Abstract
Aspirin-intolerant asthma (AIA) is a rare condition that is characterized by the development of bronchoconstriction in asthmatic patients after ingestion of non-steroidal anti-inflammatory drugs including aspirin. However, the underlying mechanisms of AIA occurrence are still not fully understood. To identify the genetic variations associated with aspirin intolerance in asthmatics, the first stage of genome-wide association study with 109,365 single nucleotide polymorphisms (SNPs) was undertaken in a Korean AIA (n = 80) cohort and aspirin-tolerant asthma (ATA, n = 100) subjects as controls. For the second stage of follow-up study, 150 common SNPs from 11 candidate genes were genotyped in 163 AIA patients including intermediate AIA (AIA-I) subjects and 429 ATA controls. Among 11 candidate genes, multivariate logistic analyses showed that SNPs of CEP68 gene showed the most significant association with aspirin intolerance (P values of co-dominant for CEP68, 6.0×10−5 to 4.0×10−5). All seven SNPs of the CEP68 gene showed linkage disequilibrium (LD), and the haplotype of CEP68_ht4 (T-G-A-A-A-C-G) showed a highly significant association with aspirin intolerance (OR = 2.63; 95% CI = 1.64–4.21; P = 6.0×10−5). Moreover, the nonsynonymous CEP68 rs7572857G>A variant that replaces glycine with serine showed a higher decline of forced expiratory volume in 1s (FEV1) by aspirin provocation than other variants (P = 3.0×10−5). Our findings imply that CEP68 could be a susceptible gene for aspirin intolerance in asthmatics, suggesting that the nonsynonymous Gly74Ser could affect the polarity of the protein structure.
Introduction
Aspirin, also known as acetylsalicylic acid, has been used as an analgesic to relieve pain and fever, as well as an anti-inflammatory medication. Up to 20% of asthmatics are sensitive to aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs). Aspirin-intolerant asthma (AIA), as a unique clinical syndrome with acute bronchospasm after ingestion of aspirin or other NSAIDs, was first described in 1922 [1], [2]. AIA is characterized by the triad of aspirin hypersensitivity, bronchial asthma, and chronic rhinosinusitis with nasal polyposis [3], [4]. AIA progression from the upper to the lower respiratory tract is accompanied by persistent asthmatic symptoms with intense eosinophilic infiltration into the upper and lower airways. AIA generally begins at about 30 years of age and occurs more frequently in women [2], [5].
Recent studies have shown that aspirin hypersensitivity is likely to be related with over-production of pro-inflammatory cysteinyl leukotrienes (CysLTs) such as LTC4, LTD4, and LTE4 [2]. CysLTs are generated from arachidonic acid via the leukotriene synthetic pathway or the 5-lipoxygenase pathway. Aspirin blocks the cyclooxygenase pathway that converts arachidonic acid into prostaglandins, and eventually diverts arachidonic acid metabolites to the 5-lipoxygenase pathway [6], [7]. Therefore, CysLT receptors and arachidonic acid cascade-related enzymes/proteins have been considered as main targets for aspirin intolerance in asthmatics. CysLT receptors are selectively antagonized by several leukotriene modifiers, such as montelukast, pranlukast, and zafirlukast [8], [9]; however, clinical studies have demonstrated that the responses to these drugs is incomplete [10], [11], suggesting the presence of an alternative pathway leading to AIA. Previous genetic studies have revealed significant associations between AIA and the polymorphisms of cysteinyl leukotriene receptor 1 [12], cysteinyl leukotriene receptor 2 [13], thromboxane A2 receptor [14], and prostaglandin E2 receptor subtype 2 [15]. The genetic polymorphisms of leukotriene C4 synthase (LTC4S) [16], [17], [18] and arachidonate 5-lipoxygenase (ALOX5) [19], [20] have been reported to be positively or negatively associated with AIA depending on populations and polymorphic loci. In addition to the genes on the arachidonate pathway, genetic variants on TNF [21], HLA [22], [23], [24], TBX21 [25], ACE [26], IL-10/TGF [27], and NLRP3 [28] genes in the pathways of immune response and inflammation are also associated with aspirin hypersensitivity in asthmatics. These data suggest that genetic variants of genes on other pathways may be more relevant to development of aspirin hypersensitivity in asthmatics than was previously thought.
Genome-wide association studies (GWAS) have recently emerged as a technology in predicting genetic variations across the genome that are associated with human diseases and clinical response to drug treatment [29], [30]. Recently, GWAS for asthma and related phenotypes have reported several susceptible genes including ORMDL3, PDE4D and IL1RL1 [31], [32], [33]. In the case of AIA, most of the recent genetic association analyses have been investigated through candidate gene approaches. Therefore, genome-wide association analysis and follow-up study of aspirin intolerance in asthmatics were performed in order to identify novel and comprehensive etiology of AIA.
Results
Characteristics of the Study Subjects
The clinical characteristics and comparisons of AIA, intermediate AIA (AIA-I), and ATA groups are summarized in Table 1. First of all, an aspirin-induced decline in FEV1 of −15% to 68% was observed in all study subjects. In both of genome-wide and follow-up studies, the % declines of FEV1 by aspirin provocation in AIA and AIA-I patients were significantly increased compared to those of ATA controls (P<0.0001), indicating that this study could reflect an association between genetic polymorphisms and aspirin hypersensitivity in asthmatics. In subjects of 2nd stage, the values of predicted FEV1 % and PC20 methacholine were significantly lower in AIA patients than those of ATA controls, whereas the total IgE level was higher in cases than in controls (P<0.01). A significant decrease of body mass index (BMI) in aspirin-intolerant asthmatics was also observed. In addition, the mean age of first medical examination was significantly lower in AIA than in the ATA group.
Table 1. Clinical profiles of aspirin-intolerant and aspirin-tolerant asthmatics.
