Abstract
The molecular mechanisms underlying retinoic acid (RA) augmentation of T cell receptor (TCR) and transforming growth factor-β (TGF-β)-induced Foxp3 transcription and inhibition of the latter by cytokines such as IL-27 were here shown to be related processes involving modifications of baseline (TGF-β-induced) phosphorylated Smad3 (pSmad3) binding to a conserved enhancer region (enhancer I). RA augmentation involved the binding of retinoic acid receptor (RAR) and retinoid X receptor (RXR) to a dominant site in enhancer I and a subordinate site in the promoter. This led to increased histone acetylation in the region of the Smad3 binding site and increased binding of pSmad3. Cytokine (IL-27) inhibition involved binding of pStat3 to a gene silencer in a second conserved enhancer region (enhancer II) downstream from enhancer I; this led to loss of pSmad3 binding to enhancer I. Thus, control of accessibility and binding of pSmad3 provides a common framework for positive and negative regulation of TGF-β-induced Foxp3 transcription.
Keywords: Foxp3, retinoic acid, TGF-beta, Smad3, Stat3, IL-27
Introduction
It is now well established that CD4+ regulatory T cells (Tregs) can be recruited from unselected CD4+ non-regulatory T cells in the peripheral (extra-thymic) lymphoid compartment; the regulatory activity of these cells augment the regulatory activity of thymus-derived Tregs, particularly at inflammatory sites. The major driving force of such recruitment is transforming growth factor-β (TGF-β) which has been shown to induce T cell receptor (TCR)-stimulated CD25− non-regulatory T cells to express the intra-cellular transcription factor that directs Treg function, Foxp3 (Zheng et al., 2002). Recently, the molecular basis of such induction was clarified, at least in part, by Tone et al who showed that the Foxp3 gene was regulated by Smad3 and NFAT transcription factors that bind to sites in an enhancer region (enhancer I) located in an intron between untranslated exon −2a and exon −1 upstream of the ATG start site of the Foxp3 gene (Tone et al., 2008). Subsequently, Smad3 is recruited to a promoter site where it forms part of a complex enhanceosome also composed of c-Rel, p65, NFAT and CREB binding to a promoter site (Ruan et al., 2010). In addition, Zheng et al., have recently reported that enhancer 1 activity is required for induced Treg development (Zheng et al., 2010).
Several other factors have also been shown to modulate regulatory cell and Foxp3 induction in addition to TGF-β and TCR stimulation. For example, all-trans retinoic acid (RA) produced by CD103+ dendritic cells in the gastrointestinal mucosa has been shown to augment TGF-β and TCR inductive effects but to have no inductive effects on its own (Coombes et al., 2007; Mucida et al., 2007). The mechanism of RA enhancement is controversial. One group of investigators maintain that the effect is indirect in that RA acts mainly to inhibit the production of pro-inflammatory cytokines that would otherwise inhibit the induction of Foxp3+ regulatory T cells (Hill JA et al., 2008). Other investigators maintain that the RA effect is direct and is not simply reversing the inhibitory activity of cytokines (Mucida et al., 2009). These different views are best resolved with a molecular analysis of RA effects, such as the analysis reported here.
Various cytokines also exert control on regulatory T cell and Foxp3 induction, both in a negative and positive direction. IL-6 and IL-27, for instance, are strongly inhibitory of TCR and TGF-β inductive effects, presumably through their shared capacity to activate Stat3. Indeed, some evidence supporting this idea has been presented, but the mechanism of inhibition is still incompletely understood as Stat3 binding sites are lacking in the Foxp3 promoter and the above mentioned enhancer I region (Neufert et al., 2007; Huber et al., 2008). IL-2, alternatively, exerts a positive effect on TCR and TGF-β induction, but, again the mechanism is unclear because the Stat5 binding site so far identified has no known relationship to previously identified Foxp3 transcriptional control regions (Yao et al., 2007. Kim and Leonard, 2007).
In the present study we explored TCR-TGF-β-RA induction of Foxp3 expression using a combined cellular and molecular approach involving the use of cell lines transfected with a Foxp3 promoter and enhancer-driven luciferase constructs, as well as ChIP analysis of transcription factor binding in CD4+ T cells. A key finding was that TCR-TGF-β induction of Foxp3 was exquisitely dependent on the generation of phosphorylated Smad3 (pSmad3) and that RA enhanced such induction by facilitating increased binding of pSmad3 to the enhancer identified by Tone et al. (Tone et al., 2008). In addition, IL-27 inhibited such induction (as well as induction by TGF-β and RA) by generating pSTAT3 which then acted as an inhibitor by binding to a conserved enhancer (enhancer II) down-stream of enhancer I, and inhibited binding of pSmad3 to enhancer I. Thus, factors that enhance and inhibit Foxp3 expression are acting reciprocally to control the binding of pSmad3 to enhancer I.
Results
An AP-1 site located in Foxp3 enhancer I region plays an important role in TCR and TGF-β induced Foxp3 expression
On the basis of the fact that both TCR and TGF-β signals induce the activation of c-Jun(activator protein-1)(AP-1) via the mitogen-activated protein kinase (MAPK) pathway we began our investigation of the molecular mechanisms governing TCR-TGF-β induction of Foxp3 with studies to determine if such induction was c-Jun(AP-1) dependent.
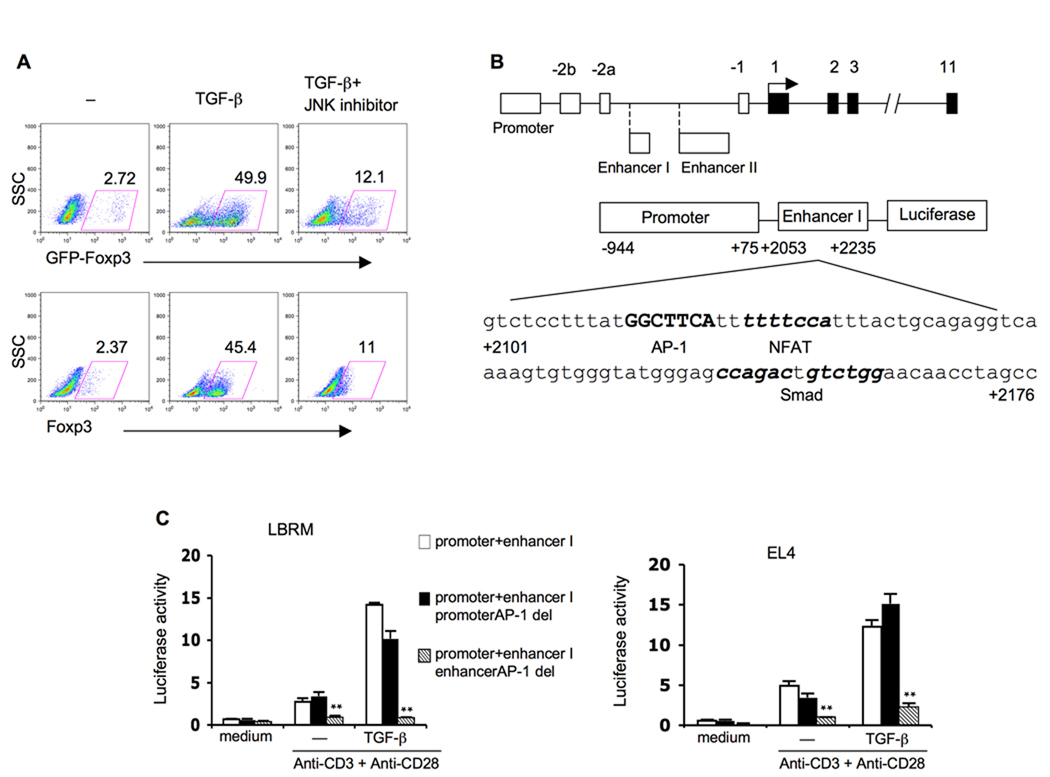
In initial studies to address this question, we subjected CD4+ T cells from Foxp3-IRES-GFP gene targeted (Foxp3-GFP) mice to anti-CD3-anti-CD28 (anti-TCR) plus TGF-β (TCR- TGF-β) stimulation in the presence (or absence) of a c-Jun N-terminal kinase (JNK) inhibitor; then, after 4 days, assayed the cells for Foxp3 expression by flow cytometry. As shown in Figure 1A, the presence of the inhibitor reduced the percentage of CD4+ T cells that express GFP (Foxp3) following stimulation from 49.9% to 12.1%. In a parallel study, we assessed Foxp3 expression in CD4+ T cells from conventional B6 mice after stimulation by enumerating cells that stain with fluorescent anti-Foxp3 antibody. In this case, the addition of JNK inhibitor reduced the percentage of Foxp3+ cells from 45.4% to 11%. In a parallel study also shown in Supplemental Figure 1 we found that p38 and extracellular signal-regulated kinase (ERK) inhibitors had only marginal blocking effects on Foxp3 induction.
