Abstract
Hox gene expression is activated by all-trans retinoic acid (RA), through binding to Retinoic Acid Receptor-Retinoid X Receptor (RAR-RXR) heterodimers bound at RA response elements (RAREs) of target genes. The RARs and RXRs each have three isotypes (α, β, and γ), which are encoded by distinct genes. Hox genes are also repressed by polycomb group proteins (PcG), though how these proteins are targeted is unclear. We used chromatin immunoprecipitation assays to investigate the association of RXRα, RARγ, cofactors, and the PcG protein SUZ12 with the Hoxa1, RARβ2, and Cyp26A1 RAREs in F9 embryonal carcinoma cells (teratocarcinoma stem cells) during RA treatment. We demonstrate that RARγ and RXRα are associated with RAREs prior to and during RA treatment. pCIP, p300, and RNA polymerase II levels increased at target RAREs upon exposure to RA. Conversely, SUZ12 was found associated with all RAREs studied and these associations were attenuated by treatment with RA. Upon RA removal, SUZ12 re-associated with RAREs. H3ac, H3K4me2, and H3K27me3 marks were simultaneously detected at target loci, indicative of a bivalent domain chromatin structure. During RA mediated differentiation, H3K27me3 levels decreased at target RAREs whereas H3ac and H3K4me2 levels remained constant. These studies provide insight into the dynamics of association of coregulators with RAREs and demonstrate a novel link between RA signaling and PcG repression.
Keywords: SUZ12, polycomb, differentiation, retinoic acid receptors, epigenetic marks
Introduction
Retinoic acid (RA) is an important regulator of vertebrate development and homeostasis because of its role in essential processes such as apoptosis, cell differentiation, and proliferation1; 2. The effects of RA are mediated through binding to the retinoic acid receptors (RARs), which are members of the nuclear receptor (NR) super family3. There are three different RARs: RARα (NR1B1), RARβ (NR1B2) and RARγ (NR1B3), which are expressed in most cell types4. Members of the NR superfamily are DNA binding transcription factors whose capacity to regulate transcription is modulated by the binding of specific ligands4; 5; 6; 7. RARs and other receptors for non-steroidal hormones, such as vitamin D receptor (VDR) and thyroid hormone receptor (TR), bind as heterodimers with one of the retinoid X receptors to hormone response elements (HRE) within the regulatory elements of target genes. In contrast, the receptors for steroid hormones, which include the estrogen (ER), androgen (AR), progesterone (PR), and glucocorticoid (GR) receptors, bind as homodimers to their respective HREs3.
In the absence of ligand, RAR-RXR heterodimers are thought to associate with RAREs and actively repress transcription through association with the corepressors (CoR) NCoR or SMRT7; 8; 9. These two proteins are found in repressor complexes containing the histone deacetylase (HDAC) HDAC310; 11. HDACs can deacetylate lysine residues of the N-terminal tails of histone proteins12. A loss of acetylation on histone tails prevents transcription through formation of a more condensed nucleosomal structure13, as well as reducing the affinity of coactivators that bind to histone tails through bromodomains14. Upon binding of the physiological agonist RA, RAR-RXR heterodimers undergo a conformational change resulting in the release of corepressor complexes3. Subsequently, RAR-RXR heterodimers can associate with coactivators15, such as the steroid receptor coactivators (p160s)16, the histone acetyl transferase (HAT) p300/CBP4, and others. Histone modifications mediated by these coactivators can create binding sites for chromatin modifiers containing specialized bromodomains17; 18. Many ATP-dependent chromatin remodelers (SWI/SNF) which use the energy of ATP hydrolysis to reposition nucleosomes contain bromodomains, and thus can be targeted to marked nucleosomes19. Acetylation of specific histone residues20 and chromatin remodeling results in the decondensation of the chromatin fiber, which is then more amenable for the recruitment of RNA polymerase II and a host of other proteins required for RA mediated transcription21; 22. Recently it has been demonstrated that the enzymes Topoisomerase II (TopoIIβ) and Poly (ADP-ribose) polymerase 1 (PARP-1) are required to form a double stranded break promoter intermediate at a RA target gene (RARβ) in order for RAR mediated transcription to occur23. Additionally, PARP-1 is required for the displacement of components of the transcriptional machinery prior to the initiation of RARβ transcription24.
Expression of the Homeobox (Hox) proteins that regulate embryonic patterning and organogenesis25 can be activated in embryonal carcinoma cells (EC) and embryonic stem (ES) cells by RA26; 27. Retinoic acid response elements (RARE) have been identified in the enhancers of a number of Hox genes26; 28; 29. Hox gene expression is also negatively regulated by Polycomb group proteins (PcG)30, though the mechanism by which PcG proteins are recruited to Hox genes is as yet undetermined. Biochemical and genetic studies indicate that PcG proteins exist in at least two separate protein complexes: Polycomb repressive complex 2/3 (PRC2) and polycomb repressive complex 1 (PRC1)30; 31. PRC2/3 is thought to be involved in the initiation of gene silencing, whereas PRC1 is implicated in the stable maintenance of gene repression. SUZ12 is a component of the PRC2/3 complex and the expression of SUZ12 mRNA and is up regulated in tumors of the colon, breast, and liver32. Additionally, the PRC2 complex contains the histone methyltransferase EZH2, which can trimethylate lysine 27 (K27) of histone H3 (H3K27me3)33. Methylation of H3K27 in vitro increases binding of the chromodomain protein Polycomb (Pc) to the chromatin33; 34; 35. Therefore it is thought that H3K27 trimethylation serves as a binding site for recruitment of PRC1.
Chromatin immunoprecipitation assays (ChIP) have been extensively used in studies monitoring the dynamics by which various NRs (most notably ER) and cofactors are recruited to regulatory elements in response to ligand36; 37; 38. However, there have been only limited studies pertaining to the dynamics of transcription complex assembly on RAREs in response to RA19; 24; 39 and much of this work was done in vitro with reconstituted chromatin templates. Therefore, we initiated experiments to monitor the association dynamics of RAR-RXR heterodimers and cofactors to RAREs in F9 EC cells during RA treatment. In particular we addressed the question of whether RAR-RXR heterodimers associate with RAREs in the absence of ligand. Additionally we monitored the kinetics with which co-activators are recruited to regulatory regions in response to RA. We also monitored histone modifications (eg. acetylation and methylation) that are usually associated with transcriptional activation at these RAREs during RA treatment. Finally, the observation that Hox genes are regulated by both RA and PcG proteins prompted us to investigate whether PcG proteins associate with RA regulatory elements, and what effect RA may have on this association.
Results
Hoxa1, Cyp26A1, and RARβ2 mRNAs are induced with similar kinetics in RA-treated F9 cells
We selected the F9 EC cell line as a model system to study all-trans-retinoic acid (RA) mediated transcriptional regulation. F9 cells respond to RA treatment by differentiating into primitive endoderm3; 40. We focused our studies on the regulation of the Hoxa1, Cyp26, and RARβ2 genes, since these three genes are transcriptionally activated by RA via well characterized RAREs26; 41; 42; 43. Wild type F9 cells were treated with 1 μM RA for various times and RNA was harvested. Expression of the selected target genes was monitored by semi-quantitative RT-PCR. The Hoxa1, Cyp26A1, and RARβ2 genes were strongly induced with similar kinetics by RA (Figure 1A). Expression of the three target genes could be detected within two hours after RA addition and expression levels continued to increase over 12 to 24 hours. To ensure that equivalent amounts of RNA from each time point were used in the RT-PCR assays, expression of the ribosomal phosphoprotein 36B4 “housekeeping gene”44 was monitored, and was shown not to fluctuate in response to RA treatment (Figure 1A).
Figure 1.
Induction of RA target genes in F9 cells as measured by RT-PCR. (A) F9 cells were treated with RA for the indicated times and RNA was harvested. Harvested RNA was assayed by semi-quantitative RT-PCR. Each experiment was repeated at least 3 times; data shown is from one experiment. Linearity of the PCR reaction was demonstrated by serial dilution of the 24 h time point template. Products were visualized by standard gel electrophoresis. (B) Quantitation of RT-PCR products by real time PCR. Results depict the mean of 3 independent experiments (±SE) with each quantitative PCR performed in triplicate. The y-axis of each graph has a different scale.
The semi-quantitative RT-PCR results were confirmed and quantitated by real time PCR analysis (Figure 1B). All three RA target genes were induced by greater than three fold following two hours treatment with RA (Figure 1B). A greater than 28 fold induction were seen for each of the RA target genes after treatment of the F9 cells with 1 μM RA for 24 hours. Real Time PCR results confirmed that the 36B4 mRNA levels did not significantly change during the RA time course treatment (Figure 1B).
