Abstract
The initial photochemistry of plant cryptochromes has been extensively investigated in recent years. It is hypothesized that cryptochrome photoexcitation involves a Trp-triad-dependent photoreduction. According to this hypothesis, cryptochromes in the resting state contain oxidized FAD; light triggers a sequential electron transfer from three tryptophan residues to reduce FAD to a neutral semiquinone (FADH•); FADH• is the presumed signaling state and it is re-oxidized to complete the photocycle. However, this photoreduction hypothesis is currently under debate. An alternative model argues that the initial photochemistry of cryptochromes involves a photolyase-like cyclic electron shuttle without a bona fide redox reaction mediated by the Trp-triad residues, leading to conformational changes, signal propagation, and physiological responses.
Introduction
Cryptochromes are photolyase-like proteins that regulate photomorphogenic development in plants and the circadian clock in plants and animals [1–3]. The photolyase/cryptochrome superfamily is classified into five families: CPD (cyclobutane pyrimidine photolyase) photolyase, 6–4 photolyase, CRY-DASH (probably a single-stranded DNA repairing enzyme), plant cryptochromes, and animal cryptochromes [4]. The animal cryptochromes are further divided into two types. Animal type 1 cryptochromes, such as Drosophila dCRY and monarch butterfly CRY1, act as photoreceptors, whereas animal type 2 cryptochromes, such as mouse cryptochromes, human cryptochromes, and monarch butterfly CRY2, act as light-independent transcription repressors [5,6]. All plant cryptochromes examined so far act as photoreceptors and they are best studied in Arabidopsis. The Arabidopsis genome encodes three cryptochromes, CRY1, CRY2, and CRY3. CRY1 and CRY2 primarily mediate light stimulation of de-etiolation and photoperiodic control of floral initiation, respectively, whereas CRY3 may act as a single-stranded DNA photolyases in mitochondria or plastids [7–11]. Cryptochromes are composed of two domains, the N-terminal PHR (Photolyase-Homologous Region) domain of about 500 residues, and the C-terminal extension CCE (Cryptochrome C-Terminal Extension) of various lengths. The PHR domain of cryptochromes bind non-covalently to the chromophore flavin adenine dinucleotide (FAD) and possibly a second chromophore, 5,10-methenyltetrahydrofolate (MTHF) [12•–14•]. The CCE domains of Arabidopsis CRY1 and CRY2 are approximately 180 and 110 residues in length, respectively, and they appear to be intrinsically unstructured, but to act as an effector domain [15–19].
Although the light-dependent catalytic mechanism of DNA photolyase, the presumed ancestor of cryptochromes, is well understood [2], the signaling process of cryptochromes, especially their photoexcitation mechanism, remains not fully understood. A cryptochrome that absorbs a photon is expected to be “photoexcited”, by which an electron of the chromophore molecule is promoted from the ground state orbital to a higher energy orbital. It is generally agreed that photoexcited cryptochromes undergo a series of biophysical and biochemical changes, including structural rearrangement, protein modifications, protein-protein interactions, and a chain of signal transduction events that leads to changes in gene expression and physiological responses of plants. However, it is not clear whether and how a light-dependent redox reaction or the photoreduction cycle is involved in the cryptochrome photoexcitation in vivo. This article attempts to discuss recent studies concerning photoexcitation mechanism of cryptochromes and different opinions about our current view of the initial phase of cryptochrome signal transduction.
The cryptochrome photoreduction
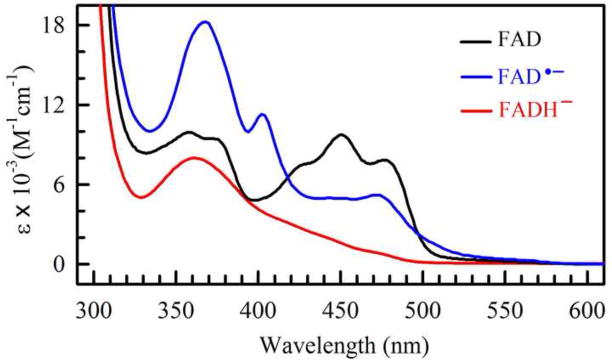
Under physiological conditions, flavins are usually synthesized in the oxidized state. But flavin bound to proteins may exist in any one of the three redox states in five different forms: oxidized (e.g. FAD), semireduced semiquinones (e.g. neutral radical FADH• or anion radical FAD•−), or fully reduced hydroquinones (e.g. FADH− or FADH2) [20•] (Fig. 1A). It is often presumed that a light-dependent flavoprotein undergoes a photocycle or photon-driven redox exchanges between the flavin chromophore and its protein environment. A possible photocycle of flavin can be analyzed in vitro by monitoring changes of absorption spectra of a purified flavoprotein, because flavoproteins of different redox states have different absorption spectra. For example, cryptochromes/photolyases containing oxidized FAD or anion radical semiquinones (FAD• −) absorb primarily UV-A and blue light; that containing a neutral radical semiquinone (FADH•) has a broad absorption spectrum extending from blue to green and red light, whereas that containing fully reduced flavin (FADH− or FADH2) absorbs little visible light (Fig. 1B). The fact that Arabidopsis cryptochromes mediate blue light responses in plants argues that oxidized flavin, which has the strongest absorption in blue regions of light, may be the resting state chromophore of these photoreceptors.