| Clinical profile | First stage of genome-wide study | Second stage of follow-up study | |||||
| AIA | ATA | Asthmatics(all subjects) | AIA | ATA | |||
| AIA | AIA-I | ||||||
| Number of subjects (n) | 80 | 100 | 592 | 102 | 61 | 429 | |
| Age of onset [year, mean (range)] | 45.06 (17.22–71.56) | 45.56 (15.41–77.13) | 46.15 (15.40–77.88) | 42.76 (18.66–72.73) ** | 43.73 (17.22–71.29) | 47.30 (15.40–77.88) | |
| Sex (n, male/female) | 27/53 | 27/73 | 206/386 | 37/65 | 22/39 | 147/282 | |
| Current Smoker (%) | 20.00 | 26.00 | 27.70 | 20.59 | 22.95 | 30.07 | |
| Height [cm, mean (range)] | 161.82±9.90 | 159.75±8.45 | 160.78±8.63 | 161.70±9.91 | 161.75±7.95 | 160.42±8.39 | |
| Weight (kg) | 61.74±10.16 | 62.41±10.32 | 62.81±10.84 | 61.64±10.39 | 60.61±10.42 | 63.40±10.97 | |
| Body mass index (kg/m2) | 23.57±3.20 | 24.44±3.39 | 24.24±3.39 | 23.56±3.37** | 23.09±3.06** | 24.58±3.39 | |
| % decline of FEV1 by aspirin provocation | 25.84±14.00*** | 0.95±2.77 | 9.27±13.24 | 33.59±13.42*** | 10.07±6.74*** | 3.54±4.85 | |
| Blood eosinophil (%) | 6.87±5.79* | 4.89±4.16 | 6.01±5.73 | 6.65±5.78 | 4.80±3.89 | 6.03±5.92 | |
| FEV1 (% predicted) | 82.53±21.90 | 82.09±20.69 | 90.54±16.97 | 85.10±16.41** | 91.73±17.12 | 91.66±16.87 | |
| PC20 methacholine (mg/ml) | 3.88±6.80 | 2.83±3.59 | 6.43±8.67 | 4.26±7.62** | 6.20±8.06 | 6.91±8.90 | |
| Log[Total IgE (IU/ml)] | 2.21±0.52 | 2.14±0.62 | 2.16±0.63 | 2.26±0.58 | 2.15±0.45 | 2.13±0.66 | |
| Positive rate of skin test (%) | 58.75 | 63.00 | 56.42 | 48.04 | 60.66 | 57.81 | |
Each clinical profile of AIA and AIA-I was compared to ATA controls, respectively.
*P<0.05;
**P<0.01;
***P<0.0001.
AIA, aspirin-intolerant asthma; AIA-I, intermediate aspirin-intolerant asthma; ATA, aspirin-tolerant asthma.
Genome-Wide Association Analyses
A total of 109,365 SNPs genotype assays were tested on the DNA samples of 80 AIA and 100 ATA subjects using the Illumina's Human-1 Genotyping BeadChip. The overall call rate of individual samples was over 98.0% after passing the genotype quality threshold, and a total of 4,515 SNPs (4.1% of 109,365 SNPs) failed to provide accurate genotype results. In further quality control analysis of the remaining 104,850 SNPs, monomorphic, X-chromosomal and Hardy Weinberg Equilibrium-departed (P<0.001) SNPs were additionally omitted; the final remaining 96,984 SNPs (88.7%) with an the average call rate of 99.9% were selected for analysis.
From results of the allelic association tests for each SNP in the co-dominant model, the 11 SNPs in the gene region that had the most significant association signals were screened for the second stage of follow-up study (Table 2).
Table 2. Top 11 Genetic variants within gene regions with lowest P value for risk of aspirin intolerance.
| SNP ID | Gene | Chr. locus | Variation | Position | Function | MAF | P value* | |
| AIA (n = 80) | ATA (n = 100) | |||||||
| rs1053744 | SBF1 | 22q13.33 | C>T | Exon39 | SET binding factor 1 | 0.500 | 0.365 | 3.0×10−7 |
| rs828616 | DCBLD2 | 3q12.1 | A>G | Exon 6 | Discoidin, CUB and LCCL domain containing 2 | 0.195 | 0.365 | 2.0×10−6 |
| rs3213729 | WDR21A | 14q24.3 | C>G | Promoter | DDB1 and CUL4 associated factor 4 | 0.230 | 0.100 | 1.0×10−5 |
| rs1932523 | FILIP1 | 6q14.1 | T>C | Intron1 | Filamin A interacting protein 1 | 0.465 | 0.290 | 3.0×10−5 |
| rs4867084 | PDZK3 | 5p13.3 | G>A | Intron1 | PDZ domain containing 2 | 0.200 | 0.365 | 4.0×10−5 |
| rs11060167 | LRRC43 | 12q24.31 | C>A | Exon8 | Leucine rich repeat containing 43 | 0.225 | 0.380 | 4.0×10−5 |
| rs6498124 | CIITA | 16p13.13 | G>T | Intron6 | Class II, major histocompatibility complex, transactivator | 0.350 | 0.535 | 4.0×10−5 |
| rs2564978 | DAF | 1q32.2 | T>C | Promoter | CD55 molecule, decay accelerating factor for complement | 0.535 | 0.390 | 5.0×10−5 |
| rs2295017 | ENPP5 | 6p12.3 | A>T | 3′UTR | Ectonucleotide pyrophosphatase/phosphodiesterase 5 | 0.090 | 0.215 | 5.0×10−5 |
| rs2252867 | CEP68 | 2p14 | A>G | Intron1 | Centrosomal protein 68 kDa | 0.415 | 0.250 | 7.0×10−5 |
| rs4245976 | C6 | 5p13 | C>T | Intron6 | Complement component 6 | 0.250 | 0.156 | 7.0×10−5 |
MAF, minor allele frequency; Chr., chromosome.
*P values of co-dominant model. UTR; untranslated region.
Follow-up Study of Second Stage
For the follow-up study, the 11 genetic variants “within gene regions” with the lowest P value (P<0.0001) for risk of aspirin intolerance were selected. Then, 150 common SNPs in 11 candidate genes with minor allele frequency of over 0.05 based on Asian population were selected from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) and genotyped in 163 AIA subjects including AIA-I group, and 429 ATA subjects. In order to include the maximum number of patients with aspirin hypersensitivity, 80 AIA and 100 ATA subjects from the first stage were included in the second stage due to the rareness of the AIA condition. The case/control associations between genotype and aspirin intolerance in asthmatics were analyzed using multivariate logistic analyses adjusted for initial diagnosed age, sex, smoking status, atopy, and BMI. Polymorphisms of the CEP68 gene showed the most significant association with AIA compared to ATA controls in co-dominant model (Table 3, P = 6.0×10−5 to 4.0×10−5). Furthermore, a nonsynonymous SNP (rs7572857G>A) of CEP68 gene that replaces glycine with serine revealed to have the lowest P value among seven SNPs of the gene (OR = 2.63; 95% CI = 1.64–4.21; P = 6.0×10−5 for co-dominant model). This rs7572857G>A was also statistically significant in the dominant model (P = 0.0005), but not in recessive model (Table S1). In further association analysis excluding the GWAS subjects in the second set, the statistically significant association still remained although reduced significances were observed (Table S2).