Figure 1. AP-1 site located in a Foxp3 enhancer region plays positive role on TCR/TGF-β induced Foxp3 gene expression.
(A) Murine CD4+ T cells from Foxp3-GFP mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without JNK inhibitor (10µM) for 5 days then collected for flow cytometry analysis.
(B) Schematic diagram of Foxp3 gene (upper) and AP-1: NFAT and Smad3 sites in Foxp3 enhancer I region (lower).
(C) Two different murine T cell lines LBRM (left) and EL4 (right) were transfected with Foxp3 promoter and enhancer I reporter construct (promoter+enhancer I) or the reporter construct with promoter AP-1 site deletion (promoter+enhancer I promoter AP-1 del) or the reporter construct with enhancer I AP-1 site deletion (promoter+enhancer I enhancer AP-1 del). Four hours later cells were split and stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) with or without TGF-β (2ng/ml). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data represent four independent experiments. **p<0.01, compared to intact construct under same stimulation conditions.
In additional studies we determined the contribution of AP-1 to Foxp3 transcription in reporter assays conducted in mouse lymphoma T cell line LBRM-33 4A2 (hereafter called LBRM cells) or a EL-4 cell line, clone LAF (hereafter called EL-4 cells). As shown in Figure 1B, the reporter construct consisted of firefly luciferase reporter driven by a 1kb Foxp3 promoter fragment (−944 to +75) linked in tandem to a previously identified 182bp enhancer region (+2053 to +2235). This enhancer region (here identified as enhancer I) has been shown to contain a functional nuclear factor of activated T cells (NFAT) binding site and a Smad binding site at +2130 to +2139 and +2144 to +2147, respectively (Tone et al., 2008). The core sequence of an NFAT site (GGAAA) is often positioned ~3 base pairs down-stream from an AP-1 site and during transcription AP-1 forms a complex with NFAT that modifies NFAT transcriptional activity (Fernando et al., 2001). Indeed, such an AP-1 site was present just up-stream of the NFAT site in the Foxp3 enhancer at +2122 to +2129 (TGAAGCC).
As shown in Figure 1C, we found that reporter activity was very low in unstimulated transfected cells but was increased upon TCR stimulation and further increased upon TCR-TGF-β stimulation, indicating the positive effect of TGF-β signaling on Foxp3 transcription in the presence of TCR stimulation. In contrast, TCR-TGF-β stimulation of cells with a construct containing a deletion of the above described AP-1 site induced little luciferase activity. Similar results were obtained with a reporter construct transfected into EL-4 cells. These data plus the flow cytometric data above (Figure 1A) clearly indicate that an AP-1 binding site adjacent to NFAT is an important factor in TGF-β-mediated induction of Foxp3 transcription.
A search of the Foxp3 gene disclosed several additional AP-1 sites including a site in the promoter region at −256 to −262 (TGACTTG) nearest to the transcription start site. To determine whether this latter AP-1 site also played a role in Foxp3 transcription we compared the luciferase signals generated by TCR-TGF-β stimulated EL-4 and LBRM cells transfected with a Foxp3 promoter-enhancer 1-driven luciferase construct with a deletion of the promoter AP-1 site with signal generated in cells transfected with the same construct without the deletion. As also shown in Figure 1C, the luciferase signals generated by the construct bearing the promoter AP-1 site deletion was not substantially reduced compared to the signals generated by the intact construct. Thus, the AP-1 site located in the promoter region nearest to the transcriptional start site appears to play no substantial role in Foxp3 transcription; however, it remains possible that other AP-1 sites in the promoter have a more important role or that AP-1 sites in the promoter act collectively to influence Foxp3 transcription.
In further studies we considered the possibility that the control of Foxp3 transcription exerted by AP-1 (or, more correctly, the NFAT-AP-1 complex) was not due to the direct effect of this complex acting as a Foxp3 transcription factor, but rather it was due to an indirect effect involving the ability of NFAT-AP-1 to regulate the binding of pSmad3 to an adjacent enhancer I site. To explore this possibility we determined pSmad3 binding to the enhancer I site by ChIP assays of primary CD4+ T cells in the presence and absence of a JNK inhibitor. These studies are discussed below in a different and more appropriate context.
TGF-βRI kinase activity is essential for TCR-TGF-β induced Foxp3 expression in murine T cells
TGF-β signaling through the heterodimeric TGF-βRI-RII results in activation of the MAPK pathway as well as the Smad pathway (Derynck and Zhang, 2003). These pathways are activated independently, the MAP kinase pathway via E3 activity of TNF receptor associated factor 6 (TRAF6) and the activation of TGF-β-activated kinase 1 (TAK1) in a TGF-βRI receptor kinase-independent manner and the Smad pathway via TGF-βRI kinase activity (Sorrentino et al., 2008). To determine whether the contribution of TGF-β stimulation to Foxp3 expression depends on the receptor kinase activity, we assessed TGF-β induction of Foxp3 in cells exposed to the TGF-βR1 kinase inhibitor, ALK5 inhibitor (SB431542) which prevents TGF-β1- induced R-Smad phosphorylation but not TRAF6, TAK1, or MAP kinase activity (Sorrentino et al., 2008). As shown in Figure 2A upper panels, the addition of ALK5 inhibitor to cultures of CD4+ T cells from Foxp3-GFP mice subjected to TCR-TGF-β stimulation led to greatly diminished induction of Foxp3-expressing cells (from 57.2% to 6.99%); similarly, as shown in Figure 2A lower panels, CD4+ T cells from B6 mice stimulated under the same conditions and assessed with fluorescent anti-Foxp3 also exhibited a greatly decreased induction of Foxp3-expressing cells (from 38.2% to 5.51%).
Figure 2. TGF-βRI kinase activity is essential for TCR-/TGF-β-induced Foxp3 expression in murine T cells.
(A) Purified CD4+ cells from Foxp3-GFP mice (upper panels) or B6 mice (lower panels) were stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without ALK5 inhibitor for 4 days then subjected for flow cytometry analysis.
(B) LBRM or EL4 cells were transfected with Foxp3 promoter and enhancer reporter construct then stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β(2ng/ml) with or without ALK5 inhibitor for 20 hrs. Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data are representative of three independent experiments. **p<0.01, compared to the luciferase activity without ALK5 inhibitor.
In related studies we determined the role of TGF-βRI kinase activity on Foxp3 promoter and enhancer transcriptional activity with the above described reporter construct. As shown in Figure 2B, TCR-TGF-β induction of luciferase activity in both LBRM and EL4 cells was reduced to the degree obtained with TCR stimulation alone in cells cultured with ALK5 inhibitor, indicating that inhibition of TGF-βRI kinase activity completely abolished the TGF-β effect on Foxp3 gene transcription. Taken together, these studies show that TGF-β-TGF-βRI signaling via the R-Smad activation pathway is necessary for TGF-β-mediated Foxp3 transcription. Whether or not TGF-β activation of the MAPK pathway and the generation of AP-1 is also a necessary function of TGF-β during its induction of Foxp3 expression is unclear since AP-1 can also be generated by TCR signaling.
