Abstract
We quantified mRNA abundance from 10 stages in the Giardia lamblia life cycle in vitro using Serial Analysis of Gene Expression (SAGE). 163 abundant transcripts were expressed constitutively. 71 transcripts were upregulated specifically during excystation and 42 during encystation. Nonetheless, the transcriptomes of cysts and trophozoites showed major differences. SAGE detected co-expressed clusters of 284 transcripts differentially expressed in cysts and excyzoites and 287 transcripts in vegetative trophozoites and encysting cells. All clusters included known genes and pathways as well as proteins unique to Giardia or diplomonads. SAGE analysis of the Giardia life cycle identified a number of kinases, phosphatases, and DNA replication proteins involved in excystation and encystation, which could be important for examining the roles of cell signaling in giardial differentiation. Overall, these data pave the way for directed gene discovery and a better understanding of the biology of Giardia lamblia.
Keywords: Giardia lamblia, transcriptome, gene expression, life cycle, SAGE 1
Giardiasis is a major contributor to the enormous burden of human diarrheal diseases, which are second only to respiratory infections as causes of mortality and morbidity worldwide [1]. Nonetheless, the pathophysiology of G. lamblia (synonyms G. intestinalis, G. duodenalis) is not well understood [2]. Although trophozoites are not invasive, Giardia is capable of causing severe and protracted diarrhea that can lead to malabsorption and failure of children to thrive. On the other hand, about half of infected people are asymptomatic and the infection frequently resolves spontaneously. Thus, both the duration and symptoms of giardiasis are highly variable in immunocompetent people, for reasons that are not understood. Although there is antigenic variation [3], there is little evidence of stable strain differences in virulence.
G. lamblia has two life cycle stages that are remarkably well-adapted to survival in very different and inhospitable environments [2]. The ovoid cyst form, which is responsible for transmission of giardiasis, persists for months in fresh water at 4°C [4]. Trophozoites colonize the human small intestine and are responsible for disease. Although the cyst was once thought to be a cryptobiotic form, it has ~15% of the oxygen consumption of trophozoites [5] and is competent to pass through the stomach and excyst in the small intestine.
We have used Serial Analysis of Gene Expression (SAGE) [6, 7] to monitor genome-wide levels of messenger RNA (mRNA) expression throughout Giardia’s life cycle in vitro (Table 1). SAGE is a powerful tool that does not depend upon construction of gene specific probes or primers, but instead simply samples any polyadenylated RNA transcript that includes a NlaIII restriction site (5’-CATG-3’). In SAGE, a short sequence tag (15 bp) from a unique position of a mRNA molecule is used to uniquely identify the source gene from within the genome. Sequence tags are isolated from the mRNA pool of a cell and are linked together to form long concatenated molecules that are cloned and sequenced. The population of tags define patterns of expression of individual genes. Quantification of all tags provides a relative measure of gene expression (i.e. mRNA abundance).
Table 1.
SAGE of the Giardia lamblia life cycle. The WB isolate, clone C6 (ATCC #50803) of Giardia lamblia was used for our transcriptome studies. To minimize genetic drift, a new clone “A11” was isolated just prior to our SAGE examination of the giardial life cycle. An ~80% confluent culture of trophozoites was used to inoculate 15 ml culture tubes at a density of 5x105 cells/ml and grown for 21 hours (log phase trophozoites). Encystation was induced in vitro as described by Sun et al. [16] from these log phase trophozoite cultures. Cells were harvested at 0 (trophozoite), 4, 12, 21, and 42 hr of encystation for total RNA isolation. Excystation was carried out by a two-step method [12] and total RNA isolated from water-resistant cysts, Stage 1 and Stage 2 of excystation, and 30 and 60 min post-excystation. Stage 1 (S1) is an acidic solution that models the stomach. Stage 2 (S2) contains trypsin at a slightly alkaline pH that mimics the small intestine. Total RNA was isolated from all cells using TRIZOL (Invitrogen) and stored at −80°C until use. Integrity of all RNA samples was checked by electrophoresis and Northern blotting for genes of known size and expression (e.g. BiP, ~2 kb, which is constitutive; IscU, 0.85 kb, down-regulated in encystation; CWP-2, 1.3 kb, upregulated in encystation). SAGE libraries were constructed using the I-SAGE kit (Invitrogen, Carlsbad, CA) using 10 μg of total RNA. Recombinant pZero clones were sequenced with an ABI 3730xl capillary DNA sequencer using the M13F primer and results analyzed with software created specifically for Giardia SAGE analysis, excluding SAGE tags containing putative sequencing error.