Table 3. Logistic analyses of SNPs in the CEP68 gene between AIA and ATA.
| Position | Amino acid change | AIA (n = 102) vs. ATA (n = 429) | [AIA+AIA-I] (n = 163) vs. ATA (n = 429) | |||||||||
| Frequencya | Co-dominant model | Frequencya | Co-dominant model | |||||||||
| AIA | ATA | OR (95% CI) | P * | Pcorr ** | [AIA+AIA-I] | ATA | OR (95% CI) | P * | Pcorr ** | |||
| SNP | ||||||||||||
| rs2302647 C>T | Promoter | 0.456 | 0.309 | 1.82 (1.32–2.50) | 0.0002 | 0.001 | 0.399 | 0.309 | 1.46(1.12–1.91) | 0.006 | 0.02 | |
| rs2252867 A>G | Intron1 | 0.461 | 0.319 | 1.78 (1.30–2.45) | 0.0004 | 0.002 | 0.414 | 0.319 | 1.50(1.15–1.96) | 0.003 | 0.01 | |
| rs12611491 A>G | Exon2 | Arg27Gly | 0.284 | 0.241 | 1.25 (0.88–1.79) | 0.22 | - | 0.285 | 0.241 | 1.28(0.95–1.74) | 0.1 | - |
| rs7572857 G>A | Exon2 | Gly74Ser | 0.176 | 0.077 | 2.63 (1.64–4.21) | 6.0×10−5 | 0.0003 | 0.126 | 0.077 | 1.71(1.11–2.62) | 0.02 | - |
| rs2723087 T>A | Intron2 | 0.461 | 0.319 | 1.78 (1.30–2.45) | 0.0004 | 0.002 | 0.414 | 0.319 | 1.50(1.15–1.96) | 0.003 | 0.01 | |
| rs6741255 T>C | Intron5 | 0.466 | 0.317 | 1.87 (1.35–2.57) | 0.0001 | 0.0005 | 0.417 | 0.317 | 1.55(1.18–2.03) | 0.002 | 0.007 | |
| rs10496123 G>A | Intron5 | 0.289 | 0.339 | 0.81 (0.58–1.14) | 0.23 | - | 0.304 | 0.339 | 0.85(0.64–1.13) | 0.27 | - | |
| Haplotype | ||||||||||||
| CEP68_ht1 | 0.240 | 0.340 | 0.60(0.42–0.86) | 0.006 | 0.276 | 0.340 | 0.73(0.54–0.97) | 0.03 | ||||
| CEP68_ht2 | 0.289 | 0.331 | 0.84(0.60–1.17) | 0.3 | 0.304 | 0.331 | 0.88(0.66–1.17) | 0.38 | ||||
| CEP68_ht3 | 0.275 | 0.218 | 1.34(0.93–1.93) | 0.12 | 0.267 | 0.218 | 1.32(0.97–1.79) | 0.08 | ||||
| CEP68_ht4 | 0.176 | 0.077 | 2.63(1.64–4.21) | 6.0×10−5 | 0.126 | 0.077 | 1.71(1.11–2.62) | 0.02 | ||||
The frequency of SNP indicates the minor allele frequency.
*P values represent the co-dominant model, which includes the additive model, adjusted for age at initial diagnosis, sex, smoking status, atopy, and body mass index.
**Pcorr values after multiple testing correction.
AIA, aspirin-intolerant asthma; AIA-I, intermediate aspirin-intolerant asthma; ATA, aspirin-tolerant asthma; OR, odds ratio; CI, confidence interval.
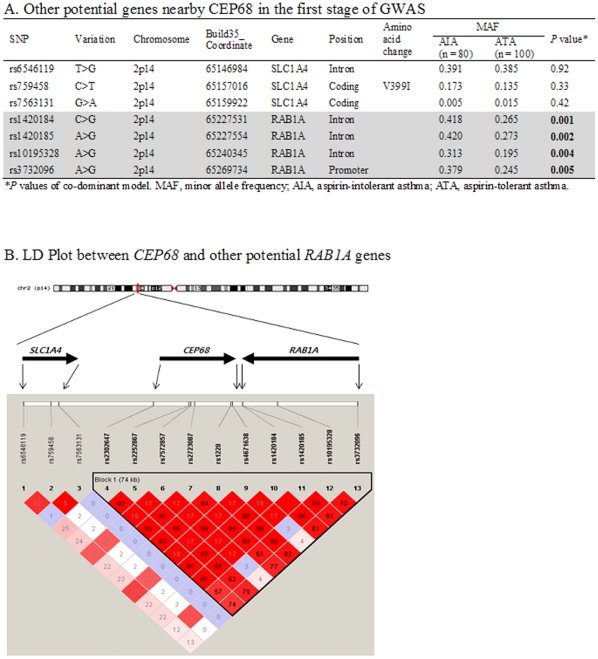
This study also observed modest associations of other genes with AIA. In particular, the intronic rs4867084G>A of the PDZK3 gene (P = 0.006 for co-dominant model) and rs11060333C>T of the LRRC43 gene (P = 0.009 for co-dominant model) showed significant associations with AIA (Table S3). In addition, several intergenic SNPs showed significant associations with AIA in our GWAS results (P<0.0001; Table 4). Two SNPs, rs139719 and rs7744030, were identified as intergenic SNPs during the first stage of GWAS (build 126 of dbSNP database version), but they have been recently listed as intronic SNPs (build 130 of dbSNP). On the other hand, as for other nearby potential genes in the region where CEP68 is located, the RAB1A gene, despite the lack of reports showing direct relations with aspirin and/or respiratory disease, was also found to be associated with AIA by composing a strong LD with CEP68 (P<0.01; Figure 1).
Table 4. Intergenic SNPs with lowest P value for risk of aspirin intolerance in the first stage of GWAS.
| SNP ID | Variation | Build 126 | Build 130 | MAF | P value* | ||
| Position | Nearby Gene | AIA (n = 80) | ATA (n = 100) | ||||
| rs139719 | C>T | Intergenic | bA9F11.1 | SGSM1 | 0.268 | 0.475 | 1.6×10−6 |
| rs7744030 | C>T | Intergenic | ENPP4 | CLIC5 | 0.081 | 0.245 | 1.1×10−5 |
| rs7963956 | T>G | Intergenic | FAM19A2 | 0.311 | 0.151 | 4.0×10−5 | |
| rs4501026 | A>C | Intergenic | C21orf94 | 0.292 | 0.485 | 6.0×10−5 | |
| rs346416 | C>A | Intergenic | IPO11 | 0.268 | 0.440 | 7.3×10−5 | |
*P values of co-dominant model. Build 126 and 130 represent the versions of dbSNP.
MAF, minor allele frequency; AIA, aspirin-intolerant asthma; ATA, aspirin-tolerant asthma.
Figure 1. Other potential candidate gene nearby CEP68.
(A) Associations of other potential genes nearby CEP68. A significant association of SNPs in RAB1A is found from the result of this GWAS (P<0.01). (B) LD Plot between CEP68 and RAB1A. LD coefficient (D') among SNPs of SLC1A4, RAB1A and CEP68 in Korean population.