RARγ and RXRα are associated with RAREs in F9 cells prior to and during RA treatment
The currently accepted model of gene regulation by nuclear receptors posits that RAR-RXR heterodimers associate with RAREs in the absence of ligand, and actively repress transcription through recruitment of corepressors such as NcoR and SMRT3; 8. As such it is thought that RAR-RXR heterodimers are constitutively associated with RAREs and that this association is not affected by exposure to ligand. We tested this hypothesis by utilizing a 2 step ChIP assay to monitor the association of RARγand RXRα with RAREs in F9 cells prior to and during RA treatment. We were unable to detect association of RARγ with target RAREs using standard ChIP assays and thus utilized a recently described, more sensitive two step crosslinking protocol45. Cells were cultured in the presence or absence of RA for various times and then subjected to the protein-protein crosslinking reagent disuccinimidyl glutarate (DSG). Cells were then formaldehyde fixed as in conventional ChIP assays, and soluble chromatin was prepared as described. Antibodies to RARγ and RXRα were used to immunoprecipitate protein-DNA complexes from soluble chromatin. Non-specific rabbit IgG antibodies were also used as a negative control in the 2 step ChIP assays.
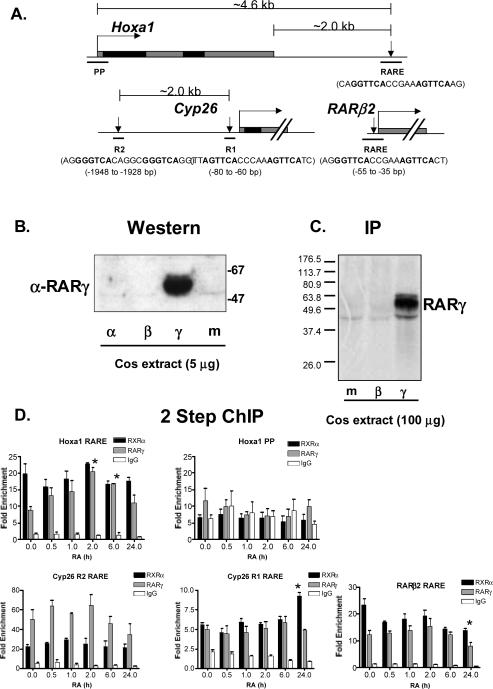
We specifically monitored the association of RARγ and RXRα with the Hoxa126 and RARβ246 RAREs, as well as the two RAREs known to regulate expression of Cyp26. The Hoxa1 RARE is located ~2kb downstream of the Hoxa1 gene, whereas the RARβ2 RARE is located ~55 bp upstream of the transcription start site (Figure 2A). Cyp26 contains a RARE ~70 bp upstream of the transcription start site denoted as R142 as well as a more recently described RARE denoted as R243, found ~1950 bp upstream of R1 (Figure 2A). Additionally, we monitored RARγ-RXRα association with the promoter proximal region of the Hoxa1 gene (PP) (Figure 2A). The specificity of the RARγ antisera was demonstrated by Western blot and immunoprecipitation (IP) analysis (Figure 2B and C). To our knowledge, our study is the first to monitor the specific association of the RARγ isotype with RAREs in living cells. As a control for the non-specific immunoprecipitation of DNA in ChIP assays, we also measured a gene free region located −18kb downstream of the Hoxb1 gene (−18kb Hoxb1). Non-specific IgG controls were also performed (Figure 2D). Levels of the aforementioned loci recovered in ChIP assays were quantitated by real time PCR assays. We define fold enrichment as the % input of a specific locus in an IP divided by the % input of the Hoxb1 −18kb 3' negative control region in the same IP (Figure 2D).
Figure 2.
Association of RARg and RXRa with RAREs in F9 cells during RA treatment. (A) Diagram of loci monitored in ChIP assays in relation to respective transcription start sites. DNA is represented by thin black line. Grey boxes denote regions of DNA transcribed into mRNA, whereas black boxes refer to regions of mRNA spliced and translated. The locations of the RAREs are indicated by downward pointing arrows, and the actual RARE sequences are shown directly below the arrows, with bold letters denoting binding sites. Bent arrows indicate transcription start sites, whereas hatchmarks signify exon/intron gene architecture, which isn't detailed. Thick black lines indicate regions of DNA amplified during ChIP assays. Schematics were drawn approximately to scale. (B) Demonstration of the specificity of the anti-RARγ antibody. Whole cell extracts (WCE) were prepared from Cos cells that were either mock transfected (m), or transfected with a plasmid expressing RARγ (γ), RARβ (β), or RARγ (γ). Five mg of each of the Cos WCEs were run on a 12% SDS-PAGE GEL followed by Western Blot analysis. Anti-RARγ blue eluate was used at a 1:200 dilution to detect RARγ. The experiment was performed 3 times. (C) Immunoprecipitation analysis also confirms specificity of the RARγ antisera. Cos cells that were either mock transfected (m), or transfected with a plasmid expressing RARβ (β), or RARγ (γ) were metabolically labeled with (35S) methionine. Cell extracts were prepared from these cells and used in IPs with the RARγ antisera (D) F9 cells were treated with RA for various times, then cells were fixed with DSG and formaldehyde and processed into soluble chromatin. Chromatin samples were IPed with antibodies to RARγ, RXRα, or IgG and bound DNA was quantitated by real time PCR. Fold enrichment is defined as % input of a specific loci in an IP divided by the % input of the Hoxb1 −18kb 3' negative control region in the same IP. Each experiment was repeated at least three times, and the quantitative PCR analyses were performed in triplicate for each sample. The data are presented as percentages of input DNA before immunoprecipitation (mean ± SE). Statistical analyses were performed with Graphpad Prism 4.0. A * symbol denotes a statistical significant difference (p < 0.05) between a time point relative to the time 0 sample for the same antibody. There were statistically significant differences (p < 0.05) between all RARγ IPs and the non-specific IgG negative controls, as well as between all RXRα IPs and the IgG negative controls at all of the RAREs assayed, whereas there was no statistically significant (p > 0.05) difference observed between both RARγ and RXRα relative to the IgG negative control at the Hoxa1 PP.
RARγ and RXRα were found associated with the Hoxa1 RARE, irrespective of the presence of RA (Figure 2D). In ChIP assays utilizing soluble chromatin prepared from untreated F9 cells, a ~19.6 fold enrichment of the Hoxa1 RARE was observed in RXRa IPs compared to a ~1.6 fold enrichment found in IPs utilizing non-specific IgG (Figure 2D, upper left hand panel, 0h). Additionally we observed an ~8.8 fold enrichment of the Hoxa1 RARE in RARγ IPs utilizing the same soluble chromatin. The association of RXRα with the Hoxa1 RARE was not affected by the presence of RA, since enrichment levels remained relatively constant during RA treatment (Figure 2D, upper left hand panel). Levels of RARγ at the Hoxa1 RARE increased as a result of RA treatment (Figure 2D, upper left hand panel, compare 0h to 2h and 6h, p value < 0.05). In contrast, RARγ and RXRα did not associate with the Hoxa1 PP region since the enrichment levels of this locus, in RARγ and RXRα IPs, were similar to the enrichment levels observed in the non-specific IgG IPs (Figure 2D, upper middle panel).
RARγ and RXRα were also associated with both the Cyp26 R1 and R2 RAREs in F9 cells prior to treatment with RA (Figure 2D, 0h). For Cyp26, the levels of RARγ and RXRα were higher at the R2 RARE compared to the R1 RARE at all time points tested. For example, there was a ~22 fold enrichment of the R2 RARE in RXRα IPs at 0 h compared to a ~5.5 fold enrichment of the R1 RARE in these same IPs (Figure 2D, lower panels, 0h). These results are consistent with a recent report demonstrating that the R2 RARE mediated higher levels of RA inducibility than the R1 RARE in transient transfection assays43. Treatment of F9 cells with RA did not change the association levels of RXRα or RARγ with the Cyp26 RAREs (Figure 2D, lower panels). Higher enrichment levels were observed for RARγ than RXRα at the Cyp26 R2 RARE at all time points.
RARγ and RXRα were associated with the RARβ2 RARE prior to RA treatment (Figure 2D, bottom right hand panel 0 h). Our results at the RARβ2 RARE are consistent with a recent report in which RAR was found constitutively bound to the RARβ2 promoter in RA treated mouse EC P19 cells24. Again we did not observe RA dependent changes in the association levels of RARγ with the RARβ2 RARE. However, the levels of RXRα at the RARβ2 RARE decreased during RA treatment (Figure 2D, compare 0h to 24h). From the results shown in Figure 2, we conclude that RARγ and RXRα associate with RAREs prior to and during RA treatment.
The co-activators p300 and pCIP are recruited similarly to RAREs during RA treatment of F9 cells
We next examined the kinetics of RA stimulated cofactor recruitment to RAREs through use of conventional 1 step ChIP assays. Specifically, we followed the association of the p160 co-activator pCIP16, as well as the histone acetyltransferase p3004, to RAREs during RA treatment of F9 cells. Prior to addition of RA, low levels of pCIP were associated with the Hoxa1 RARE, as there was a ~2.2 fold enrichment of the Hoxa1 RARE relative to the −18kb Hoxb1 negative control locus (Figure 3A, upper left hand panel, 0h). After 2 hours of RA treatment there was a 20.5 fold enrichment of pCIP at the Hoxa1 RARE relative to the −18kb Hoxb1 negative control locus. As expected, levels of the −18kb Hoxb1 “negative control” locus did not increase in ChIP assays as a result of RA treatment (Figure 3). Additionally, in ChIP assays utilizing non-specific rabbit IgG, amounts of test loci immunoprecipitated were similar to the amount of the Hoxb1 −18kb negative control region immunoprecipitated at all time points tested (data not shown in Figure 3, see Figure 4B). No increases in pCIP levels were seen at the Hoxa1 PP region (Figure 3A), indicating that the coactivator pCIP is specifically recruited to and exerts its effects from the downstream Hoxa1 enhancer.