| SAGE Library | Total Tags |
|---|---|
| Cyst | 36,385 |
| S1 Excystation | 16,141 |
| S2 Excystation | 38,586 |
| 30 min Post-Excystation | 38,633 |
| 60 min Post-Excystation | 38,669 |
| Trophozoite | 38,015 |
| 4 hour Encystation | 37,055 |
| 12 hour Encystation | 37,814 |
| 21 hour Encystation | 18,370 |
| 42 hour Encystation | 39,173 |
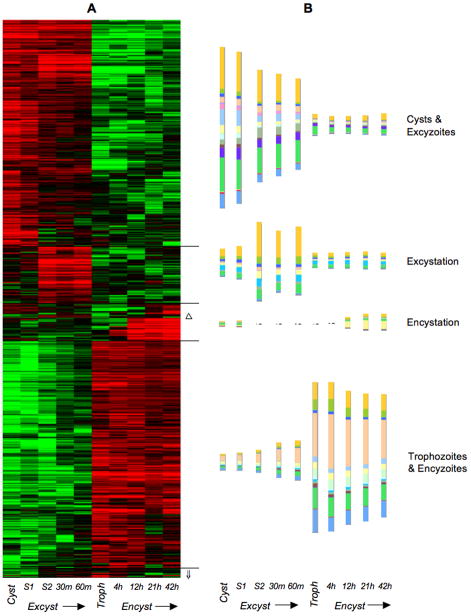
Raw SAGE data were deposited to GenBank’s GEO database under accession GSE8336. The 338,841 total tags sampled reduced to 22,807 unique 15 bp tag sequences with varying frequencies. Using the nearly complete genome sequence [8], 15,551 (68.2%) tags were assigned to a gene (Table S1), 625 (2.7%) were unresolvable due to inclusion of adenosine residues from mRNA polyadenylation, 1201 (5.3%) could not be resolved among multiple mappings to the Giardia genome, 4427 (19.4%) mapped outside predicted genes, and 1003 (4.4%) did not map to the Giardia WB-C6 genome sequence. This relative number of unassigned tags is usually seen in SAGE projects [9] and the last group could be due to contamination, sequencing errors, allelic sequence divergence, poorly assembled regions in the genome, and chimeric products. For purpose of analysis, we set aside the 7256 unassigned SAGE tags (Table S2) and focused upon transcript abundance, as represented by summed SAGE tag frequencies (sense orientation tags contributing to sense transcript frequencies and antisense orientation tags to antisense transcript frequencies) (Table S1). While not an exhaustive sampling of the giardial transcriptome due to both the number of molecules sequenced (for example, compared to Illumina sequencing) and the existence of transcripts without NlaIII sites, SAGE detected transcription of 75.6% of predicted genes in the Giardia genome. As expected from earlier studies, ~20% of transcripts detected by SAGE were in the antisense orientation [10]. We used Stekel et al.’s R-statistic [11] to score transcripts for differential expression among libraries, finding 697 transcripts with significant variation in abundance (R ≥ 8) during the Giardia lamblia life cycle (Table S1, Figure 1). This analysis is based on a new, extensive curation of the SAGE data of thousands of Giardia's transcripts. This new data is available on GiardiaDB (www.giardiadb.org) (June 2010 release) to provide the best possible transcript-level expression data (previously GiardiaDB only contained raw, uncurated SAGE data).