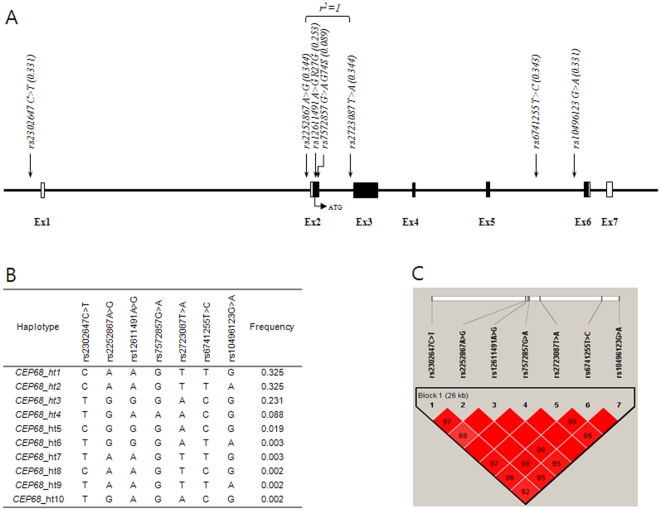
Pair-wise comparisons among SNPs of CEP68 gene showed tight LDs (Figure 2, Table S4). After haplotypes were inferred using PHASE software, results from logistic analyses for associations between haplotypes and aspirin intolerance showed that haplotype CEP68_ht4 (T-G-A-A-A-C-G) of AIA was more frequent than that of ATA as a control group, showing that the CEP68_ht4 with the nonsynonymous rs7572857 “A” allele at the fourth position was significantly associated with AIA in both the co-dominant (OR = 2.63; 95% CI = 1.64–4.21; P = 6.0×10−5, Table 3) and dominant models (OR = 2.49; 95% CI = 1.49–4.15; P = 0.0005; Table S1) when compared to that of ATA as a control group.
Figure 2. Physical Map, Haplotypes, and LD Plot of CEP68.
(A) Physical map of CEP68 and its targeted SNPs. (B) Haplotypes of CEP68. Associations of haplotypes with frequency >0.05 are shown in Table 3. (C) LD coefficient (D') among CEP68 SNPs in a Korean population.
Effect of rs7572857 on the Decline of FEV1 by Aspirin Provocation
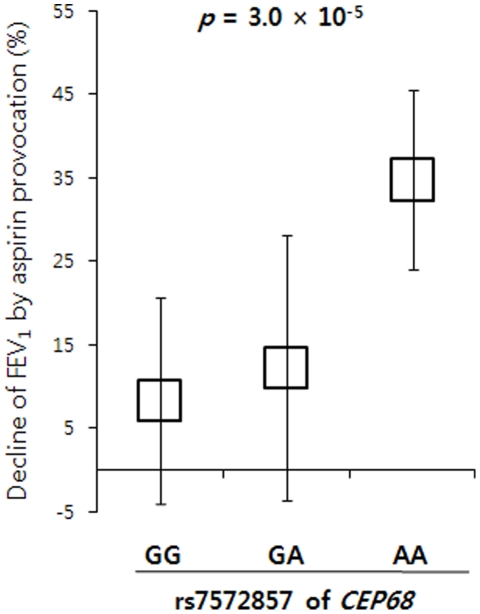
In regression analyses, the genotypes of several polymorphisms in the CEP68 gene were also significantly associated with the decline of FEV1 by aspirin provocation. Four SNPs, rs2302647, rs2252867, rs2723087, and rs6741255, showed a similar distribution pattern with about two-fold increase in the mean decline of FEV1 for the homozygous rare genotype compared to the homozygous common genotype (Table 5). In the case of rs7572857G>A, however, the nonsynonymous nucleotide substitution showed a different distribution with about a four-fold increase in the mean decline of FEV1 for the homozygote of the rare allele (Figure 3), indicating that rs7572857 could have a stronger influence on the higher decline of FEV1 by aspirin provocation than other variants. Despite our findings on the relationship between SNP genotypes and decline in FEV1 among AIA patients and then among ATA controls, results still showed that rs7572857 has an effect on AIA (P = 0.003; Table S5), but not on ATA.
Table 5. Association between genotypes of the CEP68 gene and the decline of FEV1 by aspirin provocation.
| SNP ID | C/C | C/R | R/R | P * | |||
| n | Decline of FEV1 (%) | n | Decline of FEV1 (%) | n | Decline of FEV1 (%) | ||
| rs2302647 C>T | 268 | 7.21±11.22 | 255 | 10.00±13.81 | 69 | 14.26±16.36 | 5.0×10−5 |
| rs2252867 A>G | 258 | 7.20±11.39 | 261 | 9.95±13.69 | 73 | 13.85±16.03 | 9.0×10−5 |
| rs12611491 A>G | 324 | 8.20±12.35 | 233 | 10.79±14.63 | 33 | 8.92±10.11 | 0.09 |
| rs7572857 G>A | 492 | 8.39±12.34 | 95 | 12.26±15.85 | 5 | 34.80±10.76 | 3.0×10−5 |
| rs2723087 T>A | 258 | 7.20±11.39 | 261 | 9.95±13.69 | 73 | 13.85±16.03 | 9.0×10−5 |
| rs6741255 T>C | 256 | 7.20±11.42 | 266 | 9.84±13.60 | 70 | 14.36±16.16 | 5.0×10−5 |
| rs10496123 G>A | 264 | 10.34±14.28 | 265 | 8.55±12.32 | 63 | 7.46±12.01 | 0.06 |
*P values of regression analyses represent the co-dominant model adjusted for age at initial diagnosis, sex, smoking status and atopy.
C/C, C/R and R/R indicate the homozygote of the common allele, and the heterozygote and homozygote of the rare allele, respectively.
Decline of FEV1 represents mean ± SE value.
Figure 3. Effect of rs7572857G>A in CEP68 on the Decline of FEV1 by Aspirin Provocation.
Distribution presents the mean ± SE values of % decline of FEV1 by aspirin provocation in each genotype of the nonsynonymous SNP (rs7572857G>A) in the CEP68 gene. The homozygote of the rare allele (AA) showed a significant increase in decline of FEV1.
Discussion
To our knowledge, this is the first genome-wide association study for AIA. Our results identified CEP68 as a positive risk factor for the development of aspirin intolerance in asthmatics. Two stages of genome-wide analysis and a follow-up study were used to reduce the number of false-positive SNPs and retain statistical power. In the present study, logistic and regression analyses yielded significant associations between variations of the CEP68 gene and AIA, with the lowest P value at the nonsynonymous SNP rs7572857. In addition, with a tight LD among the SNPs of the CEP68 gene, the haplotype CEP68_ht4 (T-G-A-A-A-C-G) in AIA subjects was more frequent when compared to that of ATA controls and was significantly associated with AIA. In particular, the fourth positioned “A” that was derived from rs7572857G>A was found only in haplotype CEP68_ht4. On the other hand, although we also do not rule out a possible protective effect of rs10496123G>A due to its decreased risk for AIA (OR = 0.81; Table 3), it is considered that haplotype CEP68_ht4, which includes most minor alleles of the significantly associated SNPs and the major G allele of rs10496123G>A, may mainly affect the association with AIA.