Figure 3.
Recruitment of pCIP and p300 to RAREs in F9 cells during RA treatment. F9 cells were treated with RA for various times. Cells were then fixed with formaldehyde and processed into soluble chromatin. Chromatin samples were immunoprecipitated with (A) anti-pCIP antibody or (B) anti-p300 antibody, and bound DNA was quantitated by real time PCR. For comparison each target locus was graphed with the Hoxb1 −18kb 3' negative control locus. Each experiment was repeated at least three times, and the quantitative PCR analyses were performed in triplicate for each sample. The data are presented as percentages of input DNA before immunoprecipitation (mean ± SE). Statistical analyses were performed with Graphpad Prism 4.0. Samples that were statistically (p < 0.05) different from time 0 samples were denoted with a * symbol.
Figure 4.
Recruitment of polymerase II to RAREs in F9 cells during RA treatment. F9 cells were treated with RA for various times then cells were fixed with formaldehyde and processed into soluble chromatin. Chromatin samples were immunoprecipitated with a (A) monoclonal antibody that recognizes phosphorylated serine 5 of the CTD of RNA polymerase II or (B) non-specific rabbit IgG and bound DNA was quantitated by real time PCR. For comparison each target locus was graphed with the Hoxb1 −18kb 3' negative control locus. Each experiment was repeated at least three times, and quantitative PCR analyses were performed in triplicate. The data are presented as percentages of input DNA before immunoprecipitation (mean ± SE).
The kinetics of pCIP recruitment to the Cyp26 RARE was similar to those seen at the Hoxa1 RARE, although the fold enrichment (relative to the Hoxb1 −18kb negative control locus) and induction (changes in fold enrichment from time 0) were not as large at the Cyp26 RARE (Figure 3A, 6.1 enrichment at 2 h). The Cyp26 R2 RARE was characterized43 during the preparation of this manuscript; therefore, we only monitored co-activator recruitment to the Cyp26 R1 RARE in this study. Higher basal levels of pCIP were present at the RARβ2 RARE (Figure 3A) relative to those seen at the Hoxa1 and Cyp26 RAREs, and an additional ~2.4 fold increase in pCIP levels at the RARβ2 RARE (Figure 3A, 2 h) was observed after two hours RA treatment. This result is consistent with a previous report24 which demonstrated that much of the transcriptional machinery is associated with the RARβ2 RARE in P19 EC cells prior to RA treatment.
The kinetics of RA mediated recruitment of the HAT p300 to RAREs monitored in this study were similar to the results seen for pCIP. Levels of p300 at the Hoxa1 RARE rose ~9.4 fold after two hours treatment with RA (Figure 3B). As was the case for pCIP, p300 did not associate with the Hoxa1 PP region at any point during the time course (Figure 3B). Additionally, like pCIP, p300 levels rose over time in a RA dependent manner at the Cyp26 RARE, though not to the extent observed at the Hoxa1 RARE (Figure 3B, ~3.4 fold induction at 6 h). High basal levels of p300 were found at the RARβ2 RARE (Figure 3B, ~2.5 fold enrichment at 0 h), and p300 levels increased ~2.5 fold (Figure 3B, 2 h) during RA treatment. From these results we conclude that the patterns of RA mediated recruitment of pCIP and p300 to the RAREs we tested were similar.
Association of RNA polymerase II with RAREs during treatment of F9 cells with RA
We monitored the association of RNA polymerase II (pol II) with RAREs by using an antibody that recognizes phosphorylated serine 5 of the carboxy terminal domain (CTD) of pol II47. In other systems, serine 5 phosphorylation of the CTD has been associated with transcriptional initiation48. In the absence of RA low levels of pol II was found associated with the Hoxa1 RARE (Figure 4A, 0 h). After six hours of RA treatment, pol II levels rose ~9.7 fold at the Hoxa1 RARE (Figure 4A), similar to the changes shown in Figure 3 for pCIP and p300. In contrast to pCIP and p300, pol II was found associated at the Hoxa1 PP (Figure 4A) in the absence of RA, with levels increasing ~3 fold over time as a result of RA treatment (Figure 4A). The levels of enrichment for pol II at the Hoxa1 PP over the Hoxb1 −18kb negative control region were higher than for pol II at the Hoxa1 RARE at all time points (Figure 4A). Given the qualitative and quantitative differences observed for pol II recruitment between the Hoxa1 PP and RARE region, we conclude that, at least initially, pol II is recruited to the Hoxa1 PP independently of the Hoxa1 RARE.
Similar to the Hoxa1 PP, pol II was also found associated with the RARβ2 RARE in the absence of ligand (Figure 4A), as judged by a ~6.9 fold enrichment over the −18kb Hoxb1 negative control locus. This result is consistent with two previous reports, both of which demonstrated that pol II was constitutively bound to the RARβ2 RARE in P19 EC cells24; 49. Polymerase II levels were increased ~5 fold at the RARβ2 RARE during the course of RA treatment (Figure 4A, 1 h). Our results suggest that the RA signal culminating in the expression of Hoxa1 and RARβ2 mRNAs affects a step of the transcription process subsequent to the recruitment of pol II.
In contrast to the Hoxa1 PP and the RARβ2 RARE, only low levels of pol II (Figure 4A) were found associated with the Cyp26 R1 RARE in the absence of ligand. Upon treatment of F9 cells with RA, a ~47 fold increase in the levels of pol II was observed at the Cyp26 R1 RARE. Whereas higher levels of pCIP and p300 were associated with the Hoxa1 RARE relative to the Cyp26 R1 RARE during RA treatment (Figure 3A, B), higher levels of pol II were found at the Cyp26 R1 RARE compared to the Hoxa1 RARE during RA treatment (Figure 4A). Finally, we note that even though the RAREs of the Cyp26 and RARβ2 genes are both located in close proximity to the transcription start sites (Figure 2A), the patterns of pol II association were markedly different for these two different RAREs. Pol II was constitutively bound at the RARβ2 RARE, whereas very low levels of pol II were associated with the Cyp26 RARE prior to RA treatment. Additionally, the levels of pol II rose ~10 fold higher at the Cyp26 RARE relative to the RARβ2 RARE in response to RA (Figure 4A).
When a non-specific rabbit IgG was used in ChIP assays, the amounts of test loci immunoprecipitated were very similar to the Hoxb1 −18kb negative control immunoprecipitated at all times points tested. This result demonstrates that the changes observed for enrichment levels of pCIP, p300, and pol II at our test loci during RA treatment reflected the biological response of the cells.
Acetylation and Dimethylation of histone H3 do not increase at RAREs as a result of RA treatment
Active “open” chromatin suitable for transcription is often associated with histone acetylation, as well as H3 methylation at lysine 4. Thus we conducted ChIP assays on soluble chromatin prepared from RA treated F9 cells with an antibody that recognizes acetylated lysine residue 9 (K9) and 14 (K14) of the histone H3 tail (H3K9,K14ac). Ligand dependent increases of H3K9,K14ac have been observed for androgen, estrogen, and thyroid nuclear receptor targets genes37; 50; 51. However in P19 EC cells, the RARβ2 RARE was found to contain constitutively high levels of H3K9,K14ac that did not further increase upon treatment with RA39. We also did not observe RA dependent increases in H3K9,K14ac levels at the RARβ2 RARE (Figure 5A). Additionally, H3K9,K14ac levels were not altered in a RA dependent manner at the Hoxa1 RARE or at the Hoxa1 PP region (Figure 5A After 24 hours of RA treatment only slight increases (less than two fold) in the levels of H3K9,K14ac relative to 0 h were seen at the Cyp26 R1 RARE (Figure 5A). The −18kb Hoxb1 3' locus was also associated with H3K9,K14ac, since higher levels of the −18kb Hoxb1 locus were immunoprecipitated in H3K9,K14ac ChIPs relative to ChIPs with the other antibodies used in this study. For example, −18kb Hoxb1 locus levels equivalent to ~2.5% of input were immunoprecipitated in ChIP assays with the H3K9,K14ac antibody, compared to ~0.06% of input pulled down in ChIP assays with the pCIP antibody.
Figure 5.
Acetylation and dimethylation modifications of histone H3 tails at RAREs in F9 cells do not change as a result of RA treatment. F9 cells were treated with RA for various times, and then cells were fixed with formaldehyde and processed into soluble chromatin. Chromatin samples were immunoprecipitated with an (A) anti-H3K9,K14ac antibody or an (B) anti-H3K4me2 antibody and bound DNA was quantitated by real time PCR. For comparison each target locus was graphed with the Hoxb1 −18kb 3' negative control locus. Each experiment was repeated, starting with cell culture, at least three times, and the quantitative PCR analyses were performed in triplicate. The data are presented as percentages of input DNA before immunoprecipitation (mean ± SE).
