Abstract
In the title compound, C8H9ClN2O2, the two planar fragments, i.e. the chlorophenyl and C—C(=O)—N groups, are inclined at 14.93 (17)°. In the crystal, relatively weak intermolecular N—H⋯N, C—H⋯O and N—H⋯O hydrogen bonds connect the molecules into layers. The hydrophobic parts of molecules stick outside these layers and are connected with the neighbouring layers only by van der Waals contacts and Cl⋯Cl interactions [3.406 (2) Å].
Related literature
For background to hydrazides, see: Cajocorius et al. (1977 ▶); Liu et al. (2006 ▶); Narayana et al. (2005 ▶). For related structures, see: Akhtar et al. (2009 ▶); Lokanath et al. (1998 ▶); Mahendra et al. (2004 ▶); Podyachev et al. (2007 ▶). For graph-set symbols, see: Bernstein et al. (1995 ▶). For halogen–halogen interactions, see: Pedireddi et al. (1994 ▶).
Experimental
Crystal data
C8H9ClN2O2
M r = 200.62
Monoclinic,

a = 6.444 (1) Å
b = 4.011 (1) Å
c = 35.369 (4) Å
β = 91.89 (1)°
V = 913.7 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.39 mm−1
T = 295 K
0.4 × 0.4 × 0.15 mm
Data collection
Oxford Diffraction Xcalibur (Sapphire2, large Be window) diffractometer
Absorption correction: multi-scan (CrysAlis Pro; Oxford Diffraction, 2009 ▶) T min = 0.678, T max = 0.944
2864 measured reflections
1761 independent reflections
1448 reflections with I > 2σ(I)
R int = 0.022
Refinement
R[F 2 > 2σ(F 2)] = 0.058
wR(F 2) = 0.118
S = 1.15
1761 reflections
154 parameters
All H-atom parameters refined
Δρmax = 0.22 e Å−3
Δρmin = −0.28 e Å−3
Data collection: CrysAlis Pro (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis Pro; data reduction: CrysAlis Pro; program(s) used to solve structure: SIR92 (Altomare et al., 1993 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Stereochemical Workstation Operation Manual (Siemens, 1989 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809049538/is2496sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809049538/is2496Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O4i | 0.93 (4) | 2.51 (4) | 3.160 (3) | 127 (3) |
| N1—H1B⋯O4ii | 0.89 (4) | 2.15 (4) | 3.020 (3) | 165 (3) |
| N2—H2⋯N1iii | 0.86 (3) | 2.23 (3) | 2.997 (3) | 149 (3) |
| C8—H8⋯O4iv | 0.94 (3) | 2.51 (3) | 3.376 (3) | 153 (2) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv) 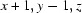 .
.
Acknowledgments
CSC thanks the University of Mysore for research facilities.
supplementary crystallographic information
Comment
Hydrazides are useful precursors in the synthesis of several heterocyclic systems (e.g., Narayana et al., 2005). Some substituted hydrazides are reported to exhibit carcinostatic activity against several types of tumors and also possess antimicrobial activity (e.g., Cajocorius et al., 1977). They are also used as intermediates in many pharmaceutically important compounds (Liu et al., 2006). A new hydrazide, 2-(4-chlorophenoxy)acetohydrazide (I, Scheme 1), C8H9ClN2O2 was synthesized and its crystal structure is reported.
The molecule of I consists of two planar fragments (Fig. 1): the phenyl ring [maximum deviation of 0.014 (2) Å] and the N—C(=O)—C group, which is planar within 0.008 (2) Å. The N1 and O6 atoms deviate significantly (by ca 0.11 Å), and in the opposite directions, from this latter plane. Overall, the molecule is only slightly bent as the dihedral angle between the planes described above is 14.93 (17)°. Even smaller values of this angle were observed in similar compounds: 5.0° in [2-methyl-4-(2-methylbenzoyl)-phenoxy]acetohydrazide (Mahendra et al., 2004), 3.6° in (2,4-dichlorophenoxy)acetohydrazide (Lokanath et al., 1998) or 5.7° in 4-tert-butylphenoxyacetohydrazide (Podyachev et al., 2007). This planar and (Z)-NCCO conformation was sometimes ascribed to the doubtful intramolecular N—H···O hydrogen bond. When the steric hindrance is present, as for instance in the structure of 2-(4-bromophenoxy)propanohydrazide (Akhtar et al., 2009), the two planar fragments become almost perpendicular, dihedral angle between them is 84.9°.