Although the inclusion of intermediate AIA in AIA case group also showed significant associations with the SNPs and haplotypes of CEP68 gene compared to ATA controls, a more correct association for genetic polymorphisms and aspirin intolerance is suggested to the comparison between AIA subjects without intermediate AIA group and ATA controls (Table 3). On the other hand, when observing the effects of polymorphisms of CEP68 on the decline of FEV1 by aspirin provocation, the homozygous AA of rs7572857G>A variant showed a different distribution and a more significant increase of FEV1 decline in AIA patients than other variants. In addition, five AIA patients were found to have the homozygote of this rare allele (AA), with 34.80% FEV1 decline; whereas no ATA controls were observed to have the AA genotype (Table S5). This observation suggests that the change from glycine to serine could have an effect on the higher decline of FEV1 by aspirin provocation.
More recently, there have been debates concerning whether GWAS can successfully detect the variants that are associated with diseases. However, GWAS not only has made it possible to predict risk factors that are associated with diseases, but also has discovered additional variants that are associated with many diseases [34], [35]. Although dense genome-wide genotyping chips containing over 600 K and as many as 1 M SNPs have been recently developed, this study used the earlier chip with about 100 K SNPs due to the starting date of our research. Significantly, ours is the first GWAS to identify genetic factors that might influence the occurrence of aspirin intolerance in asthmatics. On the other hand, while GWAS have found many common variants that have modest effects on human common diseases, albeit some may have a key role, whereas the systematic identification of rare variants that are too rare to be detected in the GWAS and confer a substantial risk of the diseases is also required [36]. In terms of our study's limitations, statistical power indicated that our GWAS contained an insufficient sample size. Statistical powers of 72% in the first GWAS stage and 52% after selecting 150 SNPs in the first stage were estimated by QpowR, an interactive power calculator for two stage association studies [37]. In addition, after Bonferroni correction for multiple comparisons, only two SNPs, rs1053744 in SBF1 and rs828616 in DCBLD2, were significantly associated with AIA. However, this study applied a modest threshold (P<0.0001) to select top 11 candidate genes because we focused on the SNPs within a gene region. Moreover, the follow-up study for 11 promising candidate genes increased the power to calculate the association between the CEP68 gene and aspirin intolerance in asthmatics.
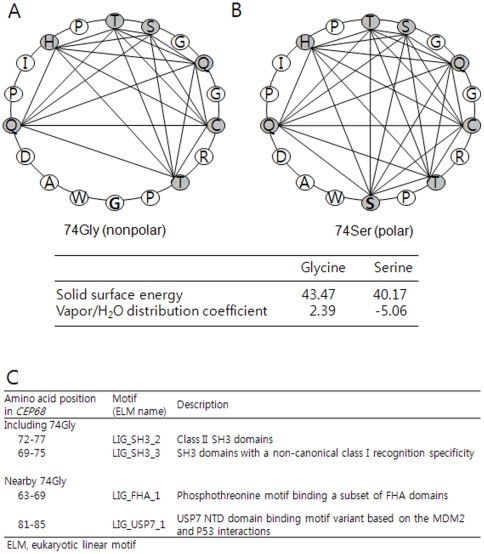
Since CEP68 gene (also referred to as KIAA0582) in the chromosome 2p14 has been recently discovered, its functions have not been fully understood, except for its role in centrosome cohesion and epidermal growth factor (EGF) signaling [38], [39]. In addition, given that smoke is a risk factor for asthma, CEP68 has been reported to be differentially expressed following exposure to environmental smoke [40]. However, the functions of the CEP68 protein are not yet fully understood. Thus, our findings provide a new insight into the relationship between CEP68 and aspirin intolerance in asthma. In particular, it is suggested that the Gly74Ser substitution in exon2 of the CEP68 gene could affect the polarity of the protein structure (Figure 4A), or affect the function of the CEP68 protein itself [41], [42]. Although in silico annotation showed that the nonsynonymous rs7572857G>A (Gly74Ser) appeared to be tolerable to diseases, this site was not highly conserved among mammals (Figure S1). Furthermore, in an additional functional site prediction, it was found that amino acids including 74Gly and its nearby positions could be targets for several kinases (Figure 4B). Since SH domains are especially unique sequences for several regulators, the change from glycine to serine might play an important role in correlations with other modulators. Therefore, phenotypic studies investigating the effects of minor A allele of rs7572857 on protein activity of CEP68 in relation to aspirin sensitivity are needed in the future.
Figure 4. Helical Wheel Projection and Properties between 74Gly and 74Ser, and Motif Search.
The helical wheel projection of Gly74Ser is presented by the Expert Protein Analysis System (ExPASy). The change from 74Gly (A) to 74Ser (B) showed an increase in polarity due to its interactions with nearby polar amino acids. The solid surface energy and the degree of vapor/H2O distribution of glycine and serine were compared. (C) Motifs including 74Gly and nearby 74Gly for the functional site were found using a prediction program (http://elm.eu.org/index.html).
Linkage disequilibrium, as a nonrandom association, has been considered to be profoundly associated with recombination hot spots, quantitative characters, and complex diseases [43]. In addition, experimental evidence has shown an association between haplotype blocks and hot spots of recombination [44]. When compared with other populations from HapMap, the asthmatic subjects in the Korean population showed a stronger LD from the selected CEP68 SNPs than other populations, with a pattern that was closest to that of the Chinese (Figure S2). Given the significant association of haplotype CEP68_ht4 with AIA, the LD of CEP68 might facilitate mapping out genetic factors that function in aspirin metabolism and/or aspirin hypersensitivity in asthma. Therefore, further replications in AIA patients from other populations are required to establish whether or not these structural variations affect AIA.
Other previous reports have suggested associations between genetic polymorphisms and AIA. In one study, 370 SNPs of 63 candidate genes that are involved in the arachidonic acid metabolic cascade were studied for associations with AIA. The results demonstrated that SNPs in the promoter region of the prostaglandin E2 receptor subtype 2 gene were significantly associated with AIA [15]. Several SNPs in the promoter of LTC4S [19], [45] and ALOX5 [20] that encode key enzymes for CysLT synthesis also showed significant associations with AIA [13], [46]. In addition, three promoter polymorphisms in the cysteinyl leukotriene receptor 1 gene were associated with AIA in males [46]. Moreover, polymorphisms of genes that are involved in the pathways of immune response and inflammation are also associated with aspirin hypersensitivity in asthma [21], [25]. This study also confirmed that several other genes, which were associated previously with AIA, showed significant association signals with AIA pathogenesis, especially in the TNF, TGF, HLA-DPB1, ALOX5 and IL-10 genes (Table S6). Despite the many studies that have shown positive associations, a comprehensive association between genetic risks and aspirin hypersensitivity has not yet been discovered. Furthermore, constant revisions to find new risk alleles are needed [35], [47]. Hence, our findings on the first genome-wide and follow-up studies of AIA could be a breakthrough in the discovery of susceptible genes using a different approach.