Fig. 1. Oxidoreduction of flavins.
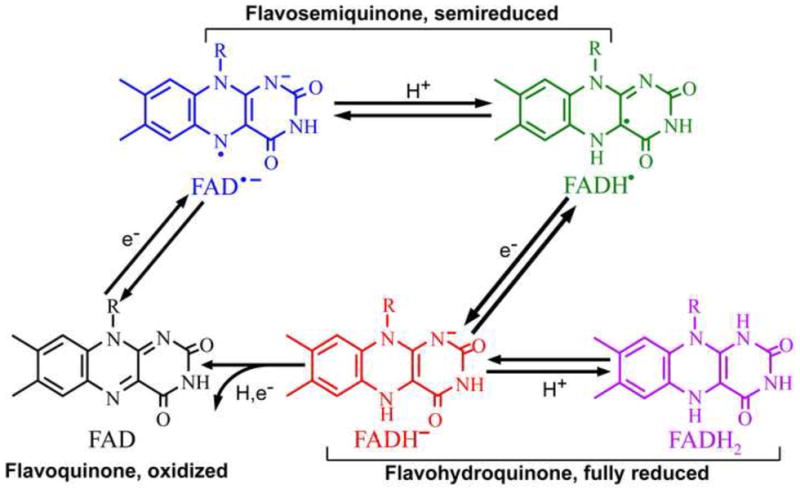
A. Five possible redox forms of flavins are shown. R indicates different side groups in different flavins. The two different forms of semiquinone radicals: anion radical (e.g. FAD• −) and neutral radical (e.g. FADH•), and two forms of reduced flavins: protonated hydroquinone (e.g. FADH2) and anionic hydroquinone (e.g. FADH−) are shown.
B. Absorption spectra and extinction coefficients (ε) of different redox forms of selected cryptochrome/photolyase [4]. Mosquito AgCRY1 (Anopheles gambiae) containing oxidized FAD (black line) or anion radical semiquinone (FAD• −, blue line), and E. coli photolyase containing neutral radical semiquinone (FADH•, green line) or fully reduced flavin (FADH−, red line). The absorption spectra of Arabidopsis cryptochromes with the respective redox forms of flavin are similar to those shown here.
Like many other flavoproteins, but in striking difference to most photolyases studied, Arabidopsis CRY1 expressed and purified from insect cells contains oxidized FAD [12•,13,21•,22]. Under anaerobic conditions, the FAD of CRY1 can be rapidly (within 2 min) photoreduced in vitro to the semireduced flavin (FADH• or FAD• −), which absorb blue and/or green lights to become fully reduced (FADH2 or FADH−) [12•,21•]. Photoreduction has also been reported for Arabidopsis CRY2 under an aerobic condition [14•]. Although CRY2 was reported to be photoreduced to only the semireduced form under aerobic condition [14•], it can be reduced, just like CRY1, to the fully reduced form under anaerobic conditions (Zhong and Lin, unpublished data). Because in vitro photoreduction is common for flavoproteins regardless of whether their function is dependent on light, it is particularly important to understand whether the photoreduction observed in vitro is physiologically relevant to the functions of cryptochrome photoreceptors in vivo.
The physiological relevance of the cryptochrome photoreduction observed in vitro was initially tested for the hypocotyl inhibition response in seedlings grown under blue, green, or UV-A light, roughly corresponding to the characteristic absorption spectra of oxidized (FAD), semireduced (FADH•), and fully reduced (FADH− or FADH2) form of CRY1 observed in vitro [8,12•,23] (Fig. 1B). When grown under continuous broadband fluorescent blue light (~410nm–520nm), green light (~480nm–580nm) or UVA-light (~300–400nm), the wild type Arabidopsis and tobacco seedlings showed fluence rate-dependent inhibition of hypocotyl elongation in all three wavelengths tested, with the apparent effectiveness of blue>UV-A>green light. The transgenic Arabidopsis and tobacco seedlings overexpressing CRY1 showed a hypersensitive growth inhibition under all three wavelengths tested [8,12•,23]. The light-dependent degradation of Arabidopsis CRY2 was also found to occur not only in blue light, but also in broadband green and UV-A light [24]. Although these earlier experiments failed to demonstrate a different physiological activities of different redox species of Arabidopsis cryptochromes, it was later found that the broadband green light has both stimulatory and inhibitory effect on the cryptochrome-dependent blue light inhibition of hypocotyl elongation, and that cryptochromes may act as sensors for the blue/green ratio in nature [25].