We also conducted ChIP assays on soluble chromatin prepared from RA treated F9 cells with an antibody that recognizes dimethylation of lysine 4 of the histone H3 tail (H3K4me2). Previous reports have demonstrated that this epigenetic histone modification is enriched at actively transcribed genes18; 52; 53. This mark has also been shown to be enriched at the promoters of androgen and estrogen target genes as the result of ligand treatment37; 51. However, as was the case for acetylated histone H3 (Figure 5A), high levels of H3K4 dimethylation were detected at all of our test loci prior to RA treatment (Figure 5B). This result is consistent with a report demonstrating broad distribution of H3K4 dimethylation with the Hox clusters in mouse primary fibroblasts52. Additionally, the histone methyltransferase Mixed Lineage Leukemia (MLL1) has been shown to bind throughout the Hox domain in U937 cells54. H3K4me2 levels did not increase as a result of RA treatment at any loci (Figure 5B). Therefore our data demonstrate that at least for the RA regulated target loci we monitored, acetylation and dimethylation of the histone H3 tails precede exposure to RA, and these levels do not change as a result of RA treatment.
SUZ12 associates with RAREs in F9 cells in the absence of RA, and this association can be attenuated by treatment with RA
Hox gene expression patterns are spatially restricted in part by the negative regulating PcG proteins34; 55; 56. We therefore investigated a) whether PcG proteins associate with the Hoxa1 RARE and PP region and b) whether such interactions are altered by the presence of RA. We conducted ChIP assays using an antibody that recognizes the PcG protein SUZ12. Kirimizis and colleagues57 purified and used this antibody in ChIP assays to demonstrate an association of SUZ12 with the promoter of the MYT1 gene, as well as with other novel binding sites, in SW480 colon cancer cells. Additionally, we attempted to conduct ChIP assays with antibodies against the PRC2/3 component EZH2 methyltranferase, but these were unsuccessful.
We did not use the −18kb Hoxb1 region as a negative control in SUZ12 ChIP assays since an RA dependent decrease in levels of this locus was observed (see Discussion). Therefore, we used the Osteopontin Vitamin D Response Element (VDRE) locus as a negative control since osteopontin is a glycoprotein most abundantly produced in osteoblasts, and the expression of this gene is regulated by calcitropic hormones and cytokines58. As expected, Osteopontin expression levels in F9 cells did not change as a result of exposure to RA, as monitored by RT-PCR (data not shown). Levels of Osteopontin VDRE DNA associated with SUZ12 also remained constant during the course of RA treatment in F9 cells (Figure 6A).
Figure 6.
SUZ12 associates with RAREs in F9 cells prior to exposure to RA, and then disassociates upon RA treatment. F9 cells were treated with RA for various times. Cells were then fixed with formaldehyde and processed into soluble chromatin. Chromatin samples were immunoprecipitated with an (A) anti-SUZ12 antibody or an (B) anti-H3K27me3 antibody and bound DNA was quantitated by real time PCR. For comparison each target locus was graphed with the Osteopontin VDRE negative control locus. The y-axis of the Cyp26 RARE graph in (A) has a different scale than the other loci graphs. Each experiment was repeated at least three times, starting with cell culture, and quantitative PCR analyses were performed in triplicate. The data are presented as percentages of input DNA before immunoprecipitation (mean ± SE).
In the absence of RA, SUZ12 was found associated with both the Hoxa1 RARE and PP region, as judged by an ~8.2 fold and a ~6.7 fold enrichment respectively (Figure 6A) of these loci over the VDRE negative control locus found upstream of the Osteopontin gene59. Upon exposure to RA, SUZ12 levels rapidly decreased at both the Hoxa1 RARE and PP region (Figure 6A from 0 h to 0.5 h). SUZ12 levels continued to decline at both these loci during the course of RA treatment (Figure 6A). A ~14.4 fold decrease in SUZ12 levels occurred at the Hoxa1 RARE after a 12 hour treatment with RA. SUZ12 levels also decreased ~11.3 fold at the Hoxa1 PP region after exposure to RA for 12 hours (Figure 6A). Therefore, we conclude that SUZ12 is associated with both the promoter and enhancer region of the Hoxa1 gene, and that this association is disrupted upon exposure to RA.
Having demonstrated that RA decreases the association of PcG proteins with the regulatory regions of the Hoxa1 gene, we wanted to determine if PcG proteins play a more global role in RA mediated transcriptional regulation. Therefore, we also monitored the association of SUZ12 with the Cyp26 R1 and RARβ2 RAREs during the course of RA treatment of F9 cells. Prior to RA addition, SUZ12 was found associated with the RARβ2 RARE, as judged by a ~6.4 enrichment of this locus over that seen for the Osteopontin VDRE (Figure 6A). As was the case for the Hoxa1 locus, the levels of SUZ12 at the RARβ2 RARE rapidly decreased upon exposure to RA, and after 12 hours of RA treatment, SUZ12 levels at the RARβ2 RARE decreased to the background levels observed for the Osteopontin control VDRE (Figure 6A).
In the absence of RA, there was a ~6.6 fold higher level of SUZ12 at the Cyp26 R1 RARE relative to the Hoxa1 RARE (Figure 6A, ~54 fold enrichment for the Cyp26 RARE at 0 h versus ~8.2 fold enrichment at 0 h for Hoxa1 RARE). Even though higher levels of SUZ12 were observed at the Cyp26 R1 RARE relative to the RARβ2 RARE and Hoxa1 regulatory regions in the absence of RA, after 12 hours of RA treatment SUZ12 levels at the Cyp26 R1 RARE also decreased to the background levels observed for the Osteopontin VDRE (Figure 6A). The higher levels of SUZ12 initially observed at the Cyp26 R1 RARE may be inversely related to the larger increase in pol II recruitment observed at this locus as a result of RA treatment (Figure 4). That SUZ12 associated with all RAREs we tested and that this association was decreased by exposure to RA indicate that PcG proteins have a more global role in transcriptional repression of RA target genes.
Trimethylation of Lysine 27 of Histone H3 at RAREs also decreases in response to RA
SUZ12 has been biochemically defined as a component of the polycomb PRC2/3 complex, which also contains the histone methyltransferase EZH233. PRC2/3 complexes maintain silencing in part through trimethylation of lysine 27 (H3K27me3) of histone H357. If SUZ12 associates with RAREs as part of a functional PRC2/3 complex, then we would expect to observe H3K27me3 marks at the aforementioned RAREs. We conducted ChIP assays on soluble chromatin prepared from RA treated F9 cells with a monoclonal antibody that specifically recognizes this histone H3 modification60. In the absence of RA, H3K27me3 was significantly higher at the Hoxa1 RARE relative to the Osteopontin control VDRE (Figure 6B). After two hours exposure to RA, levels of this epigenetic mark at the Hoxa1 RARE decreased to levels observed at the Osteopontin VDRE (Figure 6B). H3K27me3 levels at the Cyp26 R1 RARE also decreased to background levels after exposure to RA (Figure 6B), though levels of this epigenetic mark were lower at the Cyp26 RARE compared to the Hoxa1 RARE. Therefore, levels of H3K27me3 at a given locus are not strictly correlated to levels of SUZ12, since we observed significantly higher levels of SUZ12 at the Cyp26 RARE relative to the Hoxa1 RARE (Figure 6A).
These results (Figure 6B) demonstrate that in the absence of RA, H3K27me3 marks, presumably mediated through the PRC2/3 complex, were present at our test RAREs. Additionally, similar to SUZ12 at these same regions, H3K27me3 levels decreased as a result of RA treatment. This latter result suggests that H3K27me3 epigenetic marks can be rapidly removed, possibly through recruitment of an unidentified demethylase capable of reversing H3K27me3 marks18, to RAREs upon treatment with RA. In support of this proposal, a histone demethylase, lysine-specific demethylase 1 (LSD-1) has been shown to interact with a nuclear receptor (androgen) and LSD-1 is required for androgen dependent transcription61.
Upon Removal of RA, SUZ12 re-associates with the Cyp26 R1 RARE
We have shown that the association of SUZ12 with RA regulatory regions in F9 cells is decreased upon exposure to RA. This observation led us to ask whether SUZ12 can re-associate with a RARE in RA treated F9 cells upon removal of RA. We cultured F9 cells in the presence of RA for 24 hours. After the 24 hour RA treatment, media was aspirated off of cells and then these cells were rinsed 3 times with PBS. Fresh media without RA was then added back to these cell cultures, which were then incubated for various lengths of time (0.5, 1, 6, or 24 h) prior to formaldehyde crosslinking (Figure 7A). Some samples were harvested immediately after the 24 h RA treatment and rinsed 3 times with PBS (denoted by 0, rinse in Figure 7) or were crosslinked without the PBS rinses (denoted by (0) in Figure 7) to reflect the harvesting conditions for samples in Figs. 2–6. Soluble chromatin was then prepared and used in ChIP assays with antibodies to SUZ12.
Figure 7.