In conclusion, this study confers a positive association between CEP68 and aspirin-intolerant asthma, suggesting that these findings would be useful for further genetic investigations of NSAIDs metabolism as well as other inflammatory diseases. However, this study does have several limitations. First, this study included only the top 11 genes for the follow-up study due to research budget constraints. Second, our second stage replication analysis was insufficient due to the inclusion of samples from the first stage. In addition, AIA-I patients who were also referred to as positive responders to aspirin hypersensitivity [48], [49] were included in the second round. Although the significances were reduced in the further association analysis without the GWAS subjects due to the high proportion of AIA-I in the second round, the statistically significant association of the SNPs including nonsynonymous rs7572857G>A with AIA still remained (Table S2). Therefore, further replication studies in a larger number of subjects are also needed. Finally, functional studies of the nonsynonymous SNP rs7572857 in the future could provide important insights into the genetic etiology of aspirin intolerance in asthmatics.
Materials and Methods
Study Subjects
The subjects were recruited from Soonchunhyang University, Chung Ang University, Chung Nam University, Chungbuk University, and Seoul National University in Korea in a span of about 6 years (between 2003 and 2008). All subjects were Korean. All patients were diagnosed by a physician and met the definition of asthma set forth in the Global Initiative for Asthma guidelines [50]. All patients had a history of dyspnea and wheezing during the previous 12 months, plus one of the following: (1) >15% increase in FEV1 or >12% increase plus 200 mL following inhalation of a short-acting bronchodilator, (2) <10 mg/mL PC20 methacholine, and (3) >20% increase in FEV1 following 2 weeks of treatment with inhaled steroids and long-acting bronchodilators. Twenty-four common inhalant allergens were used for a skin prick test [26]. Atopy was defined as having a wheal reaction over 3 mm in diameter. Total IgE was measured by the CAP system (Pharmacia Diagnostics, Uppsala, Sweden). The asthmatic patients had experienced no exacerbation of asthma and respiratory tract infection in the 6 weeks preceding the oral aspirin challenge (OAC). The OAC was performed with increasing doses of aspirin using methods slightly modified from those described previously [26], [49]. Changes in FEV1 were followed for 5 hours after the last aspirin challenge dose. Aspirin-induced bronchospasms, as reflected by rate (%) of FEV1 decline, were calculated as the pre-challenge FEV1 minus the post-challenge FEV1 divided by the pre-challenge FEV1. OAC reactions were categorized into three groups as follows: Subjects with 20% or greater decreases in FEV1 or 15% to 19% decreases in FEV1 with naso-ocular or cutaneous reactions were designated as the AIA group, those with 15% to 19% decreases in FEV1 or naso-ocular or cutaneous reactions only were the AIA-I group, and those with less than 15% decreases in FEV1 without naso-ocular or cutaneous reactions were the ATA group. The distribution of FEV1 decline rate of subjects is shown in Figure S3. All subjects provided informed consent, and the protocols were approved by the Institutional Review Board of each hospital.
The First Stage of Genome-Wide SNP Genotyping
About 750 ng of genomic DNA from 80 AIA cases and 100 ATA controls was used to genotype each sample on the Illumina's Human-1 Genotyping BeadChip (Illumina, San Diego, USA). Genotyping was performed according to Illumina's assay manual. Briefly, each sample was processed by whole-genome amplification, fragmentation, precipitation, and resuspension in an appropriate hybridization buffer. Denatured samples were hybridized on the prepared Human-1 Genotyping BeadChip for 16 h at 48°C. Then, the processed beadchip for the single-base extension reaction was stained and imaged on an Illumina Bead Array Reader. To convert fluorescent intensities into SNP genotypes, normalized bead intensity data obtained for each sample were loaded into the Beadstudio 3.0® software (Illumina). SNP clusters for genotype calling of all SNPs were examined using Beadstudio 3.0® software. The overall call rate for all SNPs was 98.0%.
The Second Stage of Follow-up Study
Eleven genes that include the GWAS-driven top SNPs, which are positioned not at intergenic region but at the 5′-untranslated region (UTR), exons, introns, and the 3′-UTR region, were selected for the follow-up study. For the next study, a total of 150 common SNPs within 11 candidate genes, including nearby upstream region (1.5 kb) of each gene, (8 SNPs for the SBF1 gene; 12 for DCBLD2; 15 for WDR21A; 34 for FILIP1; 2 for PDZK3; 15 for LRRC43; 18 for CIITA; 5 for DAF; 7 for ENPP5; 7 for CEP68; 27 for C6, respectively) were genotyped using a total of 163 AIA cases including AIA-I subjects and 429 ATA controls. The SNPs were scanned using BeadExpress® (Illumina, San Diego, USA).
Statistics
For genome-wide analysis, associations of genotype distributions between AIA cases and ATA controls were calculated by logistic analyses. In addition to the genetic homogeneity of the study subjects as described above, a parameter (λ) of genomic control was calculated in this study by dividing median χ2 statistics by 0.456, with an estimate of 1.056 [51]. Power calculations of the first GWAS stage and after selecting candidate SNPs in the first stage were estimated using QpowR program (https://www.msu.edu/~steibelj/JP_files/QpowR.pdf). In the association analysis of the polymorphisms in CEP68, we examined Lewontin's D' (|D'|) and the LD coefficient r2 between all pairs of biallelic loci [52]. Haploview v4.1 software downloaded from the Broad Institute (http://www.broadinstitute.org/mpg/haploview) was used to determine LD of CEP68 [53]. Haplotypes were first estimated using the PHASE software [54], and then computed by logistic analyses using the Statistical Analysis System (SAS) program. Subjects harboring missing genotypes were omitted in the analysis of individual SNPs and haplotypes. Comparisons of genotype distributions between AIA and ATA were carried out with logistic analyses adjusted for initial diagnosed age, sex, smoking status, atopy, and body mass index as multivariates using SAS. The common (C) alleles were used as the referent genotype to the heterozygote and homozygote of the rare (R) allele. The effective numbers of independent marker loci in each gene were calculated to correct for multiple testing using the software SNPSpD (http://genepi.qimr.edu.au/general/daleN/SNPSpD/), which is based on the spectral decomposition (SpD) of matrices of pair-wise LD between SNPs. Significant associations were represented by P value <0.05.
Supporting Information
Comparison of co-dominant, dominant and recessive models for SNPs in the CEP68 gene between AIA and ATA.
(0.07 MB DOC)
Association analysis without GWAS samples in the second replication round analysis.
(0.05 MB DOC)
Logistic analyses of 150 SNPs in Top 11 genes between AIA and ATA.
(0.34 MB DOC)
LD coefficients (D' and r2) among CEP68 polymorphisms.
(0.03 MB DOC)
Relation of the SNP genotypes in CEP68 to the decline on FEV1 in AIA and in ATA.
(0.05 MB DOC)
List of other genes that were associated previously with AIA.
(0.05 MB DOC)
In silico annotation of nonsynonymous rs7572857G>A (Gly74Ser). Impact on protein function and conservation across species are predicted by the SNPs3D program (http://www.snps3d.org/). (A) The higher scores of entropy and Position Specific Scoring Matrix (PSSM) indicate more tolerable to diseases. (B) The 74th amino acid of CEP68 is not highly conserved among mammals.