Given that the photoreduction of Arabidopsis CRY1 in vitro produces semireduced neutral radical semiquinone (FADH•) that absorbs green light [12•], it is conceivable that a semireduced flavin may represent the signaling state of CRY1. It was reasoned that if the semireduced and green-light-absorbing CRY1 was the physiologically active form, illumination of seedlings with bichromatic green and blue lights might shift the equilibrium toward a decreased level of oxidized and semireduced CRY1 and an increased level of fully reduced CRY1 to undermine the CRY1 function [21•]. Indeed, supplement of green light to blue light partially suppressed blue light inhibition of hypocotyl elongation and blue light stimulation of anthocyanin accumulation in Arabidopsis seedlings, but no such green light effect was observed in the cry1 mutant [21•]. In addition to the CRY1-mediated responses, the antagonistic effect of green light to blue light was also shown for blue light-induced CRY2 degradation and CRY2-mediated blue light promotion of flowering in SD photoperiods [14•,21•]. In one of these experiments, it was shown that CRY2 was degraded to almost completion after eight cycles of 3-min blue light exposure followed by a 3-min dark period [21•]. In contrast, little CRY2 degradation was detected after eight cycles of 3-min blue light exposure followed by a 3-min green light pulse or eight cycles of green light treatment [21•]. However, in contrast to the far-red light reversion of most phytochrome-mediated red light responses, the antagonistic effect of green light appears relatively weak for the cryptochrome-mediated blue light responses tested [14•,21•](Yang and Lin, unpublished). Nevertheless, a photoreduction cycle was proposed for Arabidopsis cryptochromes based on these results [14•,21•]. According to this photoreduction hypothesis, cryptochromes in the dark (resting state) contains oxidized FAD, which is reduced to semireduced FDAH• upon blue light absorption. As a major photoexcitation product, the semireduced FDAH• represents the signaling state of cryptochromes, which can be oxidized to regenerate the resting state oxidized flavin via dark conversion to complete the photocycle [14•,21•]. Provided the photoreduction was involved in the initial photoexcitation of cryptochromes, what would be the electron donor for the photoreduction of flavin in cryptochromes?
The Trp-triad paradox
To address this question, researchers looked once again to photolyase for possible clues. Ironically, an answer derived from that of photolyases – three conserved tryptophan residues, referred to as Trp-triad, help transfer an electron to reduce flavin – appears to also bring skepticism to the presumed answer of the original question – whether photoreduction is the photochemical mechanism of cryptochrome photoreceptors. To better understand this paradox, one should first take a closer look at the two different photochemical reactions of photolyase. The catalytic mechanism of photolyase is peculiar in that it involves a light-dependent cyclic electron shuttle without a net gain or loss of electron [2,26]. Taking E. coli photolyase as an example, our current understanding of the photolyase catalysis may be summarized in the following. First of all, E. coli photolyase possesses fully reduced (FADH−) in vivo and uses it as the catalytically active co-factor [2,26,27••]. Photolyase binds the cyclobutane pyrimidine dimer of DNA in a light-independent manner and flips the dimer out of helix into the active site or the FAD-cavity to make contact with the flavin, producing a stable enzyme-substrate complex. Second, exposure of the photolyase-DNA complex to light initiates catalysis. In this light-dependent catalytic process, the methenyltetrahydrofolate (MTHF) chromophore (or the other type of antenna chromophore, 8-hydroxy-7,8-didemethyl-5-deazariboflavin in some other photolyases) absorbs a photon and transfers the excitation energy to the reduced flavin chromophore (FADH−) by Förster-type resonance energy transfer. The excited flavin (FADH−)* then transfers an electron to the pyrimidine dimer, generating a pyrimidine dimer radical, which undergoes bond rearrangement and breakage to yield a repaired DNA product. Before the repaired DNA dissociates from the enzyme, it transfer an electron back to the flavin-neutral radical (FADH•) to replenish the electron lost during catalysis, resulting in regeneration of the fully reduced flavin (FADH−). Because there is no net gain or loss of an electron during catalysis, photolyase catalysis is not a redox reaction per se.
Although E. coli photolyase contains fully reduced (FADH−) in vivo, it is oxidized during purification to contain primarily the semireduced neutral radical (FADH•). The purified photolyase can be photoreduced in vitro to restore the catalytically active FADH−. This in vitro phenomenon is called “pre-illumination effect” or “photoactivation”, which is distinct from the light-dependent catalysis of photolyases described above. The primary electron donor for the in vitro photoreduction/photoactivation of E. coli photolyase was found to be tryptophan 306 (W306) [28•]. The W306F mutant enzyme is incapable of photoreduction or photoactivation in vitro, but it is active in vivo and it can be chemically reduced by high concentrations of reducing agents, such as dithiothreitol (DTT), to become catalytically active in vitro [2,28•]. As the primary electron donor, W306 was expected to be in the vicinity of FAD. Surprisingly, when the crystal structure of E. coli photolyase was solved, W306 was found to locate on the surface of the enzyme, 13.4 Å away from the flavin sitting in the FAD-cavity [27••]. It was later shown by analyses of the time-resolved absorption spectroscopy and the X-ray structure of E. coli photolyase that W306 transfers an electron by three steps through two other tryptophan residues (W359 and W382) located between W306 and flavin [29]. These three tryptophan residues (W306, W359, and W382), referred to as Trp-triad (Fig. 2), are clearly required for the photoreduction of E. coli photolyase in vitro, because site-specific mutations of the tryptophan residues of Trp-triad abolished in vitro photoreduction and photoactivation of the mutant enzyme. However, none of the site-specific mutations of the three tryptophan residues of Trp-triad abolished the photolyase activity in vivo [28•,30]. Clearly, Trp-triad is not required for the photolyase function in vivo, and photoreduction is physiologically irrelevant for E. coli photolyase [2,26]. Consistent with this conclusion, an examination of the cellular redox environment of E. coli and the midpoint redox potential of E. coli photolyase indicate that E. coli photolyase needs no photoreduction to reduce its flavin in vivo. The cellular redox potential of E. coli is about −270mV [31,32], whereas the midpoint redox potential of the FADH•/FADH2 (or FADH•/FADH−) redox couple of E. coli photolyase is between 16mV to −48mV [33,34]. Therefore, E. coli photolyase can be readily reduced in vivo, presumably by cellular reductants such as glutathione or thioredoxin, without a need of photoreduction.