In the crystal structure rather long intermolecular hydrogen bonds connect molecules into three-dimensional network (Table 1). The N—H···N hydrogen bonds, for which the terminal nitrogen atom of NH2 group acts as an acceptor, make a C(3) graph-set motif (Bernstein et al., 1995) - the chain of molecules along the b axis. Two N—H···O hydrogen bonds between the NH2 group and carbonyl oxygen atoms from neighbouring molecules make antiparallel C(5) chains that are interwoven into subsequent R22(10) rings. In the crystal structure there are layers of molecules connected by these hydrogen bonded hydrophilic fragments and the hydrophobic chlorophenyl fragments stick outside the layers. There are relatively short and linear Cl···Cl contacts between these layers [Cl13···Cl13(3 - x,-1 - y,1 - z) 3.406 (2) Å, C10—Cl13···Cl13(3 - x,-1 - y,1 - z) 155.14 (13)°], suggesting that there is a possibility for "dihalogen" interactions (e.g. Pedireddi et al., 1994).
Experimental
A mixture of ethyl(4-chlorophenoxy)acetate (21.4 g, 0.1 mol) and 6.0 ml of hydrazine hydrate in 90 ml of ethanol was refluxed over water bath for 6 h. The precipitate formed was filtered and recrystallized from ethanol (m.p.: 425 K). Analysis for C8H9ClN2O2: Found (Calculated): C 47.89 (47.81), H 4.52 (4.48), N 13.96% (13.88%).
Refinement
All hydrogen atoms were freely refined.
Figures
Fig. 1.
Anisotropic ellipsoid representation of the compound I together with atom labelling scheme. The ellipsoids are drawn at the 50% probability level and hydrogen atoms are depicted as spheres with arbitrary radii.
Fig. 2.

The hydrogen bonded motifs in the crystal structure of I. Hydrogen bonds are shown as dashed lines. (a) the N—H···N chain. [Symmetry codes: (i) x, y, z; (ii) x, -3/2 + y, 3/2 - z; (iii) 2 - x, 1/2 + y, 3/2 - z; (iv) 2 - x, -1/2 + y, 3/2 - z; (v) 2 - x, -3/2 + y, 3/2 - z.] (b) the N—H···O chains and rings. [Symmetry codes: (i) x, y, z; (ii) 1 - x, 1/2 + y, 3/2 - z; (iii) 1 - x, -1/2 + y, 3/2 - z.]
Fig. 3.
Crystal packing as seen along the a axis. Hydrogen bonds and Cl···Cl contacts are shown as dashed lines.
Crystal data
| C8H9ClN2O2 | F(000) = 416 |
| Mr = 200.62 | Dx = 1.458 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 1266 reflections |
| a = 6.444 (1) Å | θ = 2.3–26.8° |
| b = 4.011 (1) Å | µ = 0.39 mm−1 |
| c = 35.369 (4) Å | T = 295 K |
| β = 91.89 (1)° | Plate, colourless |
| V = 913.7 (3) Å3 | 0.4 × 0.4 × 0.15 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Xcalibur (Sapphire2, large Be window) diffractometer | 1761 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1448 reflections with I > 2σ(I) |
| graphite | Rint = 0.022 |
| Detector resolution: 8.1929 pixels mm-1 | θmax = 26.8°, θmin = 2.3° |
| ω–scan | h = −8→6 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2009) | k = −3→4 |
| Tmin = 0.678, Tmax = 0.944 | l = −31→43 |
| 2864 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.118 | All H-atom parameters refined |
| S = 1.15 | w = 1/[σ2(Fo2) + (0.0271P)2 + 1.1206P] where P = (Fo2 + 2Fc2)/3 |
| 1761 reflections | (Δ/σ)max = 0.002 |
| 154 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.7883 (4) | 0.5362 (7) | 0.75916 (7) | 0.0346 (6) | |