Identification of a Foxp3 silencer containing a Stat3 binding site
TCR-TGF-β induced Foxp3 expression in murine T cells is inhibited by several different cytokines (such as IL-6, IL-27 and IL-21) that have in common ability to activate Stat3. The supposition that this signaling component was in fact the inhibitory factor generated by these cytokines was subsequently supported by studies showing that inhibition of Foxp3 expression by IL-27 was partially diminished in cells subject to Stat3 gene targeting with Stat3-specific siRNA (Huber et al., 2008).
To further explore this possibility, we initially determined TCR plus TGF-β-induced Foxp3 expression in Stat3-deficient mice. As shown in Figure 3A, the inhibition of TCR- TGF-β induced Foxp3 expression by IL-27 was abolished in Stat3-deficient CD4+ T cells. Conversely, as also shown in Figure 3A, IL-27 inhibition of TCR-TGF-β induced Foxp3 expression in CD4+ T cells from SOCS3-deficient mice which thus lack an endogenous inhibitor of Stat3 was greater than was observed in SOCS3-intact cells: SOC3-deficient cells SOCS3 KO: 51.3% to 4.05% vs. SOCS-intact cells: 40.4% to 8.44%. These data thus demonstrate that induction of Stat3 activation is key mechanism of cytokine inhibition of TGF-β-induced Foxp3 expression.
Figure 3. Identification of a Foxp3 silencer containing a Stat3 binding site.
(A) Purified CD4+ cells from Stat3fl/fl and Stat3fl/fl; MMTV-Cre mice(left panels); Socs3fl/fl and Socs3fl/fl; MMTV-Cre mice(right panels) were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β(5ng/ml) with or without IL-27 (20ng/ml) for 4 days then collected for Foxp3 staining and subjected to flow cytometry analysis.
(B) Alignment of human and mouse Foxp3 enhancer II region containing stat3 binding site(up) and the structure of Foxp3 promoter and enhancer I and enhancer II (contains stat3 binding site) reporter construct.
(C) Murine T cell line LBRM were transfected with Foxp3 promoter and enhancer I reporter construct or Foxp3 promoter and Enhancer I and enhancer II reporter construct or Foxp3 promoter and Enhancer I and enhancer II reporter construct with stat3 binding site deletion then stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (2ng/ml). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data are representative of four independent experiments. *p<0.05, **p<0.01, compared to intact construct under same stimulation conditions.
We then extended these findings with an investigation of the molecular basis of Stat3 suppression of Foxp3 gene transcription. An initial computer search of the Foxp3 gene for a Stat3 binding site revealed a canonical site (TTCTCGGAA) located +4364 to +4372 down-stream of the transcription start site. Then, using the Vista program, we found that this site was located within a conserved non-coding sequence of the mouse and human Foxp3 gene that was part of a regulatory region previously identified (Kim and Leonard, 2007; Tone et al., 2008.) and that could represent a second enhancer region. We therefore cloned a 973 bp fragment representing this conserved region and inserted it into the Foxp3 luciferase reporter construct described above immediately down-stream of the first enhancer region. As shown in Figure 3B, this construct consisted of the Foxp3 promoter followed by the first enhancer region (enhancer I) containing the AP-1-NFAT and Smad3 binding sites and the second enhancer (enhancer II) containing the Stat3 binding site linked to a luciferase reporter.
With these constructs in hand we then conducted studies of the activity of enhancer II in EL-4 cells and LBRM cells, recognizing that, as shown in Supplemental Figure 2A, these cells constitutively express substantial levels of pStat3 which in one case is augmented by IL-6 (EL-4 cells) and in the other by IL-27 (LBRM cells). As shown in Figure 3C, stimulation of cells transfected with the construct that includes enhancer II with TCR-TGF-β generated a luciferase signal that was decreased as compared to cells transfected with a construct lacking enhancer II. No additional decrease in luciferase signal was obtained by adding either IL-6 or IL-27 to the cultures, indicating the basal amount of pStat3 was sufficient to induce an optimal effect (data not shown). In complementary studies also shown in Figure 3C, stimulation of cells transfected with a construct containing enhancer II with a deletion of the Stat3 binding site gave rise to an enhanced luciferase signal. In all, these data strongly suggest that pStat3 binding to a Stat3 binding site in enhancer II acts as a potent silencer of Foxp3 expression. In addition, they show that in the absence of Stat3 effects on enhancer II, the latter has a positive effect on Foxp3 transcription, as predicted by the fact that this area contains binding sites for known positive transcription factors (Tone et al., 2008).
Retinoic acid enhancement of Foxp3 expression is strictly dependant on intact TGF-βRI kinase activity and Smad3
Several groups have recently shown that all-trans retinoic acid (hereafter called RA) can enhance TCR-TGF-β-induced Foxp3 expression in mouse CD4+ T cells both in vitro and in vivo (Mucida et al., 2007; Coombes et al., 2007). This effect, as shown in Figure 4A, leads to expression of Foxp3 in over 95% of T cells and thus is inducing expression in both naïve and mature T cells.
Figure 4. The positive effect of retinoic acid on Foxp3 expression completely depends on intact TGF-βRI kinase activity and Smad3.
(A) Purified CD4+ cells from Foxp3-GFP mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without retinoic acid (100nM) and JNK kinase inhibitor (10µM), cyclosporine A (20nM), IL-27 (20ng/ml) or ALK5 inhibitor (5µM) for 4 days then collected for flow cytometry analysis.
(B) Purified CD4+ cells from B6-WT littermate mice and B6-Smad3 knockout were stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (1µg/ml), TGF-β(5ng/ml), RA (100nM) with or without ALK5 inhibitor (2.5µM) for 4 days. Cells were collected and stained with APC anti-Foxp3 (eBioscience) then subjected to flow cytometric analysis.
In initial studies of the molecular mechanisms governing RA enhancement of Foxp3 expression we determined the effects of various regulatory factors on such RA enhancement. First, as shown in Figure 4A, RA enhancement was not diminished in the presence of JNK inhibitor, indicating that the RA effect renders the AP-1 effect unnecessary. Second, while the presence of cyclosporine mildly diminished baseline Foxp3 induction by TGF-β, it did not affect the enhancement of Foxp3 expression by RA, indicating that RA functions independently of NFAT. Third, the negative effect of IL-27 on TCR-TGF-β-induced Foxp3 expression overrode the positive effect of retinoic acid; this observation fits with the fact that, as shown in Supplemental Figure 2B, the presence of RA has no effect on IL-27 or IL-6 induced phosphorylation of Stat3, indicating that the RA effect is independent of these cytokines and does not enhance Foxp3 expression by merely blocking a negative signal. In addition, this correlates with previous studies showing that the RA effect is independent of Stat3 and Stat5 (Elias et al., 2008). Fourth, as shown in Supplemental Figure 3A, the RA enhancing effect was not affected by the presence of a large number of cytokines other than IL-27 and IL-6 again emphasizing that the effect was not due to positive or negative effects of cytokine regulation of Foxp3 expression; importantly, as shown in Supplemental Figure 3B, this was also true of IL-4 which suppresses baseline induction of Foxp3 by TCR-TGF-β, but not RA enhancement of baseline induction. Finally, as also shown in Figure 4A, the positive effect of RA is completely dependent on TGF-βRI kinase activity because the percentage of Foxp3+ cells decreased to baseline (3.19%) if cells were stimulated with TCR-TGF-β and RA in the presence of ALK5 inhibitor.
To further address the mechanism of RA enhancement of Foxp3 expression, we subjected CD4+ T cells from Smad3-deficient mice to TCR-TGF-β stimulation with and without RA. As shown in Figure 4B, Foxp3 induction by TCR-TGF-β was greatly diminished in B6 Smad3-deficient mice and, more importantly, RA exhibited virtually no enhancement of TCR-TGF-β-induced Foxp3 in such mice. As shown in Supplemental Figure 4A, virtually identical results were obtained with cells from a BALB/c Smad3-deficient mice indicating that the results were not strain-specific. These data clearly indicate that both TGF-β induction of Foxp3 as well as the positive effect of RA on such induction is largely dependent on Smad3.