Fig. 2. The Trp-triad of photolyases and cryptochromes.
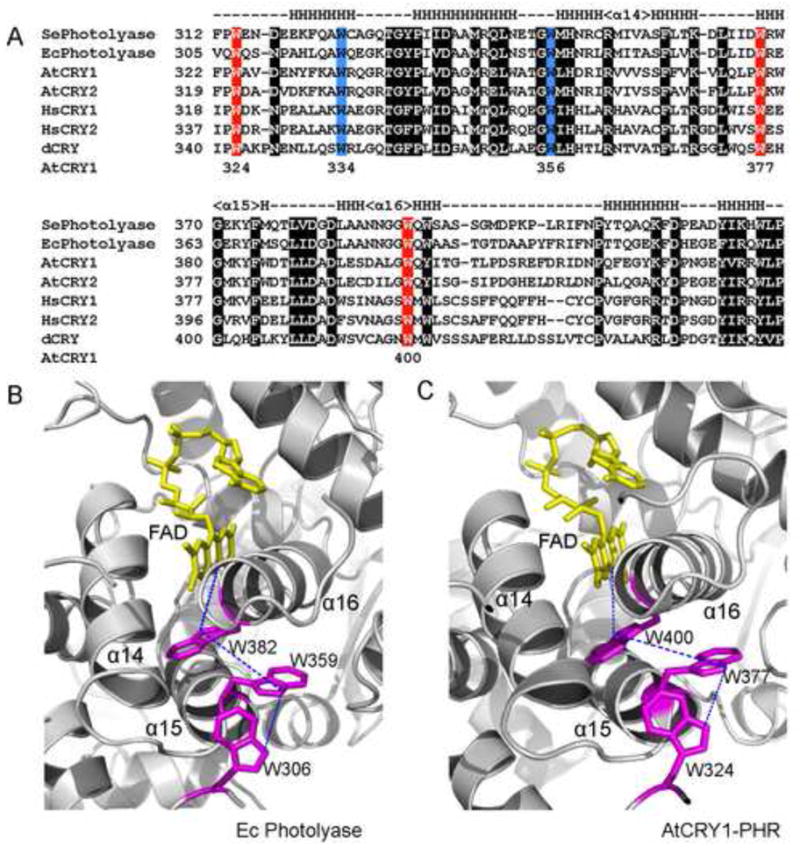
A. A partial sequence alignment of the region of photolyases and cryptochromes highly conserved and important for FAD-binding. Synechococcus elongatus (SePhotolyse) and Escherichia coli photolyases (Ecphotolyase), Arapidopsis cryptochrome 1 and 2 (AtCRY1, AtCRY2), human cryptochrome 1 and 2 (HsCRY1, HsCRY2), and Drosophila cryptochrome (dCRY) are compared. Symbols above indicate secondary structures, with dashed-lines representing loops and “H” representing alpha-helices. Numbers beneath indicate positions of the tryptophan residues of Arabidopsis CRY1. The three tryptophan residues of the Trp-triad are highlighted in red, the two tryptophan residues not part of Trp-triad are highlighted in blue.
B–C. The structure of FAD–binding pocket, FAD (yellow), the tryptophan residues of the Trp-triad (pink), and the path of electron movement from Trp-triad to FAD by photoreduction (blue dashed-line) of E. coli photolyase (B) and the PHR domain of Arabidopsis CRY1 (C) are indicated by blue dashed lines.
Paradoxically, although the Trp-triad-dependent photoreduction seems biochemically unnecessary and physiologically irrelevant to the catalytic activity of photolyases, the three tryptophan residues of Trp-triad are uniformly conserved in all photolyases and cryptochromes examined [29] (Fig. 2). The conserved Trp-triad was proposed to be the electron donor for photoreduction not only of photolyases, but also of cryptochromes [29]. A spectroscopic analysis suggests that tryptophan or tyrosine residues may be the electron donors for the in vitro photoreduction of Arabidopsis CRY1 [35]. Sequence alignment and structure comparison of photolyases and cryptochromes suggest that W324, W377, and W400 are the Trp-triad of Arabidopsis CRY1 [29] (Fig. 2). Possible involvements for two of those three tryptophan residues in CRY1 photoreduction in vitro and physiological activities in vivo have been tested [22]. Two site-specific mutant proteins, CRY1W324F and CRY1W400F, in which the tryptophan residues (W324 and W400) were replaced with phenylalanine, were expressed and purified from insect cells. Because phenylalanine is redox-inert but structurally similar to tryptophan, the CRY1W324F and CRY1W400F were expected to abolish photoreduction with minimum structural disturbances. As expected, the CRY1W324F and CRY1W400F mutant proteins failed to undergo photoreduction in vitro, confirming that the Trp-triad is required for the in vitro photoreduction of CRY1. Moreover, transgenic plants overexpressing CRY1W324F and CRY1W400F failed to rescue the cry1 mutant phenotypes such as hypocotyl inhibition and anthocyanin synthesis, suggesting that CRY1W324F and CRY1W400F were physiologically inactive [22]. It has been recently reported that human hCRY1 expressed in insect cells also exhibits a Trp-triad-dependent photocycle and light-dependent degradation similar to that of known photoreceptors, such as Arabidopsis CRY2 or Drosophila dCRY [36]. Although human cryptochromes, like mouse cryptochromes, are generally considered to be a photo-insensitive type 2 cryptochromes that act as the transcription suppressors of the central oscillators [37–40], it remains possible that they may also act as photoreceptors in the ganglion cells [41–43••]. These results are consistent with the hypothesis that photoreduction is associated with the function of cryptochromes and that Trp-triad is the electron donor(s) of cryptochrome photoreduction.