Figure 1.
A) Hierarchical cluster analysis of 697 differentially expressed (R ≥ 8) transcripts from 10 stages in the complete G. lamblia life cycle. Red indicates upregulation and green indicates downregulation relative to the median abundance (black) for each transcript. Hierarchical clustering was performed with all transcript SAGE frequencies log transformed, median centering for both transcripts and SAGE libraries, and clustering of both using centered correlation and the average linkage clustering method. Δ denotes a sub-cluster of 4 transcripts upregulated during both encystation and excystation (“Differentiation” in Table S1). ⇓ denotes a cluster of 9 transcripts downregulated in cysts and S1 (“Down Cyst, S1” in Table S1). B) Relative abundance of differentially expressed transcripts (sense only) presented as histograms color-coded according to the functional categories of their encoded proteins (see Fig. 2).
163 abundant transcripts were expressed constitutively throughout the Giardia life cycle (Table S1), most annotated as hypothetical. Proteins involved in the cytoskeleton, DNA repair, repair of oxidative damage, proton transport, transcription and its regulation, translation, protein trafficking, maturation, and degradation, as well as several kinases and phosphatases were also expressed constitutively (Table S1). An additional 6068 transcripts could not receive clear assignment as constitutively or differentially expressed (Table S1) as they were not sufficiently abundant (frequencies less than 5 in all SAGE libraries) or had uncertain expression profiles due to SAGE sampling error (R ≥ 2 and R < 8), where chance differences due to random sampling of transcripts could not be differentiated from real differences between transcriptomes [11].
Giardia takes advantage of host conditions at each step of its descent through the human gastrointestinal tract. Infection is initiated by ingestion of cysts, generally from fecally contaminated water [1, 2]. Exposure of cysts to gastric acid triggers excystation, although the “excyzoite” must not emerge from the cyst until it passes into the small intestine, or it will be killed. This is modeled by Stage 1 (S1) of the in vitro excystation protocol [12]. Upon passage of activated cysts into the small intestine, they are exposed to slightly alkaline pH and host proteases, modeled by excystation Stage 2 (S2). Emergence of the excyzoite begins in S2 and continues after transfer to complex growth medium, which models the nutrient-rich small intestine where trophozoites colonize. Excystation is a complex cellular awakening from dormancy. The emerging quadrinucleate excyzoite undergoes cytokinesis [13], producing two trophozoites with transcriptionally active nuclei [14]. Each daughter cell rapidly re-organizes its motility apparatus and ventral disc for locomotion and attachment to remain in the small intestine. Among the 284 transcripts upregulated in cysts and excyzoites were encoded large numbers of proteins unique to Giardia, many components of the cytoskeleton, surface proteins, enzymes of intermediary metabolism, and a number of kinases and phosphatases (Table S1, Figure 1). Some putative adhesion and cell cycle transcripts were also upregulated. Transcript levels during excystation of in vitro prepared cysts may not totally reflect transcription in fecal cysts, although only mature cysts survive in water and can excyst. Moreover, excystation is highly synchronous because it is initiated by the large increases in temperature and hydrogen ions.
Additional genes unique to Giardia or diplomonads were upregulated specifically during S2 and 30–60 min post-excystation, forming a distinct cluster, termed “Excystation” (Table S1, Figure 1). This cluster also included genes involved in the cytoskeleton, signal transduction, translation, and protein binding, folding, and transport. Nine transcripts formed a cluster that was down regulated in cysts and S1, but abundant in all other stages (“Down Cyst, S1”, Table S1, Figure 1). Two encoded structural proteins, with the remainder being proteins unique to Giardia or diplomonads.