SUZ12 rapidly associates with RAREs whereas RNA polymerase II disassociates from RAREs upon removal of RA. (A) Schematic diagram of the RA washout ChIP experiment protocol. F9 cells were treated with RA then rinsed with PBS, and fresh media was added for various times prior to formaldehyde crosslinking and cell harvest. The time of RA addition is denoted by +RA, and the wide grey bar. The times of the PBS rinses, followed by addition of fresh media, are denoted by the change to the white bar. The positive control (pos) was treated with RA for 48 hours and was not rinsed prior to crosslinking (x-link). The negative control (neg) was not treated with RA, nor rinsed prior to x-linking. The “0” sample was treated with RA for 24 hours and then immediately x-linked and cells harvested. The “0 rinse” sample was rinsed 3× after the 24 hour RA treatment and then medium containing 1% formaldehyde was immediately added to the culture dish. The addition of RA, and the subsequent steps were staggered so that all samples were x-linked at the same time. (B) Chromatin samples were immunoprecipitated with an anti-SUZ12 antibody or a (C) monoclonal antibody that recognizes phosphorylated serine 5 of the CTD of polymerase II, and bound DNA was quantitated by real time PCR. The percent of the total input of each locus was quantitated and normalized to the negative control locus. The osteopontin VDRE was used as a negative control for SUZ12 ChIP assays, and the Hoxb1 −18kb region was used as a negative control for pol II ChIP assays. The fold induction was then calculated relative to the 0 rinse sample. Each experiment was repeated at least three times, starting with cell culture and the quantitative PCR analyses were performed in triplicate for each experiment. The bars represent fold induction (±SE). Statistical analyses were performed with Graphpad Prism 4.0. A * symbol denotes statistical significance (p < 0.05), τ indicates a p value of 0.0685. Unless denoted otherwise, statistical significance is calculated relative to the 0 (rinse) sample.
Increases in SUZ12 levels at the Cyp26 R1 RARE were observed one hour after RA removal [Figure 7B, 1 h, ~3.2 fold induction compared to “0 (rinse)”]. The level of SUZ12 continued to increase at the Cyp26 R1 RARE in response to increased times after RA removal, such that 24 hours after RA removal there was a ~10.8 fold increase in the level of SUZ12 at this RARE relative to samples which were cultured in RA for 24 h and then harvested immediately after RA removal [Figure 7B, compare 24 h to the “0 (rinse)” sample at the Cyp26 RARE]. In the soluble chromatin prepared from cells that were harvested immediately after RA removal and weren't rinsed immediately before crosslinking (Figure 7B, “0”), the levels of SUZ12 at the Cyp26 RARE were similar to the levels of SUZ12 observed at the Cyp26 RARE in the “0 (rinse)” samples [Figure 7B, compare 0 to 0 (rinse)]. Additionally, soluble chromatin was prepared from F9 cells treated with RA for the entire 48 hours as a positive (pos) control and from F9 cells that were cultured for 48 hours without RA treatment as a negative control (neg). Higher levels of SUZ12 were observed at the Cyp26 R1 RARE in cells not treated with RA relative to cells continuously exposed to RA for 48 h (pos) (Figure 7B, compare neg to pos), as we expected. These results show that removal of RA from F9 cells results in the re-association of SUZ12 with the Cyp26 R1 RARE.
RNA polymerase II association at RAREs decreases upon removal of RA
We have demonstrated that in F9 cells treated with RA, RNA pol II is recruited to the Cyp26 R1, on a time scale of hours (Figure 4). We have also demonstrated that upon removal of RA from F9 cells, SUZ12, presumably as part of the repressive PRC2/3 complex, is recruited to the aforementioned RARE on a similar time scale (Figure 7B). Therefore, we wished to determine if the recruitment of SUZ12 to the Cyp26 R1 RARE, upon removal of RA, coincided with the disassociation of pol II from the same RARE. As such, we also monitored the association of pol II with the Cyp26 R1 RARE upon removal of RA in the experiment described above. Higher levels of pol II were associated with the Cyp26 R1 RARE in F9 cells treated with RA for 48 hr versus F9 cells that were not exposed to RA (Figure 7C, compare pos to neg), consistent with the results in Figure 4. Thirty minutes after removal of RA, a decrease in pol II levels was observed at the Cyp26 R1 RARE (Figure 7C, compare “0 rinse” to 0.5 h). Pol II levels at this RARE continued to decrease at later times after RA removal (Figure 7C). We conclude that upon removal of RA in F9 cells, the kinetics of pol II disassociation from the Cyp26 R1 RARE were inversely related to the kinetics of recruitment of SUZ12 to the same RAREs.
DISCUSSION
In this study we have monitored the association of RARγ, RXRα, cofactors, and histone modifications with RA regulatory elements in F9 cells before and during RA treatment. RARγ and RXRα were associated with RAREs before treatment with RA, and for the most part exposure to RA did not affect the association levels of these factors with RAREs (Figure 2). Additionally, the coactivators pCIP and p300 were recruited to RAREs with similar kinetics during RA treatment (Figure 3). The association of RNA pol II with RA regulatory elements varied in a gene specific manner. Low levels of pol II were found at the Hoxa1 and Cyp26 R1 RAREs prior to RA treatment and pol II levels at these RAREs increased dramatically in response to RA treatment (Figure 4). In contrast, high levels of pol II were associated with the RARβ2 RARE and the Hoxa1 PP even without RA treatment (Figure 4). We also demonstrated that the levels of certain histone modifications associated with transcription (H3K9,K14ac and H3K4me2) were constitutively high at target RAREs and did not increase during RA treatment (Figure 5).
Additionally, we demonstrated that the PcG protein SUZ12 was associated with all RAREs tested in the absence of RA, and that this association was reduced by RA treatment (Figure 6A). Moreover, an epigenetic mark (H3K27me3), presumably mediated by the PRC 2/3 complex, was detected at our target RAREs and the H3K27me3 levels at these RAREs also decreased in response to RA (Figure 6B). Finally, RA treatment, followed by RA removal, resulted in SUZ12 re-association with target RAREs and a concomitant decrease in levels of pol II associated at these same RAREs (Figure 7B, C). We have summarized the results of this study in a schematic model (Figure 8).
Figure 8.
A model for RARγ mediated transcription of three RA target genes. In the absence of RA, RARγ-RXRα heterodimers associated with RARβ2, Hoxa1, and Cyp26A1 RAREs presumably repress transcription through association with corepressors. RA target genes are also associated with SUZ12 in the absence of RA. For simplicity we have depicted SUZ12 blanketing the entire RARβ2 and Cyp26A1 loci, though this has not been experimentally determined. Pol II is prebound to the Hoxa1 proximal promoter (PP) and pol II, pCIP and p300 are pre-bound to the RARβ2 RARE. Upon ligand binding, a conformational change occurs in RARγ-RXRα heterodimers bound to RAREs. In the case of RARγ-RXRα heterodimers associated with the Hoxa1 and Cyp26A1 R1 RAREs, binding of RA results in the recruitment of pCIP/p300 and pol II. RA treatment also enables factors bound to the Hoxa1 3' RARE to interact with the Hoxa1 PP region. Furthermore, Suz12 dissociates from the three RA target genes upon exposure to RA. Lastly, RA is required to facilitate a step subsequent to pol II recruitment at the PP regions of Hoxa1 RARβ2 in order for these genes to be transcribed.
Association of RARγ and RXRα with RAREs
We have demonstrated that RARγ and RXRα are associated with RAREs prior to RA treatment, and that in general, association levels were not affected by the presence of RA. Consistent with our results, Pavri et al.24 have also demonstrated that RAR is constitutively associated with the RARβ2 RARE in P19 EC cells. Another recent study demonstrated that the non-steroidal thyroid hormone (TR) nuclear receptor is also constitutively associated with the TR response element (TRE) of a target gene (TRβ)62. However in this same study, TR binding to the TRE of another target gene (TH/bZIP) was found to dramatically increase upon TR treatment. Additionally, recruitment of the non-steroidal vitamin D nuclear receptor63 to cognate binding sites was shown to occur in a ligand specific manner. Therefore, the question arises as to what factors determine whether a non-steroidal nuclear receptor is constitutively associated with a regulatory element. The high basal levels of acetylated and methylated (K4) histones found at RAREs (Figure 5)39 may render RARE containing chromatin accessible to RAR-RXR binding prior to RA treatment. As demonstrated in this study, acetylation and methylation levels were not further increased during RA treatment (Figure 5) and thus these histone modifications may be at maximal levels prior to RA treatment. Additionally, in vitro studies have demonstrated that histone acetylation is required for RAR-RXR binding to a nucleosomal RARE64. In the case of other nuclear receptors, ligand specific increases in histone acetylation and methylation37; 51 have been reported. Thus, we speculate that the epigenetic marks associated with the chromatin embedding a hormone response element may influence the binding characteristics of the respective nuclear receptors.
In this study we specifically monitored the association of RARγ and RXRα with a number of RAREs. However there are two other isotypes of RAR (RARα, RARβ) and RXR (RXRβ, RXRγ) expressed in F9 cells3. It remains to be determined if RAR/RXR association with a given RARE occurs in an isotype specific manner. For example, RA induced expression of Hoxa1 and Cyp26 is greatly reduced in F9 RARγ −/− cells65. As such, RARγ may be the only RAR isotype associated with the Hoxa1 RARE and Cyp26 RAREs. If this is not the case, then what possible roles do RARα and RARβ have at these RAREs? Additionally, we found RARγ associated with the RARβ2 RARE (Figure 2B), even though RARβ2 expression is RA induced in F9 RARγ −/− cells65.
