—C19—C17 | 120.0 (3) |
| C7—C2—C1 | 121.1 (4) | C20—C19—H19 | 120.0 |
| C2—C3—C4 | 121.2 (5) | C17—C19—H19 | 120.0 |
| C2—C3—H3 | 119.4 | N2—C20—C19 | 121.7 (3) |
| C4—C3—H3 | 119.4 | N2—C20—C21 | 114.3 (3) |
| C5—C4—C3 | 119.8 (5) | C19—C20—C21 | 123.9 (3) |
| C5—C4—H4 | 120.1 | N1—C21—C22 | 121.8 (3) |
| C3—C4—H4 | 120.1 | N1—C21—C20 | 114.2 (3) |
| C6—C5—C4 | 120.7 (5) | C22—C21—C20 | 124.0 (3) |
| C6—C5—H5 | 119.6 | C21—C22—C23 | 120.5 (3) |
| C4—C5—H5 | 119.6 | C21—C22—H22 | 119.7 |
| C5—C6—C7 | 120.6 (5) | C23—C22—H22 | 119.7 |
| C5—C6—H6 | 119.7 | C25—C23—C22 | 116.2 (3) |
| C7—C6—H6 | 119.7 | C25—C23—C24 | 122.3 (3) |
| C2—C7—C6 | 118.9 (5) | C22—C23—C24 | 121.5 (3) |
| C2—C7—H7 | 120.6 | C23—C24—H24A | 109.5 |
| C6—C7—H7 | 120.6 | C23—C24—H24B | 109.5 |
| O4—C8—O3 | 125.0 (3) | H24A—C24—H24B | 109.5 |
| O4—C8—C9 | 119.7 (3) | C23—C24—H24C | 109.5 |
| O3—C8—C9 | 115.3 (3) | H24A—C24—H24C | 109.5 |
| C14—C9—C10 | 119.0 (4) | H24B—C24—H24C | 109.5 |
| C14—C9—C8 | 121.5 (3) | C26—C25—C23 | 121.1 (3) |
| C10—C9—C8 | 119.4 (4) | C26—C25—H25 | 119.5 |
| C11—C10—C9 | 120.3 (4) | C23—C25—H25 | 119.5 |
| C11—C10—H10 | 119.9 | N1—C26—C25 | 122.7 (4) |
| C9—C10—H10 | 119.9 | N1—C26—H26 | 118.6 |
| C10—C11—C12 | 120.0 (4) | C25—C26—H26 | 118.6 |
| C10—C11—H11 | 120.0 | Cu1—O5—H5A | 109.5 |
| C12—C11—H11 | 120.0 | H1WA—O1W—H1WB | 104.5 |
| C13—C12—C11 | 120.0 (4) | H2WA—O2W—H2WB | 104.5 |
| O3—Cu1—N1—C21 | −164.4 (2) | O4—C8—C9—C10 | −3.7 (6) |
| N2—Cu1—N1—C21 | −2.5 (2) | O3—C8—C9—C10 | 176.1 (3) |
| O1—Cu1—N1—C21 | 98.2 (2) | C14—C9—C10—C11 | 0.7 (6) |
| O3—Cu1—N1—C26 | 15.3 (3) | C8—C9—C10—C11 | 177.6 (4) |
| N2—Cu1—N1—C26 | 177.2 (3) | C9—C10—C11—C12 | −1.3 (6) |
| O1—Cu1—N1—C26 | −82.2 (3) | C10—C11—C12—C13 | 1.2 (7) |
| O3—Cu1—N2—C15 | −111.7 (4) | C11—C12—C13—C14 | −0.5 (6) |
| O5—Cu1—N2—C15 | −1.9 (3) | C10—C9—C14—C13 | 0.0 (6) |
| N1—Cu1—N2—C15 | −177.1 (3) | C8—C9—C14—C13 | −176.9 (3) |
| O1—Cu1—N2—C15 | 88.8 (3) | C12—C13—C14—C9 | 0.0 (6) |
| O3—Cu1—N2—C20 | 68.5 (4) | C20—N2—C15—C16 | 0.3 (5) |
| O5—Cu1—N2—C20 | 178.3 (2) | Cu1—N2—C15—C16 | −179.4 (3) |
| N1—Cu1—N2—C20 | 3.2 (2) | N2—C15—C16—C17 | −0.2 (6) |
| O1—Cu1—N2—C20 | −91.0 (2) | C15—C16—C17—C19 | 0.3 (5) |
| O3—Cu1—O1—C1 | 100.1 (3) | C15—C16—C17—C18 | −177.5 (4) |
| O5—Cu1—O1—C1 | 5.2 (3) | C16—C17—C19—C20 | −0.5 (5) |
| N2—Cu1—O1—C1 | −86.8 (3) | C18—C17—C19—C20 | 177.2 (4) |
| N1—Cu1—O1—C1 | −168.6 (3) | C15—N2—C20—C19 | −0.6 (5) |