Trophozoites can colonize the human small intestine for weeks to years, living by uptake of small molecules from the host with little de novo synthesis [4], as modeled by complex growth media [15]. Trophozoites derive most energy from anaerobic glycolysis or metabolism of arginine and many metabolic genes and pathways most closely resemble bacterial homologs [4, 8]. The transcripts most highly expressed by trophozoites were also present during encystation (“encyzoites”, see below) and together formed a large cluster (Figure 1, Table S1). This included pathways of energy metabolism, particularly arginine metabolism, translation, protein transport, cell structure or cell division, as well as cytoskeleton and signal transduction.
If trophozoites are carried downstream, they must encyst in order to survive outside the host [2, 4]. Crucial stimuli for encystation are transit from neutral pH and low bile concentrations near the epithelial cells to slightly alkaline pH and high bile concentration of the small intestinal lumen, as modeled in our encystation media [16]. Encystation is slower and less synchronous than excystation because not every cell may commit to differentiation immediately. Clearly, encystation is a gradual transition, with many shared transcripts (Figure 1, Table S1). Although many metabolic pathways have been identified, there is little detailed information on changes in metabolic activities during the giardial life cycle, aside from the general down-regulation of oxygen consumption during encystation [5]. Encystation is characterized by the appearance of large encystation-specific secretory vesicles (ESVs) [17] that export nascent cyst wall components, most notably three leucine-rich –repeat containing cyst wall proteins (CWPs) [16, 18, 19]. In addition, the cyst wall is composed of fibrils of poly-N-acetylgalactosamine [20]. Overall, only 42 transcripts were specifically upregulated during encystation (Table S1, Figure 1). Importantly, they included CWPs, enzymes responsible for synthesis of cyst wall poly-N-acetylgalactosamine, and high cysteine non-variant cyst protein (HCNCp) [21]. Transcripts of 13 genes unique to Giardia were also upregulated during encystation. Another three sense transcripts, encoding a kinase and two proteins unique to Giardia, were upregulated specifically during both encystation and excystation (“Differentiation”, Table S1, Figure 1).
SAGE of the Giardia lamblia life cycle highlights several key aspects of Giardia’s biology. First, the major transitions in the Giardia life cycle involve dramatic changes in the transcriptome. There appear to be major transitions in the transcriptome of Giardia between 42 hrs of encystation and the mature cyst form as well as between 60 min post-excystation and trophozoites. The changed transcription pattern at the end of encystation could be linked to the encystation-specific DNA replication step that generates a ploidy of 16N in the cyst [13]. Second, excystation or encystation in Giardia involves a relatively small number of differentially expressed genes. Many encystation genes have been characterized [16, 18, 19, 21], while a number of proteins of unknown function unique to Giardia remain to be explored for excystation. Third, differentiation in Giardia lamblia is reminiscent of meiosis, in which the genome is first replicated without division and then divided twice without DNA replication [13]. It is possible that differentiation of primitive eukaryotes into cystic forms is an ancestral form of the sexual process [22]. SAGE analysis of the Giardia life cycle identified a number of kinases, phosphatases, and DNA replication proteins involved in excystation and encystation, which could be important for examining the role of cell signaling in protistan parasite differentiation. Overall, our SAGE data provide a rich resource for examination of gene expression profiles in the context of the Giardia life cycle.