The enhancement of TCR-TGF-β- induced Foxp3 transcription by RA is not due to increased Smad3 phosphorylation under optimal TGF-β induction conditions
Based on the above results, it seemed possible that TCR-TGF-β-induced Foxp3 transcription is enhanced by RA because RA enhances the phosphorylation of Smad3 and thereby facilitates Smad3 translocation into the nucleus, as already suggested in a previous study (Xiao et al., 2008). To explore this possibility, we first determined Foxp3 expression in CD4+ T cells exposed to a wide range of TGF-β concentrations in the presence and absence of RA. As shown in Figure 5A, TGF-β induction of stable baseline levels of Foxp3+ cells was unchanged over a wide range of TGF-β concentrations (1ng/ml to 40ng/ml) and was diminished only at a very low (0.1ng/ml) TGF-β concentration. Furthermore, the addition of RA enhanced the number of Foxp3+ cells to an equal degree over the range of TGF-β concentrations that gave rise to the stable baseline levels of Foxp3 and reduced RA enhancement was only seen at the low TGF-β concentration (0.1ng/ml) that gave rise to low baseline Foxp3 expression. These data show that RA enhancement is weak until a baseline level of TCR/TGF-β-induced Foxp3 expression is reached and suggest that the main RA effect occurs after baseline TCR/TGF-β induction involving NFAT-AP-1 has occurred.
Figure 5. The enhancement of RA on TCR/TGF-β induced Foxp3 transcription is not due to increased Smad3 phosphorylation.
(A) Purified CD4+ cells from Foxp3-GFP mice was stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β at different concentrations as indicated with or without retinoic acid (200nM) for 4 days then collected for flow cytometry analysis.
(B) Purified CD4+ cells from B6 mice were stimulated with TGF-β at different concentrations as indicated with or without retinoic acid (500nM) for 2 hours. Cells were collected and cell lysates were prepared and subjected to Western-Blot (left). Intensity of bands was measured using NIH Image software and showed as the ratio between p-Smad3 and Smad3 from the same sample (right). Data are representative of two independent experiments.
In further studies along these lines, we assessed the amount of pSmad3 induced by different concentrations of TGF-β in the presence and absence of RA. As shown in the immunoblot depicted in Figure 5B (left panel) and the density analysis of this blot in Figure 5B (right panel), addition of RA was accompanied by increased phosphorylation of Smad3 only in the presence of a low concentration of TGF-β (0.1 ng/ml); in contrast, over a wide range of higher TGF-β concentrations (1–20ng/ml) addition of RA was unaccompanied by increased phosphorylation. In assessing the significance of the enhancement at 0.1 ng/ml of TGF-β, it should be noted that at this concentration both baseline (TGF-β only) and RA enhanced (TGF-β plus RA) Foxp3 levels were lower than those obtained at higher TGF-β concentrations which in fact reached a stable plateau at a TGF-β concentration of 1 ng/ml. In addition, at the 0.1 ng/ml TGF-β concentration, the amount of pSmad3 in the presence of RA was as high as that obtained at higher TGF-β concentrations, indicating that no additional phosphorylation is required to achieve a higher level amount of Foxp3 expression with higher concentrations of TGF-β. Finally, as shown in Supplemental Figure 4B, we found, as did Nolting et al., that RA induced increased amounts of Smad3 protein in the absence of increased Smad3 phosphorylation after 12 hours of culture without any effect on Foxp3 expression (Nolting et al., 2009); this could explain the apparent increase in phosphorylation induced by RA in the presence of low concentrations of TGF-β because under these conditions RA may be increasing the amount of Smad3 available for TGF-β induced phosphorylation. (whereas at higher TGF-β concentrations RA induction of Smad3 protein is redundant). Overall then, whereas increased Smad3 phosphorylation in the presence of RA may be a cause of RA enhancement at sub-optimal TGF-β concentrations, once baseline TCR/TGF-β induction of Foxp3 is obtained (i.e., at the usual inducing concentrations of TGF-β), RA enhancement of Foxp3 induction is not due to increased Smad3 phosphorylation. Summary, please…
Retinoic acid directly regulates Foxp3 promoter and enhancer activity
To study the mechanism underlying RA regulation of Foxp3 expression we analyzed RA effects on the Foxp3 reporter construct expressed in LBRM and EL4 cells as described above. Our study was based on the knowledge that the cellular effects of RA are mediated through its ligation of RAR and/or RXR followed by translocation of these factors to the nucleus and specific binding to gene target sites (Mangelsdorf et al., 1995). Indeed, as shown in Figure 6A, we found two RAR-RXR binding sites in the Foxp3 gene, one located in the Foxp3 promoter at −310 to −306 and one in enhancer I at +2611 to +2618. This mandated that we utilize cells transfected with a luciferase reporter construct containing both promoter and enhancer I components containing these sites in studies of RA regulation of the Foxp3 gene.
Figure 6. Retinoic acid directly regulates Foxp3 promoter and enhancer activity.
(A) Location of two RAR-RXR binding sites in Foxp3 promoter and enhancer I regions.
(B) LBRM (left) or EL4 (right) cells were transfected with a Foxp3 promoter and enhancer I reporter construct and stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) plus TGF-β (2ng/ml) and retinoic acid at a series of concentration as indicated. Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/renilla luciferase activity from the same well. Data are representative of two independent experiments. **p<0.01, compared to the luciferase activity without RA.
(C) EL4 cells were transfected with Foxp3 promoter and enhancer I reporter construct with deletion of the RAR-RXR binding site in either the promoter region or enhancer region (as indicated in A) or both and then stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (2ng/ml) with or without retinoic acid (500nM) as indicated. (ProRAR-RXR del: promoter RAR-RXR deletion; EnhRAR-RXR del: enhancer I RAR-RXR deletion; Pro+Enh RAR-RXR del: promoter+enhancer I RAR-RXR deletion). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well.
Data are representative of three independent experiments. **p<0.01, compared to intact construct under same stimulation conditions.
In initial studies, we determined luciferase output by both transfected EL-4 and LBRM cells subjected to TCR-TGF-β stimulation with or without RA at different concentrations. As shown in Figure 6B, addition of RA enhanced the Foxp3 promoter-enhancer I construct luciferase activity in a dose-dependent manner. Next, we cultured cells with the same stimulants but in this case used cells transfected with constructs with deleted RAR-RXR binding site in the promoter or the enhancer regions or in both regions. As shown in Figure 6C, deletion of the RAR-RXR binding site in the promoter resulted in a small decrease in reporter signal whereas deletion of the binding site in enhancer I led to a large decrease in the luciferase signal; in addition, deletion of both binding sites led to an additive decrease to a level of transcription which was only marginally higher than that obtained by TCR-TGF-β stimulation in the absence of RA. We conclude that whereas the transcriptional activity of the enhancer I RAR-RXR binding site is greater than the activity of the promoter binding site, the latter is not trivial since binding of RA to both the promoter and enhancer sites is necessary for the full effect of RA on TGF-β-induced Foxp3 transcription.