| H1A | 0.717 (5) | 0.733 (11) | 0.7541 (10) | 0.066 (11)* | |
| H1B | 0.711 (5) | 0.417 (9) | 0.7747 (9) | 0.054 (10)* | |
| N2 | 0.8013 (3) | 0.3650 (6) | 0.72417 (6) | 0.0317 (5) | |
| H2 | 0.909 (4) | 0.247 (8) | 0.7201 (8) | 0.039 (8)* | |
| C3 | 0.6568 (4) | 0.4000 (7) | 0.69716 (7) | 0.0300 (6) | |
| O4 | 0.4957 (3) | 0.5630 (6) | 0.70086 (5) | 0.0425 (5) | |
| C5 | 0.6937 (4) | 0.2401 (8) | 0.65965 (8) | 0.0341 (6) | |
| H5A | 0.573 (5) | 0.103 (8) | 0.6525 (8) | 0.047 (9)* | |
| H5B | 0.709 (4) | 0.412 (8) | 0.6420 (8) | 0.043 (8)* | |
| O6 | 0.8781 (3) | 0.0467 (5) | 0.66140 (5) | 0.0398 (5) | |
| C7 | 0.9615 (4) | −0.0520 (8) | 0.62801 (7) | 0.0342 (6) | |
| C8 | 1.1514 (4) | −0.2151 (8) | 0.63137 (8) | 0.0412 (7) | |
| H8 | 1.213 (5) | −0.241 (8) | 0.6556 (8) | 0.050 (9)* | |
| C9 | 1.2501 (5) | −0.3162 (9) | 0.59971 (9) | 0.0478 (8) | |
| H9 | 1.374 (5) | −0.431 (9) | 0.6008 (9) | 0.062 (10)* | |
| C10 | 1.1589 (5) | −0.2639 (9) | 0.56450 (8) | 0.0473 (8) | |
| C11 | 0.9690 (5) | −0.1103 (10) | 0.56080 (8) | 0.0514 (9) | |
| H11 | 0.902 (5) | −0.092 (9) | 0.5362 (9) | 0.063 (10)* | |
| C12 | 0.8682 (5) | −0.0049 (8) | 0.59270 (8) | 0.0417 (7) | |
| H12 | 0.742 (5) | 0.108 (9) | 0.5893 (8) | 0.052 (9)* | |
| Cl13 | 1.28833 (18) | −0.3900 (3) | 0.52474 (3) | 0.0838 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0321 (12) | 0.0390 (16) | 0.0327 (12) | −0.0020 (11) | 0.0021 (10) | −0.0013 (11) |
| N2 | 0.0264 (11) | 0.0380 (15) | 0.0307 (11) | 0.0052 (11) | −0.0004 (9) | −0.0004 (10) |
| C3 | 0.0266 (12) | 0.0283 (15) | 0.0352 (13) | −0.0003 (12) | 0.0016 (10) | 0.0043 (12) |
| O4 | 0.0316 (10) | 0.0512 (14) | 0.0445 (11) | 0.0149 (10) | −0.0025 (8) | 0.0003 (10) |
| C5 | 0.0307 (14) | 0.0371 (17) | 0.0342 (14) | 0.0034 (13) | −0.0026 (11) | 0.0025 (13) |
| O6 | 0.0404 (10) | 0.0479 (13) | 0.0307 (9) | 0.0165 (10) | −0.0024 (8) | −0.0001 (9) |
| C7 | 0.0363 (14) | 0.0361 (17) | 0.0301 (13) | −0.0001 (13) | 0.0013 (11) | −0.0013 (12) |
| C8 | 0.0369 (15) | 0.049 (2) | 0.0379 (15) | 0.0075 (14) | −0.0049 (12) | −0.0007 (14) |
| C9 | 0.0384 (16) | 0.051 (2) | 0.0537 (18) | 0.0085 (16) | 0.0029 (14) | −0.0046 (16) |
| C10 | 0.0562 (19) | 0.046 (2) | 0.0406 (16) | 0.0091 (16) | 0.0109 (14) | −0.0036 (14) |
| C11 | 0.063 (2) | 0.061 (2) | 0.0305 (14) | 0.0130 (19) | −0.0030 (14) | 0.0010 (16) |
| C12 | 0.0418 (16) | 0.045 (2) | 0.0375 (15) | 0.0105 (14) | −0.0040 (12) | 0.0009 (13) |
| Cl13 | 0.0958 (8) | 0.1032 (9) | 0.0543 (5) | 0.0323 (7) | 0.0293 (5) | −0.0067 (6) |
Geometric parameters (Å, °)
| N1—N2 | 1.420 (3) | C7—C12 | 1.381 (4) |
| N1—H1A | 0.93 (4) | C7—C8 | 1.389 (4) |
| N1—H1B | 0.89 (4) | C8—C9 | 1.367 (4) |
| N2—C3 | 1.319 (3) | C8—H8 | 0.94 (3) |
| N2—H2 | 0.86 (3) | C9—C10 | 1.376 (4) |
| C3—O4 | 1.237 (3) | C9—H9 | 0.92 (4) |
| C3—C5 | 1.499 (4) | C10—C11 | 1.372 (4) |
| C5—O6 | 1.419 (3) | C10—Cl13 | 1.734 (3) |
| C5—H5A | 0.98 (3) | C11—C12 | 1.387 (4) |
| C5—H5B | 0.94 (3) | C11—H11 | 0.96 (3) |
| O6—C7 | 1.372 (3) | C12—H12 | 0.93 (3) |
| N2—N1—H1A | 107 (2) | O6—C7—C8 | 115.6 (2) |
| N2—N1—H1B | 109 (2) | C12—C7—C8 | 119.8 (3) |
| H1A—N1—H1B | 107 (3) | C9—C8—C7 | 120.1 (3) |
| C3—N2—N1 | 121.3 (2) | C9—C8—H8 | 121.4 (19) |
| C3—N2—H2 | 119.6 (19) | C7—C8—H8 | 118.4 (19) |