Supplementary Material
Transcript SAGE abundance values, presented as percentages of all transcripts sampled within individual SAGE libraries. SAGE tag sequences were mapped to the Giardia lamblia genome using custom software, followed by manual curation. As mRNA transcripts can generate more than one SAGE tag due to incomplete NlaIII digestion during SAGE library construction, abundance values for transcripts were the sum of all SAGE tags along the entire transcript (sense orientation tags contributing to sense transcript frequencies and antisense orientation tags to antisense transcript frequencies). Sense transcripts included the gene from start to stop codon plus a 15 bp 3’ UTR, with manual correction of UTR length using EST sequences released with the Giardia genome project [8]. Antisense transcripts were modeled as the reverse complement of the gene from stop to start codon. Transcripts were scored for differential expression among libraries using Stekel et al.’s R-statistic [11] (variable tags have higher R values). Those with significant variation in abundance throughout the Giardia lamblia life cycle (R ≥ 8, based on Stekel et al.’s [11] large deviation calculation) were labeled for clusters of correlated expression (see Figure 1) and functional categories of their encoded products (see Figure 2). Transcripts labeled as constitutively expressed had frequencies greater than 5 in at least one SAGE library (i.e. outside of sequencing error) and R < 2 (i.e. low variability among SAGE libraries). Gene information and SAGE data can be viewed at GiardiaDB (www.giardiadb.org) (June 2010 release).
Unresolved SAGE tags, which could not be assigned to sense or antisense transcripts for any Giardia lamblia gene (see text), presented with observed frequencies within individual SAGE libraries.
Figure 2.

Key for functional categories of differentially expressed (R ≥ 8) Giardia proteins (see Table S1). Functional categories were assigned by searching UniProtKB/Swiss-Prot with BLASTP (e-values ≤ e−6) using WU-Blast2 and labeling the positive hits with the identified Swiss-Prot functional category, as well as prediction of functional domains using the Pfam profile HMM database. Included in the classification are categories for proteins unique to Giardia (no homologs detected in GenBank) or within the diplomonads (based on comparison to available Spironucleus vortens EST data), as well as for proteins with homology to hypothetical proteins found in other genomes (“Conserved Hypothetical” in Table S1).
Acknowledgments
BJD, DSR, and FDG were supported by NIH grants AI42488, GM61896, and DK35108. DP and SGS were supported by grants from the Swedish Natural Science Research Council, the Swedish Medical Research Council, and the Karolinska Institutet. AGM, SRB, SPP, and MJC were supported by NIH grant AI51089 and by the Marine Biological Laboratory’s Program in Global Infectious Diseases, funded by the Ellison Medical Foundation. Computational resources were provided by the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution (Marine Biological Laboratory) through funds provided by the W.M. Keck Foundation and the G. Unger Vetlesen Foundation. We are grateful to Mitchell Sogin and Hilary Morrison for providing genomic resources and support.
Footnotes
All SAGE data were deposited to GenBank’s GEO database under accession GSE8336, with subsequent deposit to GiardiaDB (www.giardiadb.org).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huang DB, White AC. An updated review on Cryptosporidium and Giardia. Gastroenterol Clin North Am. 2006;35:291–314. doi: 10.1016/j.gtc.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Carranza PG, Lujan HD. New insights regarding the biology of Giardia lamblia. Microbes Infect. 2010;12:71–80. doi: 10.1016/j.micinf.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Prucca CG, Lujan HD. Antigenic variation in Giardia lamblia. Cell Microbiol. 2009;11:1706–15. doi: 10.1111/j.1462-5822.2009.01367.x. [DOI] [PubMed] [Google Scholar]
- 4.Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–75. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paget TA, Macechko PT, Jarroll EL. Metabolic changes in Giardia intestinalis during differentiation. J Parasitol. 1998;84:222–6. [PubMed] [Google Scholar]