In further studies along these lines we conducted reporter construct studies in purified CD4+ T cells rather than cell lines to verify that the above results would also obtain in a more physiological intra-cellular milieu. Accordingly, purified primary CD4+ T cells were transfected with a reporter construct containing both promoter and enhancer I and enhancer II elements and then assayed for luciferase activity under various conditions. As shown in Supplemental Figure 5A, TCR activation of the cells in the presence of TGF-β led to increased luciferase activity compared to TCR activation alone, which was further augmented by the addition of RA. In addition, this increase in luciferase activity was completely reversed by the addition of anti-IL-27. The latter inhibitory effect was not seen in the cell line studies because of high baseline cytoplasmic pStat3 amounts in the cell line cells that obviate the effect of Stat3 activation by IL-27 signaling (see Discussion below). In a parallel set of studies the cells were transfected with a promoter-enhancer I reporter constructs in which the enhancer I RAR-RXR was either intact or deleted. As shown in Supplemental Figure 5B, the construct with the deleted RAR-RXR site exhibited complete loss of luciferase acivity in cells strimulated by TGF-β plus RA. Taken together, these studies in primary CD4+ T cells corroborate those with cell lines and verify that RA directly regulates Foxp3 expression via RAR-RXR binding to an enhancer site. Interestingly, the importance of RAR-RXR binding to Foxp3 expression in the primary cells was somewhat greater than in the cell lines, suggesting that in primary cells Smad3 binding to the enhancer is more dependent on RAR-RXR than in cell lines.
RA increases histone acetylation at the enhancer I region containing the NF-AT-Smad3 binding sites
Given the strict dependence of the RA enhancing effect on Smad3 activity and the proximity of the RAR-RXR binding site to the Smad3 binding site it seemed likely that the mechanism of the enhancing effect might be due to a direct effect of RA on Smad3 transcriptional activity. To test this hypothesis we first determined if RAR-RXR physically interacted with Smad3. Accordingly, we subjected CD4+ T cells subjected to TCR-TGF-β stimulation in the presence of RA to co-immunoprecipitation studies using several relevant antibodies in a variety of combinations but could find no evidence of RAR-RXR-Smad3 interaction (data not shown). Next we sought to determined whether RAR-RXR could increase the accessibility of the enhancer I to transcription factors. Since this region contains no CpG sequences we chose to do this by performing ChIP studies to assess the level of histone acetylation in these regions rather than by performing methylation studies. As shown in Figure 7A, with respect to the enhancer I region, stimulation of CD4+ T cells with anti-CD3/CD28 was associated with little or no histone acetylation, stimulation with TCR-TGF-β was associated with a mild level of histone acetylation and stimulation with TGF-β plus RA was associated with a high level of histone acetylation. In contrast, RA stimulation or TGF-β plus RA increased acetylation only slightly in the promoter region. These data indicate that the main effect of RA on the accessibility of transcription factors to gene target sites was in the enhancer 1 region.
Figure 7. RA increases histone acetylation at the Enhancer region containing NF-AT/Smad3 binding sites and facilitates increased Smad3 binding to Enhancer.
(A) Purified CD4+ cells from B6 mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without retinoic acid (200nM) for 48 hours. CHIP assay was performed using anti-acetyi histone H4 antibody or rabbit IgG. Precipitated chromatins were subjected to real time PCR using primers targeting enhancerI region (upper) or promoter region (lower). Values were shown as the percentage of corresponding input. Data are representative of three independent experiments.
(B) Purified CD4+ cells from B6 mice were treated as in (A) with or without IL-27 (20ng/ml) for 2 hours. CHIP assay was performed using anti-Smad3 antibody or rabbit IgG. Precipitated chromatins were subjected to real time PCR using primers targeting enhancer I region. Values were shown as the percentage of corresponding input. Data are representative of two independent experiments.
(C) Purified CD4+ cells from B6 mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without JNK kinase inhibitor (10µM) for 2 hours. CHIP assay was performed using anti-Smad3 antibody or rabbit IgG as described in (B).
AP-1 and RA enhance whereas IL-27 inhibits pSmad3 binding to Enhancer I
In next series of studies we first sought to determine if the increased accessibility of the Smad3 binding region induced by RAR-RXR noted above results in an actual increased binding of pSmad3 to this region. To this end we performed Smad3-specific ChIP assays on TCR-TGF-β-stimulated CD4+ T cells stimulated in the presence and absence of RA. As shown in Figure 7B, RA stimulation of cells was associated with markedly increased pSmad3 binding in the enhancer I region which contains the pSmad3 binding site.
We then sought to determine if binding of pSmad3 to the enhancer I region in CD4+ T cells is affected by IL-27 signaling which inhibits both TGF-β- and TGF-β plus RA-induced up-regulation of Foxp3 expression through activation of Stat3 and, in turn, the subsequent activation of the gene silencing site in enhancer II. Accordingly, Smad3-specific ChIP assays were performed on CD4+ T cells stimulated with TGF-β or TGF-β plus RA in the presence and absence of IL-27. As shown in Figure 7C, while RA enhanced pSmad3 binding to the enhancer I site, addition of IL-27 to the culture inhibited pSmad3 binding both in cells stimulated by TGF-β alone and with TGF-β and RA. This is in agreement with data showing that IL-27 down-regulates both TGF-β and TGF-β plus RA enhancement of Foxp3 expression.
Having thus demonstrated that both RA and IL-27 modify TCR + TGF-β induced Foxp3 expression via a common mechanism, i.e., regulation of pSmad3 binding to enhancer I, we recalled an earlier finding that RA reduces the need for AP-1 binding to enhancer I. This suggested that AP-1 (or the NFAT-AP-1 complex) acts primarily to increase pSmad3 binding to enhancer 1. To explore this possibility we performed ChIP assays of pSmad3 binding to enhancer I in primary CD4+ T cells stimulated with TCR/TGF-β in the presence and absence of JNK inhibitor. As shown in Figure 7c, the presence of the JNK inhibitor does indeed reduce pSmad3 binding. Thus, we conclude that not only RA and cytokine modification of Foxp3 expression occur via regulation of pSmad3 binding, but also that baseline Foxp3 expression depends on this mechanism.
Blockade of pSmad3 binding to its binding site in enhancer I compromises both TGF-β and TGF-β/RA Induction of Foxp3 expression
The above studies showing that TGF-β and TGF-β/RA induction of Foxp3 expression depends on generation of pSmad3 and its efficient binding to a site in enhancer I predicts that inhibition of pSmad3 binding to its enhancer site would result in reduced TGF-β- and TGF-β/RA- induced Foxp3 expression not only in cells lacking Smad3, but also in cells lacking Smad3 binding sites in enhancer 1. In initial studies to examine this prediction we transfected purified CD4+ T cells from Foxp3-GFP knockin mice with Alexa647-labeled oligonucleotides having a sequence identical to the Smad3 binding site in Foxp3 enhancer I which thus act as “decoy” oligos that block the binding to Smad3 to this enhancer site. In a parallel study we transfected the same cells with an oligonucleotide having a sequence identical to a consensus Smad3 binding sequence (Schneiders et al., 2005) to determine if blockade of the Foxp3 site was relative specific and blockade of Smad3 binding sites in other parts of the genome would have relatively little effect on Foxp3 expression. In parallel with these latter studies we determined the effect of Foxp3- specific and consensus sequence blocking oligonucleotides on expression of a luciferase reporter gene driven by a PAI promoter containing three consensus sequence of Smad3 binding site in Hep3B cells (Wrana et al., 1992). Oligonucleotides with a “scrambled” sequence were employed as controls and transfected cells were identified by labeling with Alexa647. As shown in Supplementary Figure 5C, TGF-β- and TGF-β plus RA-induced Foxp3 expression in the presence of the scrambled oligo was equivalent to that exhibited by cells stimulated in the absence of any decoy, whereas such expression in the presence of the Foxp3-specific Smad3 decoy was substantially reduced. In contrast, as also shown here, the blocking effect achieved with the consensus Smad3 decoy was substantially less and, as shown in supplemental Figure 5D, the Smad3 Foxp3-specific Smad3 decoy led to only marginal blocking of the consensus Smad3 sites. These studies provide further data supporting the concept that Smad3 binding to its binding site in the Foxp3 enhancer I is indeed an important component in TGF-β-RA induction of Foxp3 transcription, particularly in view of the fact that the Foxp3-specific Smad3 blocking decoy had only a marginal effect on Smad3 binding to consensus sequences elsewhere in the genome. In addition, inasmuch as these studies were conducted in primary CD4+ T cells with an intact Foxp3 gene, they show that Smad3 binding to its enhancer site and subsequent enhancement of Foxp3 transcription occurs under physiologic conditions.