(0.17 MB DOC)
Comparison of LD from other populations for the selected CEP68 SNPs. The LDs are constructed by seven CEP68 SNPs that are equivalent with this study from HapMap (http://hapmap.ncbi.nlm.nih.gov/index.html.en). CEU, Caucasian; CHB, Chinese, JPT, Japanese; YRI, African.
(0.12 MB DOC)
Distribution of FEV1 decline rate. Distributions are calculated from the decline rate of FEV1 by aspirin provocation and its number of subjects with 5% intervals. The distribution of AIA patients with FEV1 decline rate less than 15% is due to the responders to naso-ocular or cutaneous reactions.
(0.03 MB DOC)
Acknowledgments
The DNA samples were generously provided by the Soonchunhyang University, Bucheon Hospital Biobank, a member of the National Biobank of Korea, supported by the Ministry of Health, Welfare and Family Affairs, Republic of Korea.
Footnotes
Competing Interests: SNP Genetics, Inc. provided the Beadstation500 scanner instrument and BeadStudio 3.0 software used in the research. However, these competing interests do not alter the authors' adherence to all the PLoS ONE policies. No other relevant interests are declared.
Funding: Hyoung Doo Shin received grant number M1-0302-00-0073 from the Korea Science and Engineering Foundation (KOSEF) funded by the Korean government (MEST) (No. 2009-0080157), and a Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0093822). Choon-Sik Park received a grant from the Korea Health 21 R&D Project (A010249). SNP Genetics, Inc. provided the Beadstation500 scanner instrument and BeadStudio 3.0 software used in the research. They also were involved in the study design, data collection and analysis, decision to publish and preparation of the manuscript.
References
- 1.Widal F, Abrami P, Lermoyez J. Anaphylaxie et idiosyncrasie. 1992 [Anaphylaxis and idiosyncrasy. 1992]. Allergy Proc. 1993;14:373–376. [PubMed] [Google Scholar]
- 2.Babu KS, Salvi SS. Aspirin and asthma. Chest. 2000;118:1470–1476. doi: 10.1378/chest.118.5.1470. [DOI] [PubMed] [Google Scholar]
- 3.Szczeklik A, Stevenson DD. Aspirin-induced asthma: advances in pathogenesis, diagnosis, and management. J Allergy Clin Immunol. 2003;111:913–921. doi: 10.1067/mai.2003.1487. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski ML. Aspirin-sensitive rhinosinusitis and asthma. Clin Allergy Immunol. 2007;19:147–175. [PubMed] [Google Scholar]
- 5.Palikhe NS, Kim SH, Park HS. What do we know about the genetics of aspirin intolerance? J Clin Pharm Ther. 2008;33:465–472. doi: 10.1111/j.1365-2710.2008.00961.x. [DOI] [PubMed] [Google Scholar]
- 6.Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J Allergy Clin Immunol. 2009;124:422–427. doi: 10.1016/j.jaci.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duroudier NP, Tulah AS, Sayers I. Leukotriene pathway genetics and pharmacogenetics in allergy. Allergy. 2009;64:823–839. doi: 10.1111/j.1398-9995.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 8.Drazen JM, Israel E, O'Byrne PM. Treatment of asthma with drugs modifying the leukotriene pathway. N Engl J Med. 1999;340:197–206. doi: 10.1056/NEJM199901213400306. [DOI] [PubMed] [Google Scholar]
- 9.Leff AR. Regulation of leukotrienes in the management of asthma: biology and clinical therapy. Annu Rev Med. 2001;52:1–14. doi: 10.1146/annurev.med.52.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Park JS, Jang AS, Park SW, Lee YM, Uh ST, et al. Protection of leukotriene receptor antagonist against aspirin-induced bronchospasm in asthmatics. Allergy Asthma Immunol Res. 2010;2:48–54. doi: 10.4168/aair.2010.2.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlen SE, Malmstrom K, Nizankowska E, Dahlen B, Kuna P, et al. Improvement of aspirin-intolerant asthma by montelukast, a leukotriene antagonist: a randomized, double-blind, placebo-controlled trial. Am J Respir Crit Care Med. 2002;165:9–14. doi: 10.1164/ajrccm.165.1.2010080. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Ye YM, Hur GY, Lee SK, Sampson AP, et al. CysLTR1 promoter polymorphism and requirement for leukotriene receptor antagonist in aspirin-intolerant asthma patients. Pharmacogenomics. 2007;8:1143–1150. doi: 10.2217/14622416.8.9.1143. [DOI] [PubMed] [Google Scholar]
- 13.Park JS, Chang HS, Park CS, Lee JH, Lee YM, et al. Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet Genomics. 2005;15:483–492. doi: 10.1097/01.fpc.0000166456.84905.a0. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Choi JH, Park HS, Holloway JW, Lee SK, et al. Association of thromboxane A2 receptor gene polymorphism with the phenotype of acetyl salicylic acid-intolerant asthma. Clin Exp Allergy. 2005;35:585–590. doi: 10.1111/j.1365-2222.2005.02220.x. [DOI] [PubMed] [Google Scholar]
- 15.Jinnai N, Sakagami T, Sekigawa T, Kakihara M, Nakajima T, et al. Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach. Hum Mol Genet. 2004;13:3203–3217. doi: 10.1093/hmg/ddh332. [DOI] [PubMed] [Google Scholar]
- 16.Sanak M, Simon HU, Szczeklik A. Leukotriene C4 synthase promoter polymorphism and risk of aspirin-induced asthma. Lancet. 1997;350:1599–1600. doi: 10.1016/s0140-6736(05)64015-9. [DOI] [PubMed] [Google Scholar]
- 17.Van Sambeek R, Stevenson DD, Baldasaro M, Lam BK, Zhao J, et al. 5′ flanking region polymorphism of the gene encoding leukotriene C4 synthase does not correlate with the aspirin-intolerant asthma phenotype in the United States. J Allergy Clin Immunol. 2000;106:72–76. doi: 10.1067/mai.2000.107603. [DOI] [PubMed] [Google Scholar]
- 18.Sanak M, Szczeklik A. Leukotriene C4 synthase polymorphism and aspirin-induced asthma. J Allergy Clin Immunol. 2001;107:561–562. doi: 10.1067/mai.2001.112853. [DOI] [PubMed] [Google Scholar]
- 19.Choi JH, Park HS, Oh HB, Lee JH, Suh YJ, et al. Leukotriene-related gene polymorphisms in ASA-intolerant asthma: an association with a haplotype of 5-lipoxygenase. Hum Genet. 2004;114:337–344. doi: 10.1007/s00439-004-1082-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Bae JS, Suh CH, Nahm DH, Holloway JW, et al. Polymorphism of tandem repeat in promoter of 5-lipoxygenase in ASA-intolerant asthma: a positive association with airway hyperresponsiveness. Allergy. 2005;60:760–765. doi: 10.1111/j.1398-9995.2005.00780.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim SH, Ye YM, Lee SK, Choi JH, Holloway JW, et al. Association of TNF-alpha genetic polymorphism with HLA DPB1*0301. Clin Exp Allergy. 2006;36:1247–1253. doi: 10.1111/j.1365-2222.2006.02567.x. [DOI] [PubMed] [Google Scholar]