- 6.Knox DP, Skuce PJ. SAGE and the quantitative analysis of gene expression in parasites. Trends Parasitol. 2005;21:322–6. doi: 10.1016/j.pt.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–7. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 8.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–6. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 9.Ruzanov P, Riddle DL. Deep SAGE analysis of the Caenorhabditis elegans transcriptome. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq035. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teodoroovic S, Walls CD, Elmendorf HG. Bidirectional transcription is an inherent feature of Giardia lamblia promoters and contributes to an abundance of sterile antisense transcripts throughout the genome. Nucleic Acids Res. 2007;35:2544–53. doi: 10.1093/nar/gkm105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–61. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucher SE, Gillin FD. Excystation of in vitro-derived Giardia lamblia cysts. Infect Immun. 1990;58:3516–22. doi: 10.1128/iai.58.11.3516-3522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernander R, Palm JE, Svard SG. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol. 2001;3:55–62. doi: 10.1046/j.1462-5822.2001.00094.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu LZ, Birky CW, Jr, Adam RD. The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot Cell. 2002;1:191–9. doi: 10.1128/EC.1.2.191-199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487–8. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 16.Sun CH, McCaffery JM, Reiner DS, Gillin FD. Mining the Giardia lamblia genome for new cyst wall proteins. J Biol Chem. 2003 doi: 10.1074/jbc.M302023200. [DOI] [PubMed] [Google Scholar]
- 17.Stefanic S, Morf L, Kulangara C, Regos A, Sonda S, Schraner E, et al. Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J Cell Sci. 2009;122:2846–56. doi: 10.1242/jcs.049411. [DOI] [PubMed] [Google Scholar]
- 18.Lujan HD, Mowatt MR, Conrad JT, Bowers B, Nash TE. Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. Implications for secretory granule formation and protein assembly into the cyst wall. J Biol Chem. 1995;270:29307–13. doi: 10.1074/jbc.270.49.29307. [DOI] [PubMed] [Google Scholar]
- 19.Mowatt MR, Lujan HD, Cotten DB, Bowers B, Yee J, Nash TE, et al. Developmentally regulated expression of a Giardia lamblia cyst wall protein gene. Mol Microbiol. 1995;15:955–63. doi: 10.1111/j.1365-2958.1995.tb02364.x. [DOI] [PubMed] [Google Scholar]
- 20.Gerwig GJ, van Kuik JA, Leeflang BR, Kamerling JP, Vliegenthart JF, Karr CD, et al. The Giardia intestinalis filamentous cyst wall contains a novel beta(1-3)-N-acetyl-D-galactosamine polymer: a structural and conformational study. Glycobiology. 2002;12:499–505. doi: 10.1093/glycob/cwf059. [DOI] [PubMed] [Google Scholar]
- 21.Davids BJ, Reiner DS, Birkeland SR, Preheim SP, Cipriano MJ, McArthur AG, et al. A new family of giardial cysteine-rich non-VSP protein genes and a novel cyst protein. PLoS ONE. 2006;1:e44. doi: 10.1371/journal.pone.0000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–3. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcript SAGE abundance values, presented as percentages of all transcripts sampled within individual SAGE libraries. SAGE tag sequences were mapped to the Giardia lamblia genome using custom software, followed by manual curation. As mRNA transcripts can generate more than one SAGE tag due to incomplete NlaIII digestion during SAGE library construction, abundance values for transcripts were the sum of all SAGE tags along the entire transcript (sense orientation tags contributing to sense transcript frequencies and antisense orientation tags to antisense transcript frequencies). Sense transcripts included the gene from start to stop codon plus a 15 bp 3’ UTR, with manual correction of UTR length using EST sequences released with the Giardia genome project [8]. Antisense transcripts were modeled as the reverse complement of the gene from stop to start codon. Transcripts were scored for differential expression among libraries using Stekel et al.’s R-statistic [11] (variable tags have higher R values). Those with significant variation in abundance throughout the Giardia lamblia life cycle (R ≥ 8, based on Stekel et al.’s [11] large deviation calculation) were labeled for clusters of correlated expression (see Figure 1) and functional categories of their encoded products (see Figure 2). Transcripts labeled as constitutively expressed had frequencies greater than 5 in at least one SAGE library (i.e. outside of sequencing error) and R < 2 (i.e. low variability among SAGE libraries). Gene information and SAGE data can be viewed at GiardiaDB (www.giardiadb.org) (June 2010 release).
Unresolved SAGE tags, which could not be assigned to sense or antisense transcripts for any Giardia lamblia gene (see text), presented with observed frequencies within individual SAGE libraries.