In an second study of the prediction above regarding the consequences of inhibition of pSmad3 binding to its enhancer site, we determined the effects of deletion of the Smad3 binding site on a luciferase reporter construct driven by an intact Foxp3 gene fragment containing the Foxp3 promoter and enhancer regions. This fragment was derived from a BAC clone and consisted of 5.8kb of DNA extending from a site approximately 1 kb upstream of the Foxp3 transcription start site (containing the bulk of the Foxp3 promoter) to a site 4.8kb downstream of transcription start site (containing both enhancer I and enhancer 2 regions discussed above). As shown in Supplemental Figure 6A, TGF-β and TGF-β plus RA had the expected enhancing effects on the luciferase signal in both EL4 and LBRM cells. Perhaps more importantly, as shown in Supplemental Figure 6B (left panel), the same construct containing a deletion of the Smad3 binding site exhibited a significantly reduced signal with both TGF-β and TGFβ plus RA stimulation. In addition, in the absence of the Smad3 binding site there was no significant increase in luciferase signal in cells stimulated by TGF-β plus RA as compared to those stimulated with TGF-β alone. Thus, these studies corroborate the studies utilizing decoy oligonucleotides described above in that they show that RA enhancement of Foxp3 transcription is in fact dependent on Smad3 binding, and that such dependence occurs in the context of an intact Foxp3 gene fragment.
In related studies we also utilized the intact (Bac) Foxp3 contruct described above to verify effects of AP-1 and Stat3 binding site deletions previously studied with more conventional non-Bac reporter constructs. As shown in supplemental Figure 6B (right panel), deletion of the enhancer I AP-1 site led to decreased signal in TGF-β stimulated cells but no decreased signal in TGF-β-stimulated cells consistent with previous studies showing the lack of involvement of the enhancer I AP-site in RA enhancement; in addition, deletion of the enhancer II Stat3 binding site led to increased signal again reflecting the down-regulatory effect of this site.
In Vivo Verification of the Importance of Smad3 to Foxp3 Expression
Finally, we evaluated the role of Smad3 in the in vivo generation of Foxp3+ T cells. Here, we took advantage of previous work in which we showed that intra-nasal administration of a “tet-on” TGF-β plasmid (pTet-on-TGF-β1 that induces high amounts of TGF-β production in lamina propria cells of mice with experimental colitis, the latter necessary to activate CMV promoters in the plasmid (Kitani et al., 2003); thus administration of this plasmid recreates conditions under which Foxp3-expressing cells are induced in the intestine, particularly under the influence of endogenous retinoic acid production. Accordingly, we administered this plasmid to wild type and Smad3-deficient mice by an intra-nasal route (along with parenteral doxycycline). As shown in supplemental Figure 7, plasmid administration induced a striking increase in Foxp3 expressing cells in both colon and MLNs of wild type mice whereas in Smad3 KO it had only a marginal effect which could be attributable to effects of the induced inflammation on natural (nTreg) Foxp3+ cells. Thus, Smad3 is a necessary component of TGF-β-induced Treg (iTreg) development both in vitro and in vivo.
Discussion
The regulation of Foxp3 transcription is a multifaceted process that probably reflects the necessity of the immune system to fine-tune regulatory T cell function under a vast variety of circumstances. In the present study we focused on regulation of Foxp3 transcription occurring in relation to Tregs induced in the peripheral lymphoid system under the influence of TGF-β; other factors, some overlapping with those discussed here, may be involved in “natural” Tregs developing in the thymus (Liu et al., 2008).
The data gathered in these studies suggest that regulation of Foxp3 transcription and regulatory T cell development in the peripheral lymphoid tissues is best understood as a two stage process consisting of an initial stage in which Foxp3 transcription was initiated by T cell receptor stimulation and TGF-β signaling and a second stage in which this initial transcription was either enhanced by retinoic acid (RA) or inhibited by pro-inflammatory cytokines. Key insights into the molecular events occurring during this stage has come from a previous study by Tone et al who defined the transcriptional activity of a ~1.8 kb Foxp3 promoter fragment and a conserved downstream enhancer (located at +1988 to +2738). In these studies it was shown with reporter assays that the Foxp3 promoter appeared to have a surprisingly little role in Foxp3 transcription and the main control site was the intronic enhancer (enhancer I) which contained both NFAT and Smad3 binding sites. This conclusion arose from the fact that deletion of either NFAT or Smad3 binding sites led to compromised TCR-TGF-β-induced Foxp3 transcription. More recently, however, this group has provided evidence that during TCR-TGF-β-induced Foxp3 transcription, the Foxp3 promoter binds c-Rel, p65 and NFATc2 to a site at which a “enhanceosome” complex is formed which ultimately contains transcription elements such as Smad3 and Creb recruited from enhancer regions. Thus, the promoter does in fact play a major role in Foxp3 transcription (Ruan et al., 2010).
In our initial studies we expanded on these findings by demonstrating that TGF-β induction of Foxp3 in naïve T cells was greatly inhibited by a JNK inhibitor. This implied the existence of an AP-1 binding site in the Foxp3 promoter-/enhancer region and indeed we identified a sequence ordinarily considered a “weak” (or partial) AP-1 binding site in the enhancer region immediately upstream of the NFAT binding site. This site complemented three additional AP-1 sites in the promoter previously identified by Mantel et al. (Mantel et al., 2006). In subsequent studies employing a luciferase reporter system we confirmed the functional significance of AP-1 binding by showing that deletion of the AP-1 site in the enhancer, but not the most 5’ AP-1 site in the promoter greatly decreased Foxp3 promoter-enhancer function. However, we could not exclude the possibility that the other two AP-1 sites located at promoter region might compensate the AP-1 site or are required to act in tandem. The importance of AP-1 to TGF-β-induced Foxp3 expression is probably related to its role in facilitating the function of NFAT, a TCR-induced transcription factor which, as mentioned above, has been shown to be essential for such expression. In further studies of the role of AP-1 in Foxp3 expression we found that treatment of CD4+ T cells with a JNK inhibitor led to decreased Smad3 binding to enhancer I as determined by ChIP analysis. This suggests that one function of AP-1 and by inference, the NFAT-AP-1 complex, in TGF-β-induced Foxp3 expression is to enhance Smad3 binding. Taken together, these findings introduced the concept that a central feature of control of TGF-β-induced Foxp3 expression is the regulation of Smad3 binding to enhancer I.
In further studies supporting this latter conclusion we first showed that cells from Smad3-deficient mice cannot be induced to express Foxp3 when stimulated in the presence of either TGF-β alone or TGF-β plus RA. Since this result was contrary to that reported in a recent study by Nolting et al., in which the latter authors reported that cells from Smad3-deficient mice could be induced to express Foxp3 (Nolting et al, 2009), we performed multiple studies of cells from two independent strains of Smad3-deficient mice, one on a B6 and another on a BALB/c background (the latter identical to that used by Nolting et al.). Second we showed that inhibition of Smad3 binding to its target sequence in enhancer I with the addition of a decoy oligonucleotide that competes with the target sequence for Smad3 binding leads to substantial inhibition of both induction of Foxp3 by TGF-β alone or TGF-β plus RA. Importantly, the Foxp3-specific decoy oligo used in this study had only a marginal ability to block consensus Smad3 sequences suggesting that the blocking effect was not due to blocking of Smad3 effects on other parts of the genome. This study, conducted in CD4+ T cells with an intact and endogenous Foxp3 gene, rather than a reporter construct, provides strong evidence that for Foxp3 induction to occur, not only must activated Smad3 be generated but also that activated Smad3 must bind to a site in the Foxp3 enhancer I. Finally, we performed in vivo studies of Smad3-deficient mice in which we showed that induction of Foxp3+ cells in the colons/MLN of wild type mice with mild DSS colitis by nasal administration of a plasmid inducing high levels of TGF-β is greatly impaired in Smad3-deficient mice. This result showed that Smad3 activity is necessary for induced Foxp3 induction under in vivo conditions as well as under in vitro conditions. Taken together, these various studies establish rather definitively that induced Foxp3 expression is a Smad-dependent event.