Transcriptional coregulator levels at RAREs gradually increase during RA treatment
The recruitment dynamics of cofactors to RA regulatory elements coincided with the kinetics of RA induced expression of the Hoxa1, Cyp26, and RARβ2 mRNAs (Figure 1). In general, coregulators required for transcription were recruited within 2 h after RA treatment and then levels of these factors reached a plateau thereafter. We did not observe any distinct periodic cycles of transcription factor association with any of our test loci upon ligand treatment, as has been observed for androgen50, estrogen36; 37; 38; 66, glucocorticoid67, and vitamin D receptors63. However, similar to the results of our study, Wang et al68 demonstrated that AR and cofactor levels gradually increased over a 16 h period at regulatory regions of the PSA gene in response to ligand, and the dynamics of AR association correlated with the kinetics of PSA mRNA production. The factors governing whether NRs and cofactors are recruited to promoters in discrete cycles or more progressively are unclear at this time. However eukaryotic enhancers are modular in nature, containing binding sites for multiple transcription factors69; 70. For example, the Hoxa1 enhancer contains an evolutionarily conserved element (CE2) that is required for expression of Hoxa1 in certain tissues, and is located adjacent to the Hoxa1 RARE71. Thus it is likely that the recruitment of coregulators to regulatory elements is influenced by the binding of transcription factors to neighboring regulatory elements.
Hoxa1 RARE communication with the Hoxa1 PP region
The Hoxa1 RARE is located ~4.6 kb downstream of the Hoxa1 PP region (Figure 2), which raises a question about how factors bound to these two regulatory regions are able to communicate, allowing for initiation of Hoxa1 mRNA expression. One possible explanation is that factors bound to the Hoxa1 RARE and Hoxa1 PP region physically interact, resulting in the folding and/or looping of the intervening DNA. If such a model were correct, then factors bound at the Hoxa1 RARE should be brought into proximity to the Hoxa1 PP region, and this could be observed by ChIP assays. Park et al.72 used such an approach to demonstrate that TR bound to an upstream TRE became associated with a promoter proximal G/C box regulatory element in response to T3. We have shown that RARγ, RXRα, pCIP, and p300 associated with the Hoxa1 RARE (Figure 2, 3) but we did not observe any of these proteins at the Hoxa1 PP region. However, pol II was associated with both regions of the Hoxa1 gene, and the association of pol II with the Hoxa1 PP temporally preceded pol II association with the Hoxa1 RARE (Figure 4). Therefore, the association of pol II with the Hoxa1 RARE may have arisen through interaction with the Hoxa1 PP, or alternatively, pol II may have been recruited independently to the Hoxa1 RARE during RA treatment.
SUZ12 is associated with RAREs in F9 cells in the absence of RA
PcG proteins negatively regulate expression of Hox genes34; 55; 56 and thus we monitored whether the PcG protein SUZ12 associates with Hoxa1 regulatory regions. We demonstrated that SUZ12 associated with both the Hoxa1 RARE and PP region prior to exposure to RA, and that this association decreased during RA treatment (Figure 6). Additionally, and in contrast to the other coregulators monitored in this study, SUZ12 associated with the gene free Hoxb1 −18kb 3' region (data not shown), and this association also decreased during RA treatment. During the preparation of this manuscript, studies mapping the genome wide binding sites of SUZ12 in various cell lines were published73; 74; 75; 76; 77. Consistent with our results, SUZ12 was shown to bind to large regions covering the Hox clusters in human ES cells,74, as well as in D. melanogaster Kc cells77. However, for the majority of SUZ12 target genes SUZ12 is located at the transcription start site regions74; 76.
We also demonstrated that SUZ12 associates with the Cyp26 and RARβ2 RAREs in the absence of RA (Figure 6). A functional role for SUZ12 in the regulation of these two genes was recently reported in the human embryonic diploid fibroblast cell line TIG3,73. Downregulation of SUZ12 by siRNA in this cell line resulted in the increased expression of the Cyp26 and RARβ2 mRNAs. The fact that SUZ12 associates with several RAREs suggests that PcG proteins have a global role in the negative regulation of RA target genes.
Chromatin structure of RA regulatory regions
We observed the simultaneous presence of histone modifications associated with transcriptional repression (H3K27me3) and activation (H3K4me2) at our target RAREs (Figure 5B, 6B). During the course of RA treatment the levels of H3K27me3 decreased whereas the levels of the H3K4me2 remained constant at these RAREs. Regions of the genome containing this “bivalent domain” chromatin structure have recently been mapped and shown to coincide with highly conserved noncoding elements (HCNEs) and/or developmental genes78. We also observed that H3K9, K14ac modifications found at our target RAREs remained constant during RA treatment (Figure 5A). This result suggests that H3K9, K14ac marks contribute to the chromatin structure found at bivalent domains. Moreover, our results raise the question as to whether or not all RA target genes are embedded in bivalent domains.
RA mediated dissociation of polycomb proteins from RA regulatory regions
While homeotic gene silencing mediated by PcG proteins is mitotically inherited during development, and thus stable in nature, we have shown that the association of PcG proteins with RA target genes can be rapidly decreased by exposure to RA (Figure 6A). Additionally, we showed that the epigenetic H3K27me3 mark also rapidly decreases at RAREs during RA treatment (Figure 6B). Our results are consistent with a recent report demonstrating that SUZ12 association with target genes was decreased during RA mediated neuronal differentiation of the human EC cell line NT2/D173. We have also shown that SUZ12 can re-associate with a RAREs upon removal of RA from media (Figure 7). Thus our data indicate that association of PcG proteins with RAREs in F9 cells is the default state and that removal of PcG proteins from RAREs requires exposure to RA. How is exposure to RA mechanistically translated into the disassociation of PcG proteins from RA regulatory elements? This will be a topic for future study. Another question raised by our study is whether PcG proteins are universally associated with RARE sequences, and if so, whether PcG proteins are specifically recruited to RA target genes through RARE sequences or through some other undefined feature of these genes.
The association of PcG proteins with RA target genes may arise through interactions with RAR-RXR heterodimers. Support for such a model comes from a recent study demonstrating that the human tumor antigen PRAME (preferentially expressed antigen in melanoma) can bind to liganded RAR and repress transcription through recruitment of PcG proteins79. Additionally, the authors showed that stable expression of PRAME in F9 cells blocked RA induced differentiation and inhibited RA induced gene expression. In the context of wild type F9 cells, we posit that an as yet unidentified protein may simultaneously interact with PcG proteins and RAR-RXR heterodimers in the absence of RA, allowing for the recruitment of PcG proteins to RA target genes.
We have conducted ChIP studies to monitor the dynamics of association of RARγ, RXRα, and cofactors to RA regulatory regions during treatment with RA. These studies have revealed the patterns and kinetics by which these factors are recruited to different target genes. We have also revealed a novel link between PcG silencing and RAR signaling. Our data suggest that PcG proteins can be targeted to specific areas of the genome through association with RAREs (Figure 6). Moreover, we have demonstrated that levels of SUZ12 and repressive epigenetic marks generated by the PRC 2/3 complex at RAREs can be attenuated by exposure to RA (Figure 6). Thus, our data suggest a mechanism by which PcG mediated repression of target genes may be alleviated. More specifically, the results presented herein may explain how PcG mediated repression of Hox gene expression is relieved during activation by treatment with RA.
MATERIALS AND METHODS
Cell Culture
F9 wild type embryonal carcinoma cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Invitrogen), 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were plated in gelatin coated tissue culture plates approximately 48 hours prior to RNA harvesting (5×105 cells/60 mm dish) or formaldehyde fixation (2.5×106 cells/20 cm dish). Treatment with 1 μM retinoic acid for 0, 0.5, 1, 2, 6, 12, and 24 hours was staggered so that all time points were collected simultaneously.
Antibodies and Chemicals
all-trans Retinoic acid (RA) was obtained from Sigma-Aldrich (St. Louis, MO) and dissolved in ethanol. Anti-RARγ serum was generated by immunization of rabbits with a peptide corresponding to 15 amino acids at the carboxy terminal of RARγ. Polyclonal anti-RARγ IgG was purified from the crude serum through use of a DEAE Affi-gel Blue Gel column (Bio-Rad, Hercules, CA). Anti-RXRα (D-20, sc-553), anti-pCIP (M-397, sc-9119), anti-p300 (N-15, sc-584) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-H3K4me2 (07-030), anti-H3K9,K14ac (06-599), and anti-SUZ12 (07-379) antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-phospho Ser5 CTD of RNA polymerase II (pCTDser5) was purchased from Covance Research Products (Richmond, CA). Anti-SUZ12 (ab12201 since discontinued) and anti-H3K27me3 (mAbcam 6002) antibodies were purchased from Abcam Inc. (Cambridge, MA).