However, whether the Trp-triad-dependent photoreduction is directly associated with cryptochrome photoexcitation in vivo has been questioned. First, although the Trp-triad is conserved in all cryptochromes and the three tryptophan residues are required for the in vitro photoreduction of all photolyases and cryptochromes tested, the physiological relevance of the Trp-triad has so far been reported only for Arabidopsis CRY1. For example, the Trp-triad mutations have been prepared and tested for several insect type 1 cryptochromes (Drosophila, monarch butterfly, mosquito, and silk moth) that act as photoreceptors. All those Trp-triad mutations abolished in vitro photoreduction of the purified proteins, but none of those Trp-triad mutations abolished the in vivo physiological or photobiochemical activity of the respective cryptochromes tested, including light-dependent magnetoreception, light-dependent transcription regulation, and light-induced proteolysis [44–47]. For example, it has been shown recently that the Trp-triad-dependent photoreduction is not required for the photoentrainment of the circadian clock or blue light-dependent magnetoreception of insect cryptochromes [44]. Similarly, we have recently found that the Trp-triad-dependent photoreduction is not required for the in vivo photobiochemical or physiological activities of Arabidopsis CRY2 (Yu and Lin, unpublished). Therefore, similar to photolyase, the Trp-triad-dependent photoreduction is most likely not required for the functions of both animal and plant cryptochromes. Second, the high resolution time-resolved spectroscopic studies of photolyases and the type 1 photo-sensitive cryptochromes suggested a photolyase-like photochemistry of the insect photoreceptors [20•,48,49]. It was hypothesized that the resting state of the flavin cofactor is semireduced flavin, such as FAD•−, in insect cryptochromes. Upon photoexcitation, the excited FAD• − donates one electron to an unidentified acceptor to induce a conformational change through the primary electron transfer. Although the fate of the transferred electron remains unclear at present, this model suggests that the insect type 1 cryptochromes may have the photolyase-like photocycle that do not require the Trp-triad-dependent photoreduction for in vivo function [20•,48,49]. Third, the positive correlation of the abolished photoreduction activity in vitro and physiological activity in vivo of the CRY1W324F and CRY1W400F mutations appears insufficient to establish a causal relationship between the two activities, because it cannot exclude a possibility that the two mutant proteins may be structurally compromised. The three tryptophan residues of Trp-triad located in the structurally important FAD-binding pocket of photolyases and cryptochromes are not the only residues conserved in this highly conserved region. In addition to the three Trp-triad residues of the Trp-triad, two other tryptophan residues in this region are also perfectly conserved in proteins from E. coli photolyase to cryptochromes in Arabidopsis, Drosophila, and human (Fig. 2A). The possibility that Trp-triad may be structurally important but not photochemically critical to cryptochromes can be illustrated by a study of two frog (Xenopus laevis) cryptochromes, xCRY1 and xCRY2b [50•]. Xenopus xCRY1 and xCRY2b are photo-insensitive type 2 animal cryptochromes that showed similar activity suppressing the bMAL1/CLK-dependent transcription of the reporter gene in the COS-7 cells. The transcription suppressor activities of xCRY1 and xCRY2b are independent of light. However, mutation of any of the Trp-triad residues of xCRY2b severely reduced the transcription suppressor activity of the xCRY2b mutant proteins, whereas only one of the three mutations of xCRY1 affected its activity significantly [50•]. The fact that the light-independent activity of two type 2 cryptochromes was differentially affected by Trp-triad mutations argues strongly that those mutations simply upset the structural integrity of the respective non-photoreceptor cryptochromes rather than affected the fundamental photochemistry of cryptochromes.