Recently it has been shown that retinoic acid (RA) produced by antigen-presenting cells, particularly in mucosal tissues, enhances TCR-TGF-β-induced expansion of Foxp3 regulatory cells; however, despite considerable investigation the mechanism of this RA effect remained unclear. One possibility suggested by Kretschmer and his colleagues (Kretschmer et al., 2008) was that RA reverses the negative effect of AP-1 (induced by co-stimulation) on Foxp3 expression; this possibility, however, is unlikely in view of the positive effect of AP-1 on Foxp3 expression shown here. A second possibility is that RA promotes Smad3 phosphorylation and thus Smad3 translocation to the nucleus; in addition, RA down regulates receptors of inhibitory cytokines (Xiao et al., 2008). However, we found in extensive studies in which cells were stimulated with a wide range of TGF-β concentrations that RA induces Smad3 phosphorylation only when low (sub-optimal) TGF-β concentrations are present and under these conditions such induction may be more apparent than real since RA induces Smad3 and thus makes more Smad3 available for TGF-β-induced phosphorylation; thus, the weight of evidence suggests that while RA enhancing effects could be explained by by induction of increased Smad3 phosphorylation at low TGF-β concentrations this does not explain its enhancing effects at TGF-β concentrations likely to obtain at tissue sites. In addition, we also found that RA had no effect on the ability of inhibitory cytokines to induce pStat3 so there is no evidence to support the view that RA augmentation occurs through effects on cytokine inhibition. A third possibility, suggested by Hill et al. was that the RA effect is indirect and due to it’s ability to counter the inhibiting effect of cytokines such as IL-4, IL-21 and IFN-γ produced by non-regulatory, mature T cells (Hill et al., 2008). One problem with this possibility is that, as shown here, while IL-4 does indeed down-regulate TGF-β-induced baseline Foxp3 expression, it has little if any effect on RA augmentation of such expression. Overall, while it seems possible that RA augmentation can be due, especially in vivo, to indirect effects on inhibitory cytokine secretion, the extensive data here documenting that RA influences Foxp3 transcription would argue that its augmenting effect is mainly due to a direct effect on Foxp3 promoter-enhancer activity.
In our search for another explanation of the RA enhancing effect, we focused on the exquisite dependence of RA augmentation of TGF-β-induction of Foxp3 on Smad3 mentioned above and were thus led to the hypothesis that the RA effect occurs because it increases the level of binding of pSmad3 to its binding site in enhancer I. In fact, all of the findings reported here are in support of this view. First, the addition of RA to cultures of T cell lines bearing Foxp3 luciferase reporter constructs consistently gave rise to an increased luciferase signal; this suggested that the transcription factor resulting from RA signaling, RAR-RXR, was indeed having a direct effect on Foxp3 promoter and enhancer activity. Second, we found that the Foxp3 gene has at least two potential RAR-RXR binding sites, one in the promoter and another in enhancer I very close to the AP-1 binding site. Furthermore, deletion of the RAR-RXR binding site in enhancer I (close to the AP-1 binding site) led to a major reduction in the RA enhancing activity and deletion of both the RAR-RXR binding sites in both in enhancer I and in the promoter led to a virtual complete loss of RA enhancer activity; importantly, with respect to the RAR-RXR binding site in enhancer I, this result was verified by luciferase reporter assays conducted in primary CD4+ T cells. Third, while whereas addition of TGF-β alone to cultures of CD4+ T cells led to increased histone acetylation at the enhancer I site, addition of both RA and TGF-β led to a much greater degree of histone acetylation. This suggested that RA greatly increases the accessibility of this site to transcription factors. Finally, in ChIP assays addition of RA to TCR-TGF-β-stimulated cell cultures greatly increased binding of pSmad3 to its binding site in enhancer I. Thus, while AP-1 (or more probably NFAT-AP-1) controls initial pSmad3 binding to enhancer 1 and a basal level of Foxp3 transcription, it was evident from these studies that RA controls a second, augmented level of binding and thus a higher level of Foxp3 transcription. Finally, it should be noted that the presence of RA greatly diminished the negative effect of JNK inhibition on TCR/TGF-β-induced Foxp3 expression. We attribute this to the possibility that RAR-RXR binding in enhancer I to a great extent obviates the need for NFAT-AP-1 augmentation of pSmad3 binding.
In the present studies we also investigated the inhibition of TCR/TGF-β induction of Foxp3 or such induction augmented by RA or by cytokines such as IL-27 and IL-6 which induce Stat3 activation and showed in that such inhibition was abolished in Stat3-deficient cells and augmented in SOC3-deficient cells; we thereby verified the view that had previously been proposed that inhibition by these cytokines was mediated by Stat3 (Huber et al., 2008). It was evident in these studies that IL-27 inhibited both baseline and RA-augmented TCR-TGF-β -induced Foxp3 expression. This, however, does not obtain for IL-4, a cytokine that inhibits baseline but not RA-augmented TCR/TGF-β-induced Foxp3 expression and which induces pStat6 rather than pStat3 (Takaki et al., 2008).
In dissecting the mechanism of IL-27-induced inhibition we first sought a binding site for Stat3 and indeed found such a site in a second conserved enhancer region downstream of enhancer I which we call enhancer II. The investigation of the function of this site with our reporter system was somewhat indirect because the cells used for transfection of the reporter constructs both produced substantial amounts of endogenous pStat3, thus precluding the use of these cells as targets of cytokine inhibition via induction of activated Stat3. We overcame this difficulty by determination of the effect of the deletion of the pStat3 site on the reporter activity of a construct containing the Foxp3 promoter, enhancer I and enhancer II in tandem recognizing that the effect of deletion would be due to loss of pStat3 signaling by endogenous pStat3 and the latter would be a “proxy” for cytokine-induced pStat3. We found the deletion of the pStat3 binding site did in fact lead to augmentation of the luciferase reporter signal, indicating that Stat3 binding was acting as a potent gene silencer that affects both basal and augmented TCR-TGF-β-induced Foxp3. Interestingly, deletion of the pStat3 binding site in enhancer II generated a signal which was augmented with respect that generated by a construct that did not contain enhancer II. This revealed that in the absence of the effect of pStat3 binding enhancer II did actually function as an enhancer, possibly via the binding of other endogenous positive factors to this region such as CREB-ATF (Kim and Leonard, 2007). In a final and critical series of studies of IL-27 inhibition of TCR-TGF-β-induced Foxp3 expression we showed that in CD4+ T cells, such inhibition is associated with greatly reduced Smad3 binding to enhancer I whether or not the latter in augmented by RA. Thus, whereas the molecular mechanism of cytokine inhibition is different from that of RA augmentation, both operate through a final common pathway, namely the regulation of Smad3 binding to enhancer I.
Experimental Procedures
Mice
Specific pathogen-free, female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Foxp3-IRES-GFP knock-in mice on a C57BL/6 background were obtained from M. Oukka (Brigham and Women’s Hospital, Boston MA). Stat3fl/fl and Stat3fl/fl; MMTV-Cre mice; Socs3fl/fl and Socs3fl/fl; MMTV-Cre mice were kind gifts from Dr. John J. O'Shea (Molecular Immunology and Inflammation Branch, NIAMS/NIH). Smad3 knockout mice on a C57BL/6 background were kind gifts from Dr. Sharon Wahl (Oral Infection and Immunity Branch, NIDCR/NIH). These mice were studied at 8–14 wk of age. Animal use adhered to National Institutes of Health Laboratory Animal Care Guidelines.
Cell lines
A mouse T lymphoma cell line LBRM-33 clone 4A2 (LBRM) was obtained from ATCC. EL4 clone LAF cell (EL4) was a kind gift from Dr. Masahide Tone, University of Pennsylvania. Both cell lines were maintained in IMDM supplemented with 5% FCS, 100U/ml penicillin, 100µg/ml streptomycin.