RNA preparation and RT-PCR
F9 EC cells were plated in 60-mm dishes (5×105 cells/dish) and treated with RA as indicated. Total RNA was prepared using Trizol reagent (Invitrogen Life Technologies, Carsbad, CA) according to the manufacturer's instructions. Reverse Transcription (RT) was performed on 2 μg RNA from each timepoint using 50 ng oligo (deoxythymidine) and 50 units of Superscript II (Invitrogen Life Technologies, Carsbad, CA) reverse transcriptase as recommended by the anufacturer. Synthesized cDNAs were diluted 1:10 prior to use in semi-quantitative or real time PCR reactions. Each experiment was conducted on three independent occasions.
Western Blot and Immunoprecipitation Analysis
Whole cell extracts (WCE) were prepared from Cos cells that were either mock transfected (m), or transfected with a plasmid expressing RARα, RARβ, or RARγ. Five μg of each of the Cos WCEs were run on a 12% SDS-PAGE gel followed by transfer to a nitrocellulose membrane (0.45 μm pore size; catalog number 162-0090, Bio Rad Hercules, CA). The membrane was stained with Ponceau S (Sigma) to ensure proper transfer and equivalent loading. Primary antibody incubation was done overnight at 4° C. The Anti-RARγ blue eluate, as described above, was used at a 1:200 dilution to detect RARγ. After a 1-hour incubation with an immunoglobulin G horseradish peroxidase-conjugated secondary antibody at room temperature (anti-rabbit, 1:40,000 dilution; sc-2030, Santa Cruz Biotechnology), the membranes were developed with Supersignal Substrate (Pierce, Rockford, IL) for 5 minutes and exposed to Biomax film (Eastman Kodak, Rochester, NY). Primary and secondary antibodies were diluted in PBS containing 5% Blotto (Santa Cruz Biotechnology) and 0.1% Tween 20. For IP analysis, Cos cells either mock transfected or transfected with a plasmid expressing either RARγ or RARβ, were metabolically labeled with 100 μCuries of (35S)-methionine for 1 hour. Cos cell extract was then prepared from each of these cells and 100 μg of each of the extracts were immunoprecipitated with 25 μl of RARγ blue eluate. IP complexes were isolated by incubation with 25 μl of a 50:50 (v/v) slurry of a Protein A sepharose/TE mixture. After three washes with ChIP lysis buffer, Protein A sepharose-IP complexes were resuspended in SDS loading buffer and ran on a 12% SDS-1 PAGE gel. The gels were then fixed and subjected to autoradiography.
ChIP assays
For 2 step chromatin immunoprecipitation (ChIP) assays, 2.5 × 106 F9 EC cells were grown in a 20-cm dish and the cells were treated with 1 μM retinoic acid as indicated. Approximately (~) 36 hours after plating (~1.8–2.3×107 cells), cells were briefly rinsed with PBS, and then fixed with 10 ml of 2mM disuccinimidyl glutarate (DSG, Pierce, Rockford IL)/PBS at room temperature with shaking for 45 minutes. A 0.5 M stock concentration of DSG was prepared immediately before use by dissolving powdered DSG in 100% dimethyl sulfoxide. Cells were then briefly rinsed with PBS and subsequently fixed with 10 ml of 1% formaldehyde (J.T. Baker, Phillipsburg, NJ) /PBS at room temperature with shaking for 10 minutes. In one step ChIP experiments requiring only formaldehyde fixation, cells were fixed by directly adding formaldehyde (37%) to culture media (1% final concentration) and incubating at room temperature with shaking for 10 minutes. In both 1 and 2 step ChIP assays, formaldehyde fixation was quenched by the addition of 1.25 M glycine to a 200 mM final concentration. Cells were then washed with cold phosphate buffered saline (PBS) and lysed by adding 350 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.5× Complete Mini protease inhibitors, catalog number 11836153001 [Roche Molecular Biochemicals, Mannheim, Germany]) to the cell pellet. The amount of lysis buffer added to samples was adjusted to normalize for differences in cell numbers between plates. DNA was sheared by sonicating for 2×15 seconds (setting 3) on a Branson 150 Sonifier, and cell debris was eliminated by centrifugation at 14,000 revolutions per minute (rpm) for 10 min at 4°C. For each immunoprecipitation (IP), 50 μl of soluble chromatin (usually out of a total volume of ~350 μl) was diluted tenfold with lysis buffer and pre-cleared with 25 μl of a 50% protein A-sepharose/PBS slurry, catalog number 17-0780-01 (Amersham Biosciences, Uppsla Sweden) for 1–2 h at 4°C.
Immunoprecipitations were performed at 4°C overnight with 2 μg of each specific antibody. Complexes were collected by incubation with 50 μl of 50% protein ASepharose/PBS slurry catalog number 17-0780-01 (Amersham Biosciences) for 2 h at 4°C. For immunoprecipitations with anti-pCTD antibodies, 2.5 μg of anti-IgM (M8644-1MG, Sigma-Aldrich) was added 1h prior to addition of beads. Beads were washed twice for 5 minutes at room temperature with lysis buffer. Two more washes were carried out with ChIP wash buffer (50 mM Tris-HCl [pH 8.5], 500 mM LiCl, 5mM EDTA, 1% NP-40, 1% sodium deoxycholate) followed by two washes in TE buffer (10mM Tris-HCl [pH 8.0], 1 mM EDTA). Immunocomplexes were eluted from beads by incubation with 100 μl elution buffer (50 mM Tris-HCl [pH 8.0], 1% SDS, 1 mM EDTA) at 65°C for 10 minutes, followed by vortexing for 15 seconds. After a brief centrifugation step, eluted protein-DNA complexes were transferred to new tubes, NaCl was added to a final concentration of 200 mM, and samples were incubated at 65°C overnight to reverse crosslinking.
DNA was purified using a Qiaquick Spin Kit (Qiagen Sciences, MD) and assayed by either real time PCR or semi-quantitative PCR. For input samples, 25 μl (5.5% of amount used for IP with specific antibody) was added to 75 μl elution buffer, NaCl was added to a 200 mM final concentration, and reverse crosslinked at 65°C overnight. Each ChIP experiment was carried out on at least three independent occasions, starting with cell culture.
RA washout ChIP experiments
F9 EC cells (2.5 × 106) were grown in 20 cm dishes and treated with RA for 24 hours (pulse). Media were then aspirated off of plates, and the cells were rinsed with PBS 3 times. Fresh media lacking RA was then added back to the plates and the cultures were incubated at 37°C for various lengths of time (chase). Treatment with RA was staggered so that all plates were crosslinked at the same time (Figure 7A). For example, the -RA 24 h sample was initially exposed to RA 18 hours earlier than the -RA 6 h sample. Consequently the -RA 24 h sample was rinsed with PBS 18 hours earlier than the -RA 6 h sample. The positive control sample was exposed to RA for 48 h, whereas the negative control did not receive treatment with RA.
Semi-quantitative and real time PCR
Semi-quantitative PCRs were performed with Taq polymerase in 20 μl reactions containing PCR buffer (final concentration of 100 mM Tris-HCl [pH 8.3], 100 mM KCl, 1.8 mM MgCl2), 0.1 mM concentration of each deoxynucleoside triphosphate, 0.1 μM concentration of each primer (Table 1), and 3 μl of template. Reactions were performed in a MJ Research PTC-200 thermal cycler using a touchdown protocol as follows: following a 15 second 94°C denaturation step, the initial annealing/extension temperature started at 70°C (30 seconds) and was reduced by 0.5°C for each of the first 20 cycles. The number of additional cycles (2–13) utilizing a 60°C annealing/extension step (30 seconds) varied for each primer pair. PCR products were electrophoresed in a 2% agarose gel and visualized with ethidium bromide (0.3 mg/ml). Amplification in the linear range was demonstrated by 3 fold serial dilution of templates. Real Time PCR assays were carried out on a MJ Research Opticon DNA Engine using the same reaction mixture as described above, supplemented with a final concentration of 5% DMSO and 1:20,000 dilution of SYBR green reagent (Invitrogen Life Technologies, Carsbad, CA). The touchdown protocol describe above was used for amplification with the following modifications: the initial annealing/extension step of 70°C was decreased 1.0°C for 10 cycles, followed by an additional 35 cycles utilizing a 60°C annealing/extension step. Fluoresence was monitored during a 5 second 77°C step (temperature was modified depending on primer pair) immediately after the 60°C annealing/extension steps. Real Time data were quantitated through use of a standard curve generated by serial dilution of input samples. Standard curves were shown to be linear in the range of 30% to 0.001% of the input. ChIP samples were quantitated by measuring CT values and interpolating the % input on the standard curve. Each experiment was repeated at least 3 times, and quantitative PCRs were performed in triplicate. Results are presented as the mean of triplicates, along with error bars corresponding to the standard error. Statistical analyses of ChIP assays were performed by a 2-way analysis of variance using the software GraphPad Prism 4.0. The UCSC in silico PCR program (http://genome.ucsc.edu/cgi-bin/hgPcr) was used to ensure primers designed for ChIP generated a single amplicon from the appropriate target loci.
TABLE 1.