A redox potential problem
In addition to the photoreduction controversy, there seems another redox puzzle remaining unsolved for the cryptochrome photoreceptors. This puzzle concerns the resting state of flavin. We have asserted in the previous section that for cryptochromes to absorb blue light, the chromophore flavin should be in its oxidized form, because oxidized FAD is the redox species of flavin that absorbs blue light most effectively [12•]. This assumption, however, has never been proven. Many factors affect the redox states of a flavoprotein, including the midpoint redox potentials of the respective flavoprotein and redox potential of the cell. For example, the midpoint potential for the FADH•/FADH− redox couple of E. coli photolyase has been measured to be between 16mV to −48mV [33,34,51]. Therefore, photolyase should contain mostly reduced flavin (FADH−) in a cellular environment with a redox potential more negative (reduced) than −48mV. Indeed, it has been known for a long time that E. coli photolyase exists as the fully reduced form in vivo without a need of photoreduction [2], presumably because E. coli cells have the redox environment of about −280mV [32]. The midpoint redox potentials of Arabidopsis CRY1 have also been measured, by at least two independent experiments, to be approximately −143mV to −153mV for the FAD/FADH• couple and −161mV to −181mV for the FADH•/FADH2 couple [12•,34]. It means that in a solution of about −143/−153mV, approximately half of the CRY1 molecules contain semireduced FADH• (or FAD• −) and the other half oxidized FAD. Only in a solution that is less negative than −143/−153mV would more than half of CRY1 contain the blue light- absorbing oxidized FAD. By the same token, in a solution that is more negative than (or FADH−) that absorbs −161/−181mV, more than half of CRY1 would contain FADH2 little blue light. It is difficult to accurately measure the cellular redox potential, because cells, especially plant cells undergoing photosynthesis, are usually not in the equilibrium state and it is difficult to estimate the net potentials of all redox species inside the cell. However, the cellular redox potentials have been roughly approximated from the redox potentials of a few primary redox regulators, such as glutathione, NAD(P), ascorbate, or thioredoxin couples [52,53]. For example, based on the redox potentials of glutathione or thioredoxin, the redox potentials of cultured mouse cytosol and E. coli cells were estimated to be approximately −260mV and −280mV, respectively [31,32]. The redox potential of Arabidopsis cells (cytosol) has been recently measured in vivo, using transgenic plants expressing redox-sensitive GFP, to be approximately −320mV [54,55]. This estimation seems consistent with the redox potentials of mammalian (ca. −260mV), bacterial (ca. −280mV), or plant cells (ca. -300mV) measured previously by other methods [31,32,56,57](G. Noctor, personal communication). Assuming the redox potential of the nucleus is also about −300mV, which is significantly more negative than the middle point potential of the FADH•/FADH2 couple, the flavin chromophore of an Arabidopsis cryptochrome should theoretically exist in vivo as the fully reduced form (FADH2) that absorbs little blue light! This apparent discrepancy between the theoretical redox equilibrium of cryptochromes under the presently known cellular redox environment and our expectation that cryptochromes should contain flavin in the oxidized state to absorb blue light presents an inescapable challenge to the study of cryptochrome photochemistry. It should be pointed out that the in vivo absorption spectrum of cryptochromes is likely to involve not only flavin but also the putative second chromophore, which is likely MTHF (5,10-methenyltetrahydrofolate). However, MTHF does not seem to be very helpful in this issue, because it also absorbs little blue light. One potential explanation of this problem may be that cryptochromes in the nucleus interact with other proteins to form a protein complex to provide a more oxidative microenvironment such that the ground-state cryptochromes may exist as a less reduced form, such as FAD or FAD−, to absorb blue light.
An alternative photocycle of cryptochromes
In summary, the prevailing Trp-triad-dependent photoreduction hypothesis of cryptochrome photochemistry (Fig. 3A) has several difficulties. First, it does not explain why elimination of flavin photoreduction fails to abolish the photobiochemical or photophysiologically activities of photolyases and most cryptochrome photoreceptors studied in vivo. Second, the tryptophan residues of Trp-triad required for photoreduction in vitro are only three of the five conserved tryptophan residues in the FAD-binding region of photolyases/cryptochromes; and they may be more important for the structural integrity than for the photochemistry per se of photolyase/cryptochrome. Third, the redox potential of plant cells appears unfavorable for the oxidized FAD to exist as the resting-state chromophore of cryptochromes.
Fig. 3. Alternative models of cryptochrome photochemistry.
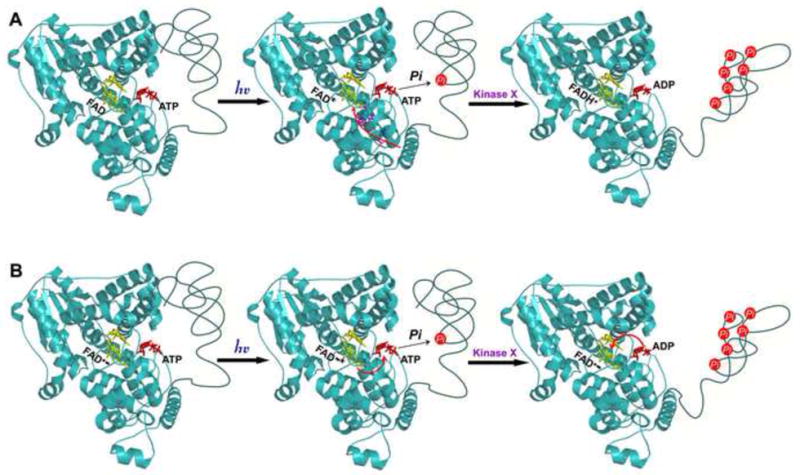
A. The Trp-triad-dependent photoreduction model. In this model, the resting state of a cryptochrome contains the oxidized FAD. Upon photon absorption, the excited FAD* gains an electron from tryptophan residues of Trp-triad (see Fig. 2) and a proton from an unknown source to become the active neutral radical semiquinone, FADH•. How the Trp-triad-dependent photoreduction triggers autophosphorylation and conformational change is not clear.
B. The photolyase-like cyclic electron shuttle model. In this model, the resting state of a cryptochrome contains the anion radical semiquinone (FAD−). Upon photon absorption, the excited FAD−* transfers an electron to ATP, triggering phosphotransfer and autophosphorylation of the cryptochrome. The electron is subsequently transferred back to flavin to complete the cycle. Autophosphorylation of cryptochromes facilitates further phosphorylation by an unidentified kinase (kinase X), resulting in separation of the CCE domain from the PHR domain and formation of the open conformation. Cryptochromes with the open conformation interacts with signaling partners to trigger signal transduction. The locations of FAD and ATP are indicated by the yellow and red structures, respectively. The putative locations of phosphorous group (red circle) and electron transfer path (red arrows) are indicated.
If the Trp-triad-dependent photoreduction mechanism did not explain the photochemistry of cryptochromes, what are alternatives mechanisms? In this regard, the very similar structure of photolyases and cryptochromes and the unusual U-shaped configuration of FAD (Fig. 2B) commonly found in photolyase and cryptochromes might offer some clues. We propose that the photochemistry of cryptochromes involves a photolyase-like photocycle that is dependent on a cyclic electron shuttle but not on the Trp-triad-dependent photoreduction (Fig. 3B). According to this proposition, the resting state cryptochrome contains anion radical semiquinone FAD• −, which apparently absorbs blue light with the efficiency of at least half that of FAD (Fig. 1B), but it is more likely to exist than FAD under reducing redox environment of the cell (see above). Absorption of a photon excites flavin to trigger an intermolecular electron transfer from flavin to an acceptor, such as ATP bound to cryptochromes or an unknown residue of cryptochromes. Given that pyrimidine-dimer is the electron acceptor for photolyase and that the location of the ATP-binding site at cryptochrome is equivalent to the pyrimidine-dimer binding site in photolyases [58], it is conceivable that ATP may act as an electron acceptor for cryptochromes. Electron transfer to ATP may promote a phosphotransfer reaction to “autophosphorylate” the cryptochrome protein, leading to further phosphorylation and conformational change of the photoreceptor. Upon phosphotransfer, the electron is shuttled back from ATP to flavin to complete the photocycle. The phosphorylated cryptochrome adopts an open conformation to interact with signaling proteins, resulting in gene expression changes and physiological responses [17,59•]. One attractive feature of the photolyase-like model is that it explains why the overall structure of the PHR domain of cryptochromes has deviated little from that of photolyases during evolution and what triggers the light-dependent autophosphorylation of cryptochromes. On the other hand, the photolyase-like hypothesis has its own problems. For example, it does not seem to help solve the discrepancy between the redox potentials of cryptochromes and the cellular environment. Continuous investigation of both the Trp-triad-dependent photoreduction and the photolyase-like hypotheses will shed more light to the photoexcitation mechanism of this arguably most widely distributed photoreceptors in nature.
Acknowledgments
The authors thank Drs. Graham Noctor and Alfred Batschauer for helpful discussions about this manuscript, Dr. Elaine Tobin for reading and editing of the manuscript, and Drs. Xuhong Yu, Bin Liu, Ya-Ting Kao, and John Klejnot for figure preparations. Works in authors’ laboratories are supported in part by National Institute of Health (GM56265 to CL) and (GM074813 to DZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 2.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Shalitin D. Cryptochrome Structure and Signal transduction. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 4.Kao YT, Saxena C, He TF, Guo L, Wang L, Sancar A, Zhong D. Ultrafast dynamics of flavins in five redox states. J Am Chem Soc. 2008;130:13132–13139. doi: 10.1021/ja8045469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H, Yuan Q, Briscoe AD, Froy O, Casselman A, Reppert SM. The two CRYs of the butterfly. Curr Biol. 2005;15:R953–954. doi: 10.1016/j.cub.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 6.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24 :948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 8.Lin C, Ahmad M, Gordon D, Cashmore AR. Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc Natl Acad Sci U S A. 1995;92:8423–8427. doi: 10.1073/pnas.92.18.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 10.El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 11.Kleine T, Lockhart P, Batschauer A. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 2003;35:93–103. doi: 10.1046/j.1365-313x.2003.01787.x. [DOI] [PubMed] [Google Scholar]
- 12•.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269:968–970. doi: 10.1126/science.7638620. This report identified the FAD chromophore of cryptochromes, it also first reported the in vitro photoreduction of cryptochrome and the midpoint potential of Arabidopsis CRY1. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 14•.Banerjee R, Schleicher E, Meier S, Munoz Viana R, Pokorny R, Ahmad M, Bittl R, Batschauer A. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. This reports proposed the Trp-triad-dependent photoreduction hypothesis for Arabidopsis CRY1. [DOI] [PubMed] [Google Scholar]
- 15.Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 16.Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci U S A. 2007;104:7289–7294. doi: 10.1073/pnas.0701912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 19.Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- 20•.Kao YT, Tan C, Song SH, Ozturk N, Li J, Wang L, Sancar A, Zhong D. Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J Am Chem Soc. 2008;130:7695–7701. doi: 10.1021/ja801152h. The report proposed an universal photoexcitation mechanism for photolyases and cryptochromes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. This reports proposed the Trp-triad-dependent photoreduction hypothesis for Arabidopsis CRY2. [DOI] [PubMed] [Google Scholar]
- 22.Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K, Ahmad M. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 23.Lin C, Ahmad M, Cashmore AR. Arabidopsis cryptochrome 1 is a soluble protein mediating blue light– dependent regulation of plant growth and development. Plant J. 1996;10:893–902. doi: 10.1046/j.1365-313x.1996.10050893.x. [DOI] [PubMed] [Google Scholar]
- 24.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellaro R, Crepy M, Trupkin SA, Karayekov E, Buchovsky AS, Rossi C, Casal JJ. Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis. Plant Physiol. 2010;154:401–409. doi: 10.1104/pp.110.160820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 27.Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 28••.Li YF, Heelis PF, Sancar A. Active site of DNA photolyase: Tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochem. 1991;30:6322–6329. doi: 10.1021/bi00239a034. The first report that tested the role of tryptophan and photoreduction hypothesis of DNA photolyase, which also demonstrated that photoreduction is not required for photolyase activity in vivo. [DOI] [PubMed] [Google Scholar]
- 29•.Aubert C, Vos MH, Mathis P, Eker AP, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. This report proposed the Trp-triad-dependent photoreduction for photoexcitation of cryptochromes. [DOI] [PubMed] [Google Scholar]
- 30.Kavakli IH, Sancar A. Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry. 2004;43:15103–15110. doi: 10.1021/bi0478796. [DOI] [PubMed] [Google Scholar]
- 31.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert H. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 33.DeRosa MC, Sancar A, Barton JK. Electrically monitoring DNA repair by photolyase. Proc Natl Acad Sci U S A. 2005;102:10788–10792. doi: 10.1073/pnas.0503527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balland V, Byrdin M, Eker AP, Ahmad M, Brettel K. What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. J Am Chem Soc. 2009;131:426–427. doi: 10.1021/ja806540j. [DOI] [PubMed] [Google Scholar]
- 35.Giovani B, Byrdin M, Ahmad M, Brettel K. Light–induced electron transfer in a cryptochrome blue-light photoreceptor. Nat Struct Biol. 2003;5:5. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- 36.Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, Berndt A, Wolf E, Bittl R, Ahmad M. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 38.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 39.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 40.Tamanini F, Chaves I, Bajek MI, van der Horst GT. Structure function analysis of mammalian cryptochromes. Cold Spring Harb Symp Quant Biol. 2007;72:133–139. doi: 10.1101/sqb.2007.72.066. [DOI] [PubMed] [Google Scholar]
- 41.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 42.Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc Natl Acad Sci U S A. 2000;97:14697–14702. doi: 10.1073/pnas.260498597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Gelder RN, Wee R, Lee JA, Tu DC. Reduced pupillary light responses in mice lacking cryptochromes. Science. 2003;299:222. doi: 10.1126/science.1079536. [DOI] [PubMed] [Google Scholar]
- 44••.Gegear RJ, Foley LE, Casselman A, Reppert SM. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature. 2010;463:804–807. doi: 10.1038/nature08719. This report demonstrated that the Trp-triad-dependent photoreduction is not photochemically required for the function of Drosophila cryptochrome in both the circadian clock and magnetoreception. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song SH, Ozturk N, Denaro TR, Arat NO, Kao YT, Zhu H, Zhong D, Reppert SM, Sancar A. Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J Biol Chem. 2007;282:17608–17612. doi: 10.1074/jbc.M702874200. [DOI] [PubMed] [Google Scholar]
- 46.Froy O, Chang DC, Reppert SM. Redox potential: differential roles in dCRY and mCRY1 functions. Curr Biol. 2002;12:147–152. doi: 10.1016/s0960-9822(01)00656-x. [DOI] [PubMed] [Google Scholar]
- 47.Ozturk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 48.Saxena C, Wang H, Kavakli IH, Sancar A, Zhong D. Ultrafast dynamics of resonance energy transfer in cryptochrome. J Am Chem Soc. 2005;127:7984–7985. doi: 10.1021/ja0421607. [DOI] [PubMed] [Google Scholar]
- 49.Kao YT, Saxena C, Wang L, Sancar A, Zhong D. Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci U S A. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Zhu H, Green CB. A putative flavin electron transport pathway is differentially utilized in Xenopus CRY1 and CRY2. Curr Biol. 2001;11:1945–1949. doi: 10.1016/s0960-9822(01)00601-7. This report demonstrated that the conserved Trp-triad is structurally important rather than photochemically critical for the function of cryptochromes. [DOI] [PubMed] [Google Scholar]
- 51.Gindt YM, Schelvis JP, Thoren KL, Huang TH. Substrate binding modulates the reduction potential of DNA photolyase. J Am Chem Soc. 2005;127:10472–10473. doi: 10.1021/ja051441r. [DOI] [PubMed] [Google Scholar]
- 52.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 53.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- 55.Schwarzlander M, Fricker MD, Muller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ. Confocal imaging of glutathione redox potential in living plant cells. J Microsc. 2008;231:299–316. doi: 10.1111/j.1365-2818.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 56.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 57.Foyer CH, Bloom AJ, Queval G, Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol. 2009;60:455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- 58.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. This report identified the first blue light-dependent CRY-interacting protein in Arabidopsis. [DOI] [PubMed] [Google Scholar]