| N1—N2—H2 | 119.0 (19) | C8—C9—C10 | 120.0 (3) |
| O4—C3—N2 | 123.6 (2) | C8—C9—H9 | 123 (2) |
| O4—C3—C5 | 118.5 (2) | C10—C9—H9 | 117 (2) |
| N2—C3—C5 | 117.8 (2) | C11—C10—C9 | 120.5 (3) |
| O6—C5—C3 | 110.6 (2) | C11—C10—Cl13 | 120.3 (2) |
| O6—C5—H5A | 111.3 (19) | C9—C10—Cl13 | 119.1 (3) |
| C3—C5—H5A | 108.6 (17) | C10—C11—C12 | 120.0 (3) |
| O6—C5—H5B | 108.7 (18) | C10—C11—H11 | 120 (2) |
| C3—C5—H5B | 107.4 (19) | C12—C11—H11 | 120 (2) |
| H5A—C5—H5B | 110 (3) | C7—C12—C11 | 119.5 (3) |
| C7—O6—C5 | 118.1 (2) | C7—C12—H12 | 122.2 (19) |
| O6—C7—C12 | 124.6 (3) | C11—C12—H12 | 118.1 (19) |
| N1—N2—C3—O4 | −4.8 (4) | C7—C8—C9—C10 | −1.6 (5) |
| N1—N2—C3—C5 | 173.5 (2) | C8—C9—C10—C11 | 0.1 (6) |
| O4—C3—C5—O6 | −176.2 (3) | C8—C9—C10—Cl13 | 179.3 (3) |
| N2—C3—C5—O6 | 5.4 (4) | C9—C10—C11—C12 | 0.4 (6) |
| C3—C5—O6—C7 | −164.9 (2) | Cl13—C10—C11—C12 | −178.9 (3) |
| C5—O6—C7—C12 | −6.7 (4) | O6—C7—C12—C11 | 178.9 (3) |
| C5—O6—C7—C8 | 174.5 (3) | C8—C7—C12—C11 | −2.4 (5) |
| O6—C7—C8—C9 | −178.4 (3) | C10—C11—C12—C7 | 0.8 (6) |
| C12—C7—C8—C9 | 2.8 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O4i | 0.93 (4) | 2.51 (4) | 3.160 (3) | 127 (3) |
| N1—H1B···O4ii | 0.89 (4) | 2.15 (4) | 3.020 (3) | 165 (3) |
| N2—H2···N1iii | 0.86 (3) | 2.23 (3) | 2.997 (3) | 149 (3) |
| C8—H8···O4iv | 0.94 (3) | 2.51 (3) | 3.376 (3) | 153 (2) |
Symmetry codes: (i) −x+1, y+1/2, −z+3/2; (ii) −x+1, y−1/2, −z+3/2; (iii) −x+2, y−1/2, −z+3/2; (iv) x+1, y−1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2496).
References
- Akhtar, T., Khawar Rauf, M., Ebihara, M. & Hameed, S. (2009). Acta Cryst. E65, o441. [DOI] [PMC free article] [PubMed]
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst 26, 343–350.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Cajocorius, J., Cojocarius, Z. & Niester, C. (1977). Rev. Chim 28, 15–18.
- Liu, F., Stephen, A. G., Adainson, C. S., Gousset, K., Aman, M. J., Freed, E. O., Fisher, R. J. & Burke, T. R. Jr (2006). Org. Lett. 8, 5165–5168. [DOI] [PMC free article] [PubMed]
- Lokanath, N. K., Sridhar, M. A., Shashidhara Prasad, J., Nagaraja, H. S. & Mohan Rao, P. (1998). Acta Cryst. C54, 669–670.
- Mahendra, M., Doreswamy, B. H., Sridhar, M. A., Prasad, J. S., Khanum, S. A., Shashikanth, S. & Sudha, B. S. (2004). Struct. Chem 15, 211–214.
- Narayana, B., Ashalatha, B. V., Vijayaraj, K. K., Fernandes, J. & Sarojini, B. K. (2005). Bioorg. Med. Chem 13, 4638–4644. [DOI] [PubMed]
- Oxford Diffraction (2009). CrysAlis Pro. Oxford Diffraction Ltd, Yarnton, England.
- Pedireddi, V. R., Reddy, D. S., Goud, B. S., Craig, D. C., Rae, A. D. & Desiraju, G. R. (1994). J. Chem. Soc. Perkin Trans. 2, p. 2353–2360.
- Podyachev, S. N., Litvinov, I. A., Shagidullin, R. R., Buzykin, B. I., Bauer, I., Osyanina, D. V., Avvakumova, L. V., Sudakova, S. N., Habicher, W. D. & Konovalov, A. I. (2007). Spectrochim. Acta Part A, 66, 250–261. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siemens (1989). Stereochemical Workstation Operation Manual. Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536809049538/is2496sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809049538/is2496Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