DNA sequences of oligonucleotides used for ChIP and RT-PCR
| Loci | Sense primer (5'–3') | Antisense primer (5'–3') | Product size (bp) |
|---|---|---|---|
| ChIP primers | |||
| Hoxa1 rare | TCTTGCTGTGACTGTGAAGTCG | GAGCTCAGATAAACTGCTGGGACT | 268 |
| Hoxa1 pp | ATTGGCTGGTAGAGTCACGTG | GAAAGTTGTAATCCCATGGTCAGA | 276 |
| CYP26 R1 rare | CCCGATCCGCAATTAAAGATGA | CTTTATAAGGCCGCCCAGGTTAC | 87 |
| CYP26 R2 rare | TTCACTGAGATGTCACGGTCC | TTCCCAATCCTTTAGCCTGA | 64 |
| RARβ2 rare | TGGCATTGTTTGCACGCTGA | CCCCCCTTTGGCAAAGAATAGA | 284 |
| −18kb Hoxb1 3' | ACTCCAGCTCCCATTTCCCACTT | CTGCCTGCCTCTGCCTCACA | 411 |
| Osteopontin | GTATTCCAGTCTCACAAACTGCTTG | CATACTGTGTTCCAGGTCAGTTGG | 336 |
| RT-PCR primers | |||
| Hoxa1 a | TGGAGGAAGTGAGAAAGTTGGC | ATGGGAGTCGAGAGGTTTCC | 484 |
| b | TTCCCACTCGAGTTGTGGTCCAAGC | 147 | |
| CYP26 | GAAACATTGCAGATGGTGCTTCAG | CGGCTGAAGGCCTGCATAATCAC | 272 |
| RAR β 2 | GATCCTGGATTTCTACACCG | CACTGACGCCATAGTGGTA | 247 |
| 36B4 | AGAACAACCCAGCTCTGGAGAAA | ACACCCTCCAGAAAGCGAGAGT | 448 |
| Osteopontin | TGACGAATCTCACCATTCGGATGA | TTTCCAGACTTGGTTCATCCAGCT | 338 |
Primer used in semi-quantitative PCR analysis
Primer used in real time RT-PCR analysis
Acknowledgements
We thank Gene Bryant for help and advice with ChIP assays. We are grateful to Nigel Mongan for critical reading of this manuscript and helpful discussion. This work was supported by National Institutes of Health (NIH) grants RO1CA043796(LJG) and DRO1DK454560 (partial support for RG; P.I. L. Freedman).
Abbreviations
- RA
retinoic acid
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- NR
nuclear receptor
- RARE
retinoic acid response element
- HAT
histone acetyl transferase
- pol II
RNA polymerase II
- Hox
homeobox
- EC
embryonal carcinoma
- PcG
polycomb group protein
- PRC
polycomb repressive complex
- ChIP
chromatin immunoprecipitation
- ER
estrogen receptor
- CTD
carboxy terminal domain
- WCE
whole cell extract
- DSG
disuccinimidyl glutarate
- IP
immunoprecipitation
- CTY
cycle threshold
- PP
promoter proximal
- VDRE
vitamin D response element
- TR
thyroid receptor
- AR
androgen receptor
- PSA
prostate specific antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–80. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 2.Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–33. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- 3.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol. 2005;19:1418–28. doi: 10.1210/me.2005-0125. [DOI] [PubMed] [Google Scholar]
- 5.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–64. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 6.Olefsky JM. Nuclear receptor minireview series. J Biol Chem. 2001;276:36863–4. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- 7.Rochette-Egly C. Dynamic combinatorial networks in nuclear receptor-mediated transcription. J Biol Chem. 2005;280:32565–8. doi: 10.1074/jbc.R500008200. [DOI] [PubMed] [Google Scholar]
- 8.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 9.Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA. The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep. 2005;6:445–51. doi: 10.1038/sj.embor.7400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–57. [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? Embo J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–8. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 15.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–54. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 16.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 17.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–75. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 18.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 19.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 20.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 21.Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–54. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 22.Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 23.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 24.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 26.Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3' of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38:217–27. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 27.LaRosa GJ, Gudas LJ. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol Cell Biol. 1988;8:3906–17. doi: 10.1128/mcb.8.9.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3' of the Hox A and Hox B homeobox gene clusters. Functional analysis. J Biol Chem. 1997;272:2167–75. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 29.Mainguy G, In der Rieden PM, Berezikov E, Woltering JM, Plasterk RH, Durston AJ. A position-dependent organisation of retinoid response elements is conserved in the vertebrate Hox clusters. Trends Genet. 2003;19:476–9. doi: 10.1016/S0168-9525(03)00202-6. [DOI] [PubMed] [Google Scholar]
- 30.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 31.Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–46. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–21. [PubMed] [Google Scholar]
- 33.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–64. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 35.Czermin B, Imhof A. The sounds of silence--histone deacetylation meets histone methylation. Genetica. 2003;117:159–64. doi: 10.1023/a:1022927725945. [DOI] [PubMed] [Google Scholar]
- 36.Metivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, Gannon F. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. Embo J. 2004;23:3653–66. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 38.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre B, Ozato K, Lefebvre P. Phosphorylation of histone H3 is functionally linked to retinoic acid receptor beta promoter activation. EMBO Rep. 2002;3:335–40. doi: 10.1093/embo-reports/kvf066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang SY, LaRosa GJ, Gudas LJ. Molecular cloning of gene sequences transcriptionally regulated by retinoic acid and dibutyryl cyclic AMP in cultured mouse teratocarcinoma cells. Dev Biol. 1985;107:75–86. doi: 10.1016/0012-1606(85)90377-x. [DOI] [PubMed] [Google Scholar]
- 41.de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–80. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 42.Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–97. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- 43.Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241–8. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba H, Clifford J, Metzger D, Chambon P. Specific and redundant functions of retinoid X Receptor/Retinoic acid receptor heterodimers in differentiation, proliferation, and apoptosis of F9 embryonal carcinoma cells. J Cell Biol. 1997;139:735–47. doi: 10.1083/jcb.139.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–25. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 46.Sucov HM, Murakami KK, Evans RM. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc Natl Acad Sci U S A. 1990;87:5392–6. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–27. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 48.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270:3859–70. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre B, Brand C, Lefebvre P, Ozato K. Chromosomal integration of retinoic acid response elements prevents cooperative transcriptional activation by retinoic acid receptor and retinoid X receptor. Mol Cell Biol. 2002;22:1446–59. doi: 10.1128/mcb.22.5.1446-1459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol. 2004;18:2633–48. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Xia X, Fondell JD, Yen PM. Thyroid hormone-regulated target genes have distinct patterns of coactivator recruitment and histone acetylation. Mol Endocrinol. 2006;20:483–90. doi: 10.1210/me.2005-0101. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244–53. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 56.Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–6. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 57.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noda M, Yoon K, Prince CW, Butler WT, Rodan GA. Transcriptional regulation of osteopontin production in rat osteosarcoma cells by type beta transforming growth factor. J Biol Chem. 1988;263:13916–21. [PubMed] [Google Scholar]
- 59.Noda M, Vogel RL, Craig AM, Prahl J, DeLuca HF, Denhardt DT. Identification of a DNA sequence responsible for binding of the 1,25-dihydroxyvitamin D3 receptor and 1,25-dihydroxyvitamin D3 enhancement of mouse secreted phosphoprotein 1 (SPP-1 or osteopontin) gene expression. Proc Natl Acad Sci U S A. 1990;87:9995–9. doi: 10.1073/pnas.87.24.9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25:1041–53. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 62.Buchholz DR, Paul BD, Shi YB. Gene-specific changes in promoter occupancy by thyroid hormone receptor during frog metamorphosis. Implications for developmental gene regulation. J Biol Chem. 2005;280:41222–8. doi: 10.1074/jbc.M509593200. [DOI] [PubMed] [Google Scholar]
- 63.Vaisanen S, Dunlop TW, Sinkkonen L, Frank C, Carlberg C. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1alpha,25-Dihydroxyvitamin D3. J Mol Biol. 2005;350:65–77. doi: 10.1016/j.jmb.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 64.Lefebvre P, Mouchon A, Lefebvre B, Formstecher P. Binding of retinoic acid receptor heterodimers to DNA. A role for histones NH2 termini. J Biol Chem. 1998;273:12288–95. doi: 10.1074/jbc.273.20.12288. [DOI] [PubMed] [Google Scholar]
- 65.Boylan JF, Lufkin T, Achkar CC, Taneja R, Chambon P, Gudas LJ. Targeted disruption of retinoic acid receptor alpha (RAR alpha) and RAR gamma results in receptor-specific alterations in retinoic acid-mediated differentiation and retinoic acid metabolism. Mol Cell Biol. 1995;15:843–51. doi: 10.1128/mcb.15.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 67.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20:560–72. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 68.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 69.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 70.Michelson AM. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc Natl Acad Sci U S A. 2002;99:546–8. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson JR, Chen SW, Ho L, Langston AW, Gudas LJ. An evolutionary conserved element is essential for somite and adjacent mesenchymal expression of the Hoxa1 gene. Dev Dyn. 1998;211:97–108. doi: 10.1002/(SICI)1097-0177(199801)211:1<97::AID-AJA9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 72.Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–53. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 73.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang Y, Faria TN, Chambon P, Gudas LJ. Identification and characterization of retinoic acid receptor beta2 target genes in F9 teratocarcinoma cells. Mol Cancer Res. 2003;1:619–30. [PubMed] [Google Scholar]
- 76.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 77.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–9. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 78.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 79.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–47. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]