- 22.Dekker JW, Nizankowska E, Schmitz-Schumann M, Pile K, Bochenek G, et al. Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin Exp Allergy. 1997;27:574–577. [PubMed] [Google Scholar]
- 23.Choi JH, Lee KW, Oh HB, Lee KJ, Suh YJ, et al. HLA association in aspirin-intolerant asthma: DPB1*0301 as a strong marker in a Korean population. J Allergy Clin Immunol. 2004;113:562–564. doi: 10.1016/j.jaci.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Park HS, Kim SH, Sampson AP, Lee KW, Park CS. The HLA-DPB1*0301 marker might predict the requirement for leukotriene receptor antagonist in patients with aspirin-intolerant asthma. J Allergy Clin Immunol. 2004;114:688–689. doi: 10.1016/j.jaci.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 25.Akahoshi M, Obara K, Hirota T, Matsuda A, Hasegawa K, et al. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet. 2005;117:16–26. doi: 10.1007/s00439-005-1285-0. [DOI] [PubMed] [Google Scholar]
- 26.Kim TH, Chang HS, Park SM, Nam BY, Park JS, et al. Association of angiotensin I-converting enzyme gene polymorphisms with aspirin intolerance in asthmatics. Clin Exp Allergy. 2008;38:1727–1737. doi: 10.1111/j.1365-2222.2008.03082.x. [DOI] [PubMed] [Google Scholar]
- 27.Kim SH, Yang EM, Lee HN, Cho BY, Ye YM, et al. Combined effect of IL-10 and TGF-beta1 promoter polymorphisms as a risk factor for aspirin-intolerant asthma and rhinosinusitis. Allergy. 2009;64:1221–1225. doi: 10.1111/j.1398-9995.2009.01989.x. [DOI] [PubMed] [Google Scholar]
- 28.Hitomi Y, Ebisawa M, Tomikawa M, Imai T, Komata T, et al. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J Allergy Clin Immunol. 2009;124:779–785 e776. doi: 10.1016/j.jaci.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 30.Roses AD. Pharmacogenetics in drug discovery and development: a translational perspective. Nat Rev Drug Discov. 2008;7:807–817. doi: 10.1038/nrd2593. [DOI] [PubMed] [Google Scholar]
- 31.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 32.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 35.Kraft P, Hunter DJ. Genetic risk prediction–are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 36.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 37.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian asian men. PLoS One(In Press) 2010 doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Low TY, Choong LY, Ray RS, Tan YL, et al. Phosphoproteomics identified Endofin, DCBLD2, and KIAA0582 as novel tyrosine phosphorylation targets of EGF signaling and Iressa in human cancer cells. Proteomics. 2007;7:2384–2397. doi: 10.1002/pmic.200600968. [DOI] [PubMed] [Google Scholar]
- 40.Rouse RL, Boudreaux MJ, Penn AL. In utero environmental tobacco smoke exposure alters gene expression in lungs of adult BALB/c mice. Environ Health Perspect. 2007;115:1757–1766. doi: 10.1289/ehp.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinshilboum R, Wang L. Pharmacogenomics: bench to bedside. Nat Rev Drug Discov. 2004;3:739–748. doi: 10.1038/nrd1497. [DOI] [PubMed] [Google Scholar]
- 42.Maheshwari R, Dhathathreyan A. Investigation of surface properties of amino acids: polarity scale for amino acids as a means to predict surface exposed residues in films of proteins. J Colloid Interface Sci. 2004;277:79–83. doi: 10.1016/j.jcis.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 43.Slatkin M. Linkage disequilibrium–understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 45.Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, et al. Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirin-induced asthma. J Allergy Clin Immunol. 2002;109:936–942. doi: 10.1067/mai.2002.124466. [DOI] [PubMed] [Google Scholar]
- 46.Kim SH, Oh JM, Kim YS, Palmer LJ, Suh CH, et al. Cysteinyl leukotriene receptor 1 promoter polymorphism is associated with aspirin-intolerant asthma in males. Clin Exp Allergy. 2006;36:433–439. doi: 10.1111/j.1365-2222.2006.02457.x. [DOI] [PubMed] [Google Scholar]
- 47.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson DD. Aspirin-sensitive Asthma. In: Barnes PJ, Grunstein MM, Leff AR, Woolcock AJ, editors. Asthma. New York: Lippincott-Raven; 1997. pp. 2025–2040. [Google Scholar]
- 49.Cormican LJ, Farooque S, Altmann DR, Lee TH. Improvements in an oral aspirin challenge protocol for the diagnosis of aspirin hypersensitivity. Clin Exp Allergy. 2005;35:717–722. doi: 10.1111/j.1365-2222.2005.02261.x. [DOI] [PubMed] [Google Scholar]
- 50.Global Initiative for Asthma (GINA) Global strategy for asthma management and prevention. NHLBI/WHO workshop report. Bethesda, MD: National Institutes of Health, National Heart, Lung, and Blood Institute, 1995, NIH publication, 1995; pp. 95–3659. [Google Scholar]
- 51.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 52.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 54.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of co-dominant, dominant and recessive models for SNPs in the CEP68 gene between AIA and ATA.
(0.07 MB DOC)
Association analysis without GWAS samples in the second replication round analysis.
(0.05 MB DOC)
Logistic analyses of 150 SNPs in Top 11 genes between AIA and ATA.
(0.34 MB DOC)
LD coefficients (D' and r2) among CEP68 polymorphisms.
(0.03 MB DOC)
Relation of the SNP genotypes in CEP68 to the decline on FEV1 in AIA and in ATA.
(0.05 MB DOC)
List of other genes that were associated previously with AIA.
(0.05 MB DOC)
In silico annotation of nonsynonymous rs7572857G>A (Gly74Ser). Impact on protein function and conservation across species are predicted by the SNPs3D program (http://www.snps3d.org/). (A) The higher scores of entropy and Position Specific Scoring Matrix (PSSM) indicate more tolerable to diseases. (B) The 74th amino acid of CEP68 is not highly conserved among mammals.
(0.17 MB DOC)
Comparison of LD from other populations for the selected CEP68 SNPs. The LDs are constructed by seven CEP68 SNPs that are equivalent with this study from HapMap (http://hapmap.ncbi.nlm.nih.gov/index.html.en). CEU, Caucasian; CHB, Chinese, JPT, Japanese; YRI, African.
(0.12 MB DOC)
Distribution of FEV1 decline rate. Distributions are calculated from the decline rate of FEV1 by aspirin provocation and its number of subjects with 5% intervals. The distribution of AIA patients with FEV1 decline rate less than 15% is due to the responders to naso-ocular or cutaneous reactions.
(0.03 MB DOC)