| O5—Cu1—O3—C8 | −13.1 (3) | Cu1—N2—C20—C19 | 179.2 (2) |
| N2—Cu1—O3—C8 | 96.2 (4) | C15—N2—C20—C21 | 176.9 (3) |
| N1—Cu1—O3—C8 | 160.0 (3) | Cu1—N2—C20—C21 | −3.3 (3) |
| O1—Cu1—O3—C8 | −104.0 (3) | C17—C19—C20—N2 | 0.7 (5) |
| Cu1—O1—C1—O2 | −0.6 (5) | C17—C19—C20—C21 | −176.6 (3) |
| Cu1—O1—C1—C2 | 179.9 (2) | C26—N1—C21—C22 | 1.6 (5) |
| O1—C1—C2—C3 | 26.3 (5) | Cu1—N1—C21—C22 | −178.6 (2) |
| O2—C1—C2—C3 | −153.2 (4) | C26—N1—C21—C20 | −178.2 (3) |
| O1—C1—C2—C7 | −155.6 (4) | Cu1—N1—C21—C20 | 1.5 (3) |
| O2—C1—C2—C7 | 24.9 (5) | N2—C20—C21—N1 | 1.2 (4) |
| C7—C2—C3—C4 | 0.0 (6) | C19—C20—C21—N1 | 178.7 (3) |
| C1—C2—C3—C4 | 178.2 (4) | N2—C20—C21—C22 | −178.7 (3) |
| C2—C3—C4—C5 | −0.4 (7) | C19—C20—C21—C22 | −1.2 (5) |
| C3—C4—C5—C6 | 1.1 (8) | N1—C21—C22—C23 | −1.2 (5) |
| C4—C5—C6—C7 | −1.3 (9) | C20—C21—C22—C23 | 178.6 (3) |
| C3—C2—C7—C6 | −0.2 (7) | C21—C22—C23—C25 | −0.3 (5) |
| C1—C2—C7—C6 | −178.3 (4) | C21—C22—C23—C24 | 178.9 (3) |
| C5—C6—C7—C2 | 0.8 (8) | C22—C23—C25—C26 | 1.4 (5) |
| Cu1—O3—C8—O4 | −5.3 (6) | C24—C23—C25—C26 | −177.8 (3) |
| Cu1—O3—C8—C9 | 174.9 (2) | C21—N1—C26—C25 | −0.5 (5) |
| O4—C8—C9—C14 | 173.1 (4) | Cu1—N1—C26—C25 | 179.8 (3) |
| O3—C8—C9—C14 | −7.0 (5) | C23—C25—C26—N1 | −1.0 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2W—H2WB···O2i | 0.85 | 2.51 | 3.195 (8) | 138 |
| O2W—H2WA···O1W | 0.85 | 2.24 | 2.821 (10) | 125 |
| O1W—H1WB···O4 | 0.85 | 2.26 | 2.932 (6) | 136 |
| O1W—H1WA···O2Wii | 0.85 | 2.28 | 2.937 (10) | 134 |
| O5—H5A···O4 | 0.82 | 1.93 | 2.642 (4) | 145 |
| O2—H2···O5 | 0.82 | 1.86 | 2.636 (5) | 158 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZQ2009).
References
- Bruker (2005). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Deschamps, J. R., Hartshorn, C. M. & Chang, E. L. (2002). Acta Cryst. E58, m606–m608.
- Dong, G.-Y., Cui, G.-H. & Lin, J. (2006). Acta Cryst. E62, m628–m630.
- Feng, H., Hu, D.-C., Guo, H.-X., Zha, F. & Hu, C.-Q. (2007). Acta Cryst. E63, m2538.
- Lin, S.-H., Yang, Y.-Y. & Ng, S. W. (2008). Acta Cryst. E64, m1076. [DOI] [PMC free article] [PubMed]
- Qian, H.-F. & Huang, W. (2006). Acta Cryst. C62, m349–m351. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Willett, R. D., Pon, G. & Nagy, C. (2001). Inorg. Chem. 40, 4342–4352. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809043189/zq2009sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809043189/zq2009Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