In Vitro Cell Stimulation
Murine CD4+ T cells were cultured in 1ml of IMDM supplemented with 10% FCS, 100U/ml penicillin, 100µg/ml streptomycin, and 5 × 10−3 M mercaptoethanol. Cells were stimulated with plate-bound anti-CD3 (10µg/ml) and soluble anti-CD28 (2µg/ml). Cytokines, neutralizing antibodies and other reagents were added to cultures at the following concentrations: rTGF-β1 (5 ng/ml, R&D), IL-27 (20 ng/ml, Peprotech), cyclosporin A (CsA) (20nM), ALK5 inhibitor (5µM, EMD) and JNK kinase inhibitor (5µM, EMD 420116). Neither CsA, nor the ALK5 or JNK kinase inhibitors affected cell viability at the concentrations used (data not shown).
Flouresence staining
For flow cytometric analysis cells were fixed and permeabilized in cytofix/permeablization solution (BD PharMingen) and stained with PE or APC anti-Foxp3 (eBioscience).
Construction of reporter plasmids and luciferase assay
A 1019 bp fragment of Foxp3 promoter was amplified from genomic DNA by PCR and cloned into pGL4.15 vector (Promega) between Xho I and Hind III sites. Mlu I and Acl I sites were introduced into pGL4-Foxp3 promoter vector by Site-Directed Mutagenesis PCR using QiuckChange XL Kit from Strategene. A 182 bp fragment of Foxp3 enhancer were amplified by PCR and cloned into HindIII and Mlu I sites and a 973 bp fragment of silencer were amplified by PCR and cloned into Mlu I and Acl I sites. AP-1, Stat3 and RAR binding sites were deleted by Site-Directed Mutagenesis PCR. All the plasmids were sequenced to verify the insertions and deletions. Luciferase assay were performed in LBRM and EL4 cells. We transfected 4 × 106 cells by Amaxa nuclear transfection kit (program C-009) using 8 µg firefly luciferase reporter plasmid and 30 ng phRL-SV40 (for LBRM) or 50ng phRL-TK (for EL4) Renilla luciferase plasmid as an internal control. 4 hours after transfection, cells were split and stimulated with plate bound anti-CD3 (7µg/ml), soluble anti-CD28 (2µg/ml), rTGF-β (2ng/ml) and all trans retinoic acid as indicated. 24 hours later luciferase activity was analyzed by Dual-Glo Luciferase Reporter Assay System (Promega). Data were shown as the absolute ratio of firefly luciferase activity / Renilla luciferase activity from the same well.
Chromatin Immunoprecipitation (ChIP) Assay
Mouse CD4+ T cells were stimulated with plate bound anti-CD3 (7µg/ml), soluble anti-CD28 (2µg/ml) with rTGF-β (5 ng/ml), all trans retinoic acid (500nM) or both. Formaldehyde (1% final concentration) was then added to cross-link proteins and DNA. Cells were then washed and lysed in SDS lysis buffer containing PMSF and proteinase inhibitor cocktail (active motif). Nuclei were sonicated (Branson Sonifier, 11% amplitude) for 10 seconds 6 pulses to shear DNA to 200–1500bp. Sheared chromatins were 10 times diluted and pre-cleaned with Salmon Sperm DNA/protein A agarose. A proportion (2.5%) of the diluted chromatins were kept as "input." The rest of the chromatins were incubated with 5ug antibody or isotype IgG overnight followed by additional incubation with Salmon Sperm DNA/protein A agarose for 1 hour and then washed 5 times. The bead-bound protein-DNA complexes were eluted in 1% SDS, 0.1 M NaHCO3, and cross-links were reversed at 65 °C and treated with proteinase K at 55°C. Precipitated DNA was subjected to real time PCR using power SYBR green PCR kit (Applied Biosystem). Following primer pairs were used: promoter region: forward: 5'-gggcactcagcacaaacatgatg-'3, reverse: 5'-gaggcttccttctgctccaaac-'3, enhancer I region: forward: 5'-caggctgacctcaaactcacaaag-'3, reverse: 5'-catacccacacttttgacctctgc-'3; forward: 5'- gcttctgtgtatggttttgtgt-'3, reverse: 5'-atcatcacagtacatacgagga-'3. Values were normalized to corresponding input control.
Supplementary Material
Acknowledgements
We thank J. J. O'Shea (Molecular Immunology and Inflammation Branch, NIAMS/NIH) for kindly providing us Stat3fl/fl and Stat3fl/fl; MMTV-Cre mice; Socs3fl/fl and Socs3fl/fl; MMTV-Cre mice. We thank S. Wahl (Oral Infection and Immunity Branch, NIDCR/NIH) for Smad3-deficient mice. We thank M. Oukka (Harvard Medical School) for kindly providing us Foxp3-IRES-GFP gene targeted mice. We thank M. Tone (University of Pennsylvania) for EL4 clone LAF cells. This research was supported by the Intramural Research Program of NIAID and NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Elias KM, Laurence A, Davidson TS, Stephens G, Kanno Y, Shevach EM, O’Shea JJ. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–1020. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Steinwald V, Guralnik A, Brüstle A, Kleemann P, Rosenplänter C, Decker T, Lohoff M. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20:223–234. doi: 10.1093/intimm/dxm139. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182:6648–6652. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani A, Fuss I, Nakamura K, kumaki F, Usui T, Strober W. Transforming growth factor (TGF)-b1-producing regulatory T cells induce Smad-mediated interleukin 10 secretion that facilitates coordinated immunoregulatory activity and amelioration of TGF-b1-mediated fibrosis. J Exp Med. 2003;198:1179–1188. doi: 10.1084/jem.20030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer K, Apostolou I, Verginis P, von Boehmer H. Regulatory T cells and antigen-specific tolerance. Chem Immunol Allergy. 2008;94:8–15. doi: 10.1159/000154846. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Macián F, López-Rodríguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Mantel PY, Ouaked N, Rückert B, Karagiannidis C, Welz R, Blaser K, Schmidt-Weber CB. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Mucida D, Pino-Lagos K, Kim G, Nowak E, Benson M, Kronenberg M, Noelle R, Cheroutre H. Retinoic Acid Can Directly Promote TGF-β-Mediated Foxp3+ Treg Cell Conversion of Naive T Cells. Immunity. 2009;30:471–472. doi: 10.1016/j.immuni.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufert C, Becker C, Wirtz S, Fantini MC, Weigmann B, Galle PR, Neurath MF. IL-27 controls the development of inducible regulatory T cells and Th17 cells via differential effects on STAT1. Eur. J. Immunol. 2007;37:1809–1816. doi: 10.1002/eji.200636896. [DOI] [PubMed] [Google Scholar]
- Nolting J, Daniel C, Reuter S, Stuelten C, Li P, Sucov H, Kim BG, Letterio JJ, Kretschmer K, Kim HJ, von Boehmer H. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J. Exp. Med. 2009;206:2131–2139. doi: 10.1084/jem.20090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2010;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiders D, Heger J, Best P, Michael Piper H, Taimor G. SMAD proteins are involved in apoptosis induction in ventricular cardiomyocytes. Cardiovasc Res. 2005;67:87–96. doi: 10.1016/j.cardiores.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Schoenborn J, Sekimata M, Weaver W, Wilson C. Transfection of primary T cells for stimulation-dependent cytokine enhancer assays. Nat Protocols. 2007 DOI: 10.1038/nprot.2007.236. [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;76:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B, Kuchroo VK. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181:2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, Hennighausen L, Wu C, O’Shea JJ. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SG, Gray JD, Ohtsuka K, Yamagiwa S, Horwitz DA. Generation ex vivo of TGF-beta-producing regulatory T cells from CD4+CD25-precursors. J Immunol. 2002;169:4183–4189. doi: 10.4049/jimmunol.169.8.4183. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.