Abstract
Agouti related protein (AgRP) and agouti signaling protein (ASIP) are homologs that play critical roles in energy balance and pigmentation, respectively, by functioning as antagonistic ligands at their cognate melanocortin receptors (MCRs). Signaling specificity is mediated in part through receptor binding selectivity brought about by alterations in the cysteine-rich carboxy-terminal domains of the ligands. AgRP binds with high affinity to the melanocortin 3 and melanocortin 4 receptors (MC3R and MC4R), but not to the MC1R, whereas ASIP binds with high affinity to all three receptors. This work explores the structural basis for receptor selectivity by studying chimeric proteins developed by interchanging loops between the cysteine-rich domains of ASIP and AgRP. Binding data demonstrate that MC4R responds to all chimeras, and is therefore highly tolerant of gross loop changes. By contrast, MC1R responds primarily to those chimeras with sequence close to wild type ASIP. Further analysis of binding and functional data suggests that the ASIP C-terminal loop – a six amino acid segment closed by the final disulfide bond – is essential for high affinity MC1R binding and inverse agonism. Comparison with previously published molecular models suggests that this loop makes contact to the first extracellular loop (EC1) of MC1R through a series of key hydrophobic interactions.
Complex multicelluar organisms require a broad spectrum of receptors to adjust to environmental changes and maintain homeostasis. Thermal regulation, UV light protection, pigmentation, energy balance and sexual behavior are regulated, to a significant extent, by the melanocortin system, which is composed of the five melanocortin receptors (MCRs) and an intricate set of ligands.1; 2; 3; 4; 5 The MCRs belong to the G-protein coupled receptor (GPCR) super family of proteins. High affinity agonist binding within the extracellular MCR pocket introduces a conformational change that initiates an intracellular signal cascade. The native MCR agonists are derived from pro-opiomelanocortin peptide (POMC), which is processed into numerous fragments including α-melanocyte stimulating hormone (α-MSH) and adrenocorticotropic hormone (ACTH).6 Among the GPCR super family, MC1R, MC3R and MC4R are unique since they respond not only to agonists, but also to the endogenous antagonists, Agouti signaling protein (ASIP) and Agouti related protein (AgRP).7; 8; 9; 10 ASIP and AgRP compete with α-MSH for binding to MC1R, MC3R and/or MC4R. Beyond competitive antagonism, these proteins also induce a conformational change that reduces intracellular cAMP and thus function as inverse agonists.11; 12
α-MSH binding to MC1R stimulates the production of eumelanin, a dark brown or black pigment.13 ASIP has the opposite effect, simulating the production of the yellow pigment pheomelanin. In mice with so-called Agouti-colored coats, individual hairs have eumelanin at the top and bottom of the shaft, while the middle segment contains pheomelanin, a pattern arising from regulated expression of ASIP.14 Mice that lack ASIP have hairs composed entirely of eumelanin, while mice that lack a functional MC1R have hairs composed almost entirely of pheomelanin.15
Feeding circuits controlled by neurons in the hypothalamus are regulated by a parallel system.4; 9; 16 Here, α-MSH binding to the MC3 and MC4 receptors elevates cAMP levels and suppresses feeding, while AgRP decreases cAMP levels at both receptors and promotes feeding.8 Specificity of ASIP and AgRP action is brought about by a combination of receptor binding selectivity and tissue-specific expression. AgRP binds with high affinity to the MC3R and MC4R but not to the MC1R; by contrast, ASIP binds with high affinity to the MC1R, MC3R and MC4R. This pattern of receptor binding selectivity is reflected in the unusual phenotype of a regulatory ASIP mutation known as lethal yellow (Ay), in which a genomic rearrangement causes ASIP to be expressed abnormally throughout the body, leading to a yellow coat, hyperphagia, and obesity.8; 17; 18
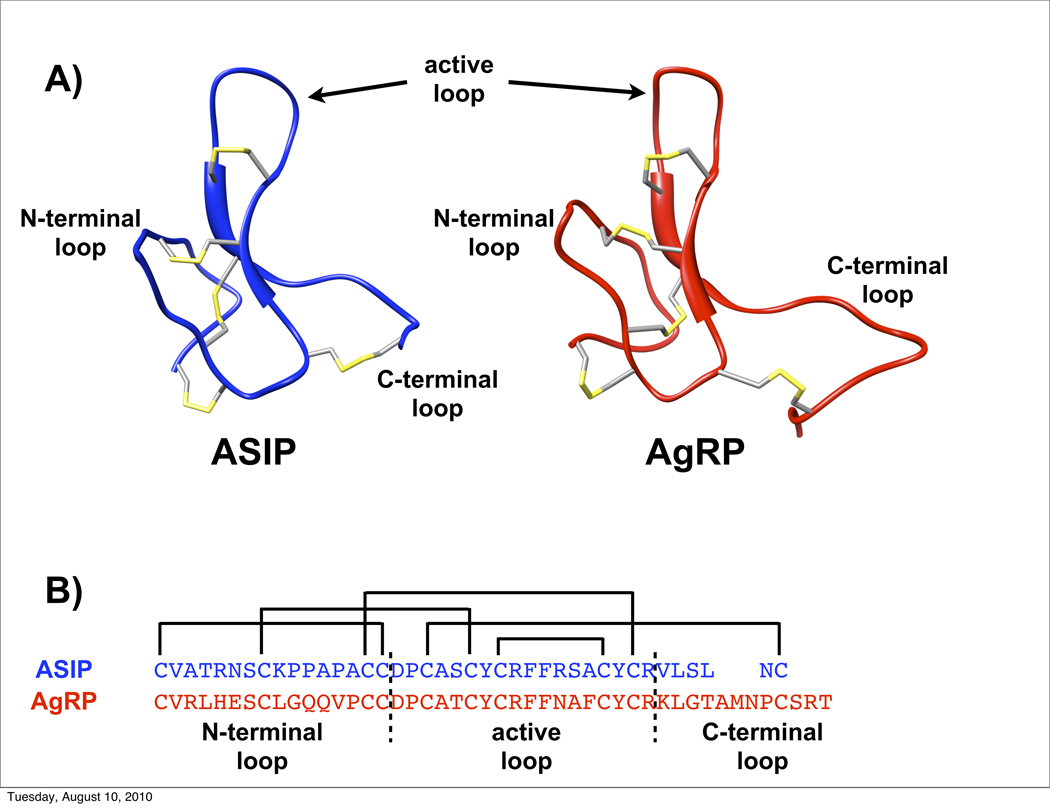
Here we explore the structural basis for ASIP and AgRP specificity, focusing on the Cys-rich regions of ASIP or AgRP, which mediate receptor binding. NMR structures, determined by our labs, find these regions possess similar folds, with ten conserved cysteine residues forming a scaffold of five disulfide bonds (Figure 1).10; 19; 20; 21 Three of these disulfides possess the local spatial arrangement and threading of an inhibitor cystine knot (ICK), or knottin, motif.10; 19; 21 Emerging from the disulfide core, are three polypeptide loops, referred to as the N-terminal loop, the active loop and the C-terminal loop.21 The active loop is so named because it carries the Arg-Phe-Phe (RFF) triplet that is essential for MCR binding and activity.22 23 24 Distance restraints determined from NMR experiments further demonstrated that the N-terminal and active loops are well structured, whereas the C-terminal loops are less ordered, especially in AgRP.20; 21 Given that the greatest sequence differences between AgRP and ASIP are in their respective C-terminal loops, we proposed that MC3/4R contacts in AgRP are localized to the N-terminal and active loops. To test this hypothesis, we developed a minimized AgRP (AgRP(87–120, C105A)) that contains only the N-terminal and active loops. Pharmacological testing showed that this mini-AgRP retains the full MCR affinity and receptor subtype specificity of the entire AgRP cysteine-rich domain.20
Figure 1.
A) Structure of ASIP (in blue) and AgRP (in red), and identification of the three loops interchanged in chimera constructs. Disulfide bonds are shown in yellow. B) The sequences of AgRP (above) and ASIP (below) showing disulfide connectivities and the delineation of the three interchanged regions.
We also constructed variants of mini-AgRP designed to act as agonists rather than antagonists, by substituting the core α-MSH sequence His-Phe-Arg-Trp (HFRW) for the Arg-Phe-Phe triplet in the active loop.25 Several designed mini-AgRPs did exhibit potent agonist function at MC3R and MC4R but, surprisingly, all of the designed mini-AgRPs also bound to MC1R with relatively high affinity. These results suggest that the mechanism, whereby the cysteine rich domain of AgRP interacts with MC3R and MC4R, is substantially different from ASIP’s interaction with MC1R. Understanding the structural basis for these apparent differences could have important therapeutic implications since selective activation of MC3R and MC4R is a potential treatment for obesity and metabolic syndrome,26 whereas selective activation of MC1R is a potential defense against skin damage and skin cancer susceptibility caused by sun exposure.27
Here we take a new approach to investigate the basis for AgRP and ASIP receptor selectivity using a family of chimeric proteins made by loop interchange between the respective cysteine-rich domains. By examining the full cysteine-rich domain as opposed to a mini-AgRP scaffold, we aim to identify polypeptide segments in ASIP required for MC1R antagonist function. We consider ASIP and AgRP as composed of three modules: the N-terminal loop (beginning with the first Cys residue), the active loop and the C-terminal loop. In addition to segment interchange, we also consider mini-protein constructs lacking the C-terminal segments. Particular emphasis is placed on evaluating the C-terminal loop, the segment exhibiting the greatest sequence variation between ASIP and AgRP. Pharmacology and functional analyses identify critical features required for MC1R recognition. Consistent with our hypothesis of an accessible MC1R binding pocket, our findings demonstrate that all three of ASIP’s loops are required for function at MC1R.
Results
Naming Scheme and Folding
Pharmacological studies of the ASIP and AgRP structured, Cys-rich domains show that these domains alone are sufficient for high affinity binding and activity at respective MCRs.19; 21; 28 The ASIP and AgRP three-dimensional structures, along with sequences and disulfide connectivities, are shown in Fig. 1A and 1B. The Cys-rich domains are considered as three key segments: the N-terminal loop, the active loop and the C-terminal loop. In ASIP, there is an additional 13 amino acids preceding the first Cys that was originally included to improve solubility in aqueous solution.19 AgRP possesses a natural cleavage point following residue 82, thus leaving only four residues before the first Cys in the cysteine-rich domain.28; 29 These segments were included when necessary to overcome solubility issues.
Fig 1B delimits the boundaries of three loop modules described above. Chimeras were developed by interchange of these three segments. Each chimera is identified by a numerical code specific for the segment at these three positions (Table 1). (Some peptides have been renamed from previous literature to facilitate comparison). A ‘+1’ indicates a segment from ASIP, whereas a ‘-1’ indicates a segment from AgRP. A ‘0’ denotes absence of the specific segment, and a ‘+0.5’ indicates ASIP’s C-terminal loop, but with serines replacing the cysteines that would otherwise anchor the loop through a disulfide.
Table 1.
Sequences of ASIP, AgRP and chimera proteinsa
| N-terminus | N-terminal loop | active loop | C-terminal loop | |
|---|---|---|---|---|
| ASIP chi(+1 +1 +1)b | KKVVRPRTPLSAP | CVATRNSCKPPAPACC | DPCASCYCRFFRSACYCR | VLSLNC |
| AgRP chi(−1 −1 −1) | CVRLHESCLGQQVPCC | DPCATCYCRFFNAFCYCR | KLGTAMNPCSRT | |
| chi(−1 −1 0)c | CVRLHESCLGQQVPCC | DPAATCYCRFFNAFCYCR | ||
| chi(+1 +1 0) | CVATRNSCKPAAAACC | DPAASCYCRFFRSACYCR | ||
| chi(+1 −1 0) | CVATRNSCKPAAAACC | DPAATCYCRFFNAFCYCR | ||
| chi(−1 +1 0)d | CVRLHESCLGQQVPCC | DPAATCYCRFFRSACYCR | ||
| chi(+1 +1 −1) | CVATRNSCKPPAPACC | DPCASCYCRFFRSACYCR | KLGTAMNPCSRT | |
| chi(−1 −1 +1) | SSRR | CVRLHESCLGQQVPCC | DPCATCYCRFFNAFCYCR | VLSLNC |
| chi(+1 −1 +1) | KKVVRPRTPLSAP | CVATRNSCKPPAPACC | DPCASCYCRFFNAFCYCR | VLSLNC |
| chi(+1 +1 +0.5) | KKVVRPRTPLSAP | CVATRNSCKPPAPACC | DPSASCYCRFFRSACYCR | VLSLNS |
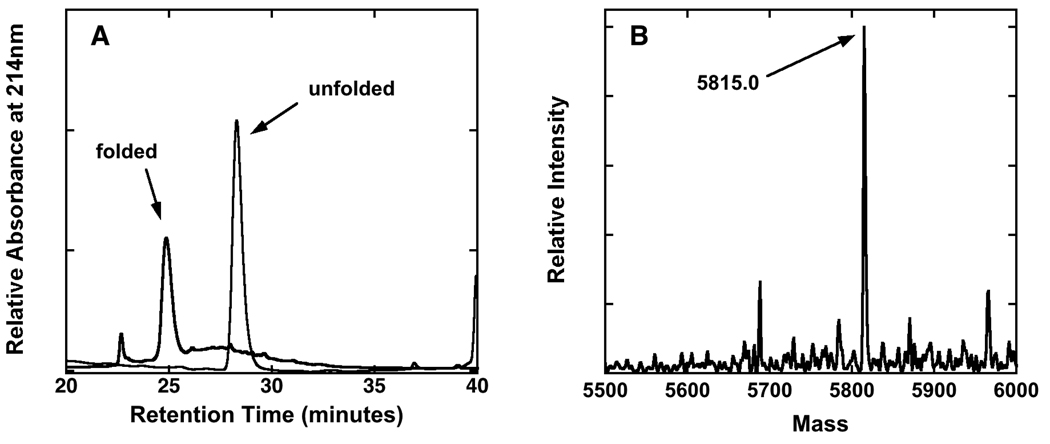
All constructs containing the ASIP active loop possess two Xaa→Tyr mutations to enhance stability.19 When incorporated into wild type C-terminal ASIP, to give the protein “ASIP-YY,” these mutations improve folding yield to nearly 100%, but without affecting MCR binding or selectivity. The AgRP and ASIP-YY C-terminal domains fold as cooperative units, consistent with other known ICK proteins.30; 31 When monitored by HPLC under oxidizing conditions, the peak arising from the unfolded material diminishes over several hours, followed by the concomitant emergence of a new peak from the folded protein (mass spectrometry gives a loss of ten AMUs, consistent with five disulfide bonds). For each new construct studied here, we used HPLC to monitor folding behavior as described above, and mass spectrometry (MS) to determine the degree of disulfide bond formation (Fig. 2). Much like C-terminal AgRP, mini-AgRP and ASIP-YY, oxidizing conditions produced uniform and fully folded products, consistent with the example shown in Fig. 2.
Figure 2.
Oxidative folding and analysis of an example chimera. A) Reverse phase HPLC of chi(+1 +1 +0.5) at 2.0 hours and 24 hours showing conversion to the folded species. B) Deconvoluted electrospray mass spectrum of the major peak collected at t = 24 hours. The mass of the species corresponding to this peak reflects a loss of 8.0 amu relative to the unfolded species, consistent with the formation of 4 disulfide bonds.
Binding Assays
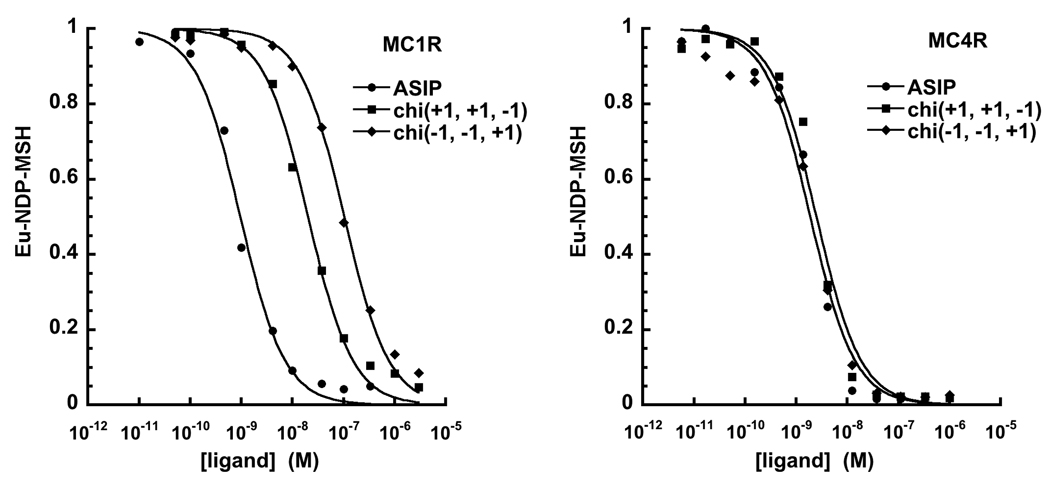
The receptor binding affinity of each new construct was determined by measuring its ability to displace Eu-labeled NDP-MSH from HEK 293 cells transfected with human MC1R or human MC4R (Table 2).20; 25; 32; 33; 34 Fig. 3 and Table 2 reveal a remarkable contrast between peptide binding affinities at MC1R and MC4R. (Note that only select, representative curves are shown in the figure to avoid overlap. Curves not shown here are available in the Supplement.) All chimeras bind to MC4R with reasonably similar affinities, given by Ki values ranging from 0.21 nM to 11 nM. For example, the chimeras chi(+1 +1 0) with chi(−1 +1 0), which have the ASIP active loops but interchanged N-terminal loops, exhibit MC4R Ki values of 0.90 nM and 5.4 nM, respectively. These inhibitory constants are close to the values for C-terminal ASIP (chi(+1 +1 +1)) and AgRP (chi(−1 −1 −1)), as well as mini-AgRP (chi(−1 −1 0)).
Table 2.
Ligand displacement Ki values
| MC1R | MC4R | |||
|---|---|---|---|---|
| Peptide | Ki (nM) | fold diff. | Ki(nM) | fold diff. |
| ASIP [chi(+1 +1 +1)] | 0.70 ± 0.06 | 1 | 1.3 ± 0.3 | 1 |
| AgRP [chi(−1 −1 −1)]a | 47% | n.a.d | 11 ± 1 | 8.5 |
| chi(−1 −1 0)a | 26% | n.a.d | 6.1 ± 0.5 | 4.7 |
| chi(+1 +1 0) | 23 ± 3 | 33 | 0.90 ± 0.19 | 0.69 |
| chi(−1 +1 0)b | 870 ± 12 | 1242 | 5.4 ± 0.3 | 4.2 |
| chi(+1 −1 0)c | n.d. | n.d. | ||
| chi(+1 +1 −1) | 4.8 ± 0.3 | 6.8 | 1.1 ± 0.2 | 0.85 |
| chi(−1 −1 +1) | 32 ± 3.6 | 46 | 1.5 ± 0.3 | 1.1 |
| chi(+1 −1 +1) | 4.9 ± 1.1 | 7.0 | 2.1 ± 0.3 | 1.6 |
| chi(+1 +1 +0.5) | 0.44 ± 0.03 | 0.63 | 0.21 ± 0.04 | 0.16 |
Figure 3.
Representative NDP-MSH displacement curves at MC1R and MC4R. (Note: curves not shown here are available in the Supplement.) Transfected HEK 293 cells pre-incubated with europium labeled NDP-MSH are treated with increasing concentrations of C-terminal ASIP, AgRP or various chimera ligands. Bound NDP-MSH, reported on the vertical axis, is determined through relative Eu fluorescence. A rightward shift (higher concentration) corresponds to a ligand of lower affinity. Ki values determined from these curves are reported in Table 2.
In contrast to the uniform affinities for all constructs at MC4R, binding affinities at MC1R vary significantly. As determined in previous work, C-terminal AgRP (chi(−1 −1 −1)) and mini-AgRP (chi(−1 −1 0)) bind very weakly at MC1R. At high concentration, these ligands only partially displace agonist, suppressing NDP-MSH binding by 47% and 26%, respectively.20 In contrast, C-terminal ASIP (chi(+1 +1 +1)) binds with high affinity (Ki = 0.7 nM). The chimeric proteins tested in this study display MC1R binding affinities that fall between these two extremes, with Ki values ranging over two to three orders of magnitude (Fig. 3, Table 2).
Probing for systematic binding differences among the peptides, we note that peptides lacking a C-terminal loop (chi(+1 +1 0), chi(−1 −1 0), chi(−1 +1 0)) exhibited the weakest MC1R binding. Among this group, chi(+1 +1 0), with the native ASIP N-terminal and active loops, possesses the highest affinity (but with a 33-fold lower affinity than ASIP). Comparison of chi(−1 −1 0) and chi(−1 −1 +1) further underscores the role of the ASIP C-terminal loop. Of these constructs, the former fails to fully displace agonist, whereas the latter exhibits a moderate binding affinity, characterized by a low dissociation constant (Ki = 32 nM). Interestingly, elimination of the disulfide bond that anchors the C-terminal loop, in chi(+1 +1 +0.5), does not reduce MC1R affinity relative to wildtype ASIP.
The N-terminal loop also affects MC1R binding, as illustrated by pair wise comparison of chi(+1 +1 0) to chi(−1 +1 0), and chi(+1 −1 +1) to chi(−1 −1 +1). For these two pairs of peptides, the ASIP N-terminal loop increases binding 38-fold and seven-fold, respectively. The active loop also plays a role, as seen with chi(+1 −1 +1), which yields an approximate 7-fold lower affinity compared wildtype ASIP.
Functional Analysis
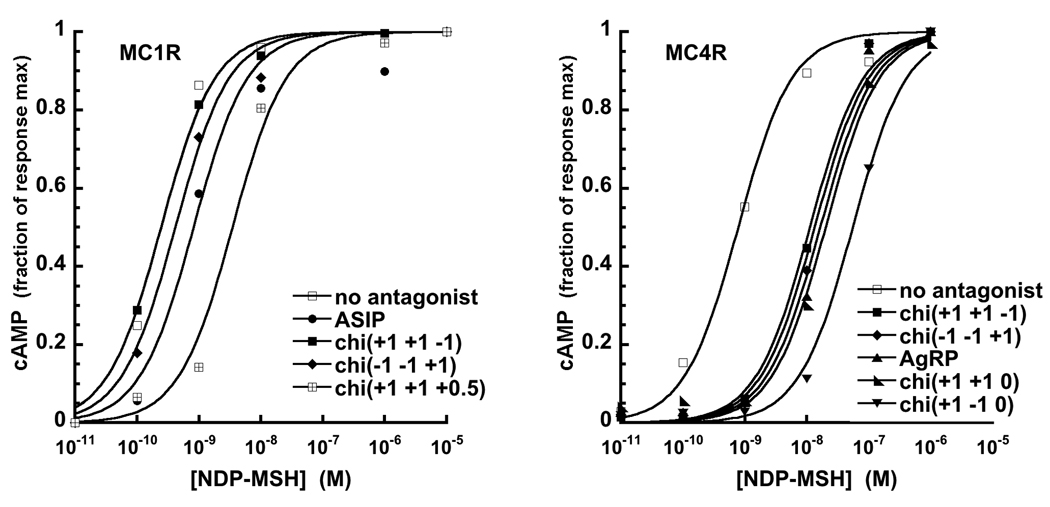
To examine antagonist activity, each chimera was evaluated for its ability to suppress NDP-MSH stimulated cAMP production. HEK 293 cells expressing either MC1R or MC4R were pre-incubated with 100 nM ASIP (chi(+1 +1 +1)), AgRP (chi(−1 −1 −1)) or chimera proteins, and treated with increasing concentrations of NDP-MSH, as shown in Figure 4. A rightward shift relative to NDP-MSH alone (labeled “no antagonist”) indicates suppression of NDP-MSH signaling. Resulting EC50s are reported in Table 3. Consistent with the binding assays above, there are significant differences between the responses at MC1R and MC4R. At MC4R, all chimeras are antagonists (Table 3), with most exhibiting EC50 values greater than wildtype ASIP. The sole exception is chi(−1 +1 0), which still retains antagonist function but is slightly weaker than either ASIP or AgRP.
Figure 4.
Representative curves demonstrating that C-terminal ASIP, AgRP and various chimera proteins inhibit NDP-MSH stimulated cAMP production at MC1R and MC4R. (Note: curves not shown here are available in the Supplement.) Experiments were performed on transfected HEK 293 cells pretreated with the ligand of interest. A rightward shift relative to NDP-MSH alone (no antagonist) reflects antagonist function. EC50 values are reported in Table 3. At MC1R, several chimeras stimulated cAMP release, thus functioning as agonists, and are not shown.
Table 3.
EC50 values for inhibition of NDP-MSH stimulated cAMP production
| MC1R | MC4R | |
|---|---|---|
| Peptide | EC50 (nM) | EC50 (nM) |
| no ligand | 0.25 ± 0.04 | 0.79 ± 0.01 |
| ASIP [chi(+1 +1 +1)] | 0.83 ± 0.02 | 6.4 ± 2.6 |
| AgRP [chi(−1 −1 −1)] | no binding | 17 ± 3 |
| chi(−1 −1 0) | no binding | 13 ± 3 |
| chi(+1 +1 0) | agonist | 20 ± 3 |
| chi(−1 +1 0) | agonist | 1.4 ± 0.4 |
| chi(+1 −1 0) | agonist | 56 ± 7 |
| chi(+1 +1 −1) | no shift | 11 ± 2 |
| chi(−1 −1 +1) | 0.42 ± 0.06 | 14 ± 2 |
| chi(+1 −1 +1) | 1.1 ± 0.1 | 19 ± 5 |
| chi(+1 +1 +0.5) | 3.5 ± 0.7 | 61 ± 8 |
In contrast, MC1R data show significant variation. Only those chimeras with an ASIP C-terminal loop suppress cAMP production close to the level observed for ASIP (chi(+1 +1 +1)). Several of the remaining chimeras elicit release of cAMP, without addition of NDP-MSH, and thus function as agonists. Among the chimeras that possess antagonist function, chi(+1 −1 +1) and chi(+1 +1 +0.5) were more inhibitory than ASIP (chi(+1 +1 +1)). These data show that the C-terminal loop not only enhances binding affinity but also is essential for antagonism of NDP-MSH signaling at MC1R.
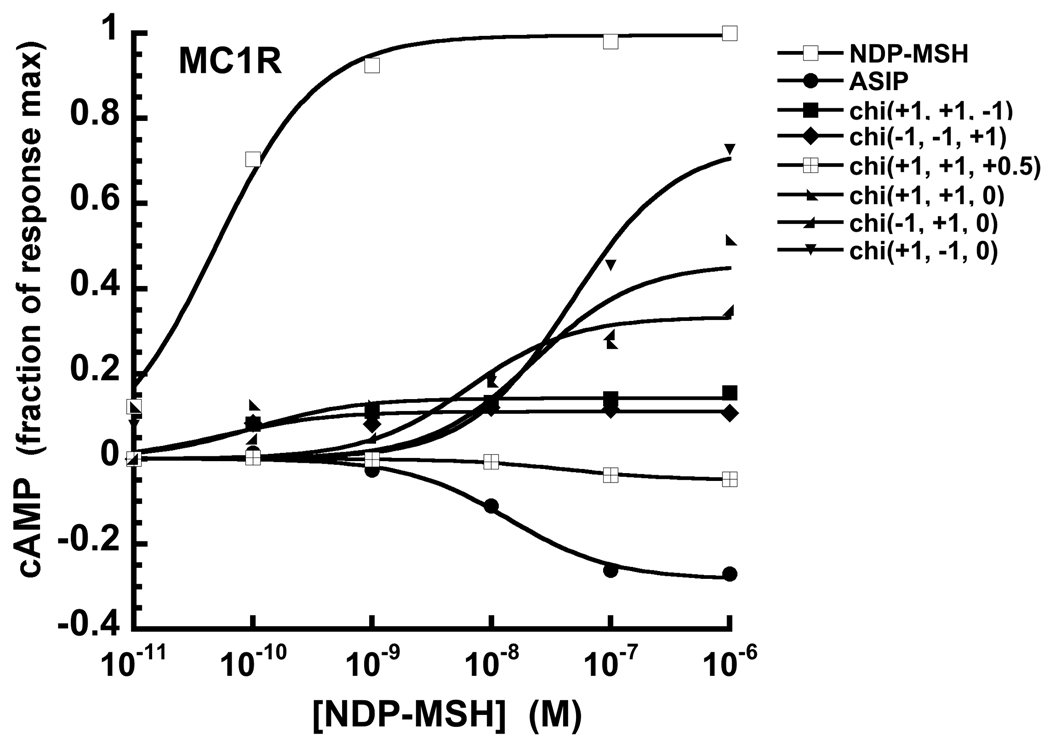
Beyond competitive antagonism, C-terminal ASIP (chi(+1 +1 +1)) and AgRP (chi(−1 −1 −1)) act as inverse agonists, suppressing constitutive cAMP production at MC1R and MC4R, respectively.12; 35 Whereas a competitive antagonist blocks the action of an agonist, an inverse agonist also suppresses constitutive receptor activity. Chimeras that exhibited antagonist function at MC1R were further tested for inverse agonism and compared to NDP-MSH and ASIP, as shown in Fig. 5. These experiments were performed in the absence of NDP-MSH or other competing species. Basal response is taken as the zero point and, as expected, NDP-MSH and ASIP at concentrations beyond 1.0 nM drive MC1R signaling in opposite directions, as measured through cAMP production. All chimeras with a C-terminal loop, tested out to 2.0 – 3.0 nM, fall in between, eliciting little influence on cAMP output. Beyond 1.0 nM, chi(+1 +1 −1) and chi(−1 −1 +1) cause a slight increase in cAMP, to a level of about 10% of that of NDP-MSH. In general, however, these constructs behave as neutral antagonists, blocking NDP-MSH action but without altering constitutive activity. In contrast, chimeras lacking a C-terminal loop are partial agonists at concentrations beyond 10 nM (noted in Table 3), as reported for other small molecules and AgRP derived peptides at high concentration.12; 34
Figure 5.
Effects of NDP-MSH, C-terminal ASIP and select chimera constructs on cAMP production at MC1R, which displays constitutive activity. NDP-MSH is an agonist elevating cAMP, whereas ASIP is an inverse agonist, as demonstrated by suppression of constitutive cAMP production. Interestingly, chimeras lacking a C-terminal loop (chi(+1 +1 0), chi(−1 +1 0) and chi(+1 −1 0)) are agonists at concentrations beyond 10 – 100 nM.
Discussion
The melanocortin receptors MC1R and MC4R diverged from a common ancestor early in vertebrate evolution, but are similar in their respective responses to most ligands. However, a key difference between these receptors is that MC4R is inhibited by both ASIP and AgRP, whereas MC1R responds only to ASIP. Understanding the differential response of MC1R and MC4R is significant as it provides insight into the evolution of the melanocortin system and guidance for the development of pharmaceuticals that distinguish between these receptor subtypes. The chimera proteins developed here offer a new perspective by elucidating the requirements for MC1R antagonism. A striking finding is that the ASIP C-terminal loop is essential for inhibitory action at MC1R.
Previous work from our lab and others suggest that ASIP and AgRP interact with the MC4R in ways that are structurally very similar. The active loops of both ligands are thought to bind within the receptor transmembrane pocket,36 and the N-terminal loops of both ligands interact with receptor extracellular loops (EC) 2 and 3.37; 38 The latter conclusion is based in part on the observations (1) that mini-AgRP (chi(−1 −1 0)) binds to MC4R with an affinity that is equivalent to that of the full AgRP C-terminal domain but considerably greater than a cyclic peptide composed of the active loop alone;20 and (2) that mini-ASIP (chi(+1 +1 0)) behaves similarly to mini-AgRP in binding and pharmacologic studies described here.
However, results from additional aspects of the current study suggest a very different picture for ASIP’s interaction with MC1R. High affinity binding is found primarily for those chimeras that possess the ASIP C-terminal loop. Elimination of the C-terminal loop decreases affinity for MC1R by at least a factor of ten, whereas addition of the ASIP C-terminal loop to the mini-AgRP scaffold (which generates chi(−1 −1 +1)) converts a low affinity peptide into one of moderately high affinity (Ki = 32 nM). Moreover, even though chi(+1 +1 0) and chi(−1 +1 0) both bind to the MC1R (Ki = 23 nM and 870 nM, respectively), they no longer function as antagonists. Instead, they are agonists, stimulating release of cAMP. The one exception is chi(+1 +1 −1), which is the ASIP knottin domain but with the AgRP C-terminal loop. This chimera binds with high affinity (Ki = 4.8 nM), but does not stimulate significant release of cAMP or suppress NDP-MSH action.
Taken together, these data suggest that, in addition to the N-terminal and active loops, the ASIP C-terminal loop constitutes a primary contact to MC1R that is required for antagonist function. These results are in accord with previous work from our lab suggesting a broad MC1R binding interface with only partial overlap of the agonist and antagonist sites.25 Without exact placement of the ASIP active loop, enforced by the full ASIP structure, the active loop potentially redirects to the agonist site. Similar results were obtained from a peptide derived from the AgRP active loop, which acts as an MC1R agonist.34 The ASIP C-terminal loop must therefore make direct contact with MC1R in a way that directs the active loop to the appropriate site that stabilizes the receptor in the inactive conformation.
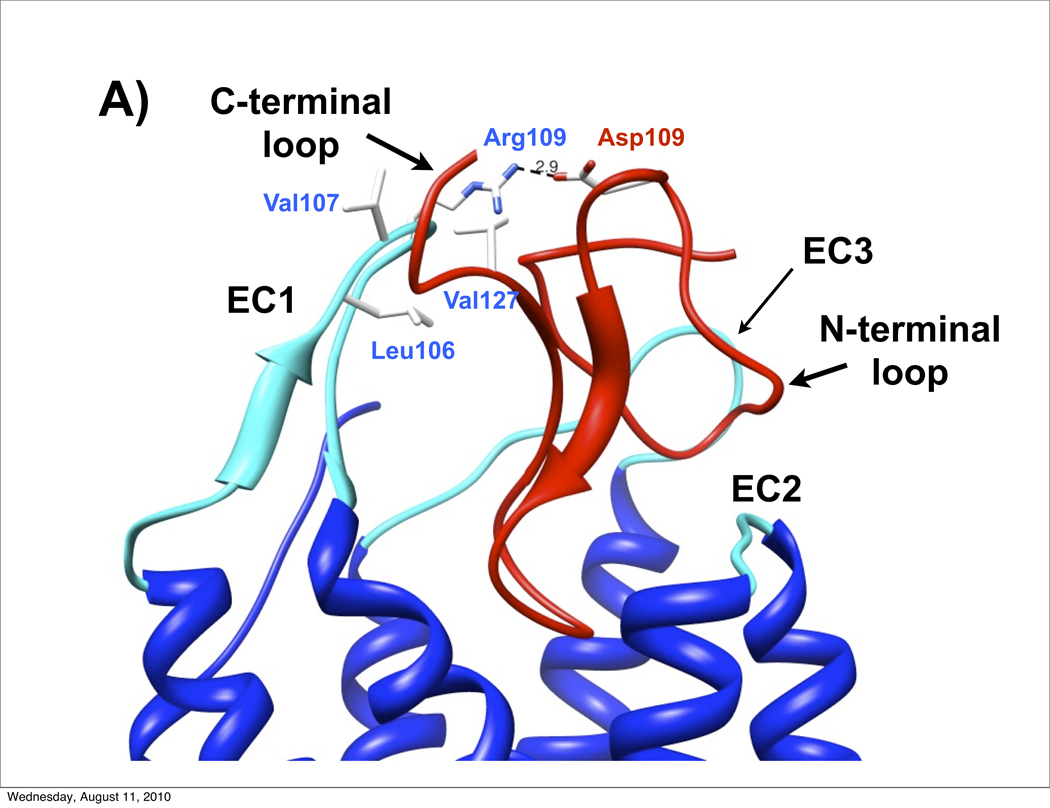
Both Wilczynski et al.38 and Chai et al.39 developed AgRP-MC4R molecular models based on extensive receptor mutagenesis experiments. As noted above, the AgRP active loop interacts deeply within the MC4R transmembrane pocket and the N-terminal loop bridges between the second and third extracellular loops (EC2 and EC3). The large, 12 residue AgRP C-terminal loop points away from the extracellular surface and does not make extensive receptor contacts. Chai et al. further developed ASIP-MC1R models and found analogous ligand-receptor placement.39 However, the shorter six residue C-terminal loop in ASIP emerges proximal to the first extracellular loop (EC1), forming a third primary contact point. The crest of EC1 (residues 106–111, sequence LVARAA) and ASIP C-terminal loop (VLSLN) are dominated by non-polar amino acids, suggesting stabilization through a significant hydrophobic interaction. Fig. 6A shows the relevant extracellular segment of ASIP-MC1R molecular model, with emphasis on these hydrophobic contacts between EC1 and the ASIP C-terminal loop. Also apparent is the interaction between the ASIP N-terminal loop and extracellular loops 2 and 3. Interestingly, within this model, the single positive charge in EC1, arising from Arg109, forms a salt bridge with Asp109 in ASIP. A negative charge (Asp or Glu) at this position in ASIP is found in all vertebrate ASIP sequences (except for rat, opossum and chicken, which have Asn).28 Moreover, an Asp→Ala mutation in the mouse sequence (position 108) reduces MC1R affinity by approximately a factor of ten 23. Other than the Arg-Asp residues that form the putative salt bridge, the respective C-terminal loop and EC1 hydrophobic residue clusters are strongly conserved in both ASIP28 and MC1R.40; 41 Our results are consistent with these modeling studies above, suggesting that contact between EC1 and the ASIP C-terminal loop is key for MC1R antagonism, and explains, in part, why wildtype AgRP fails to bind to MC1R.
Figure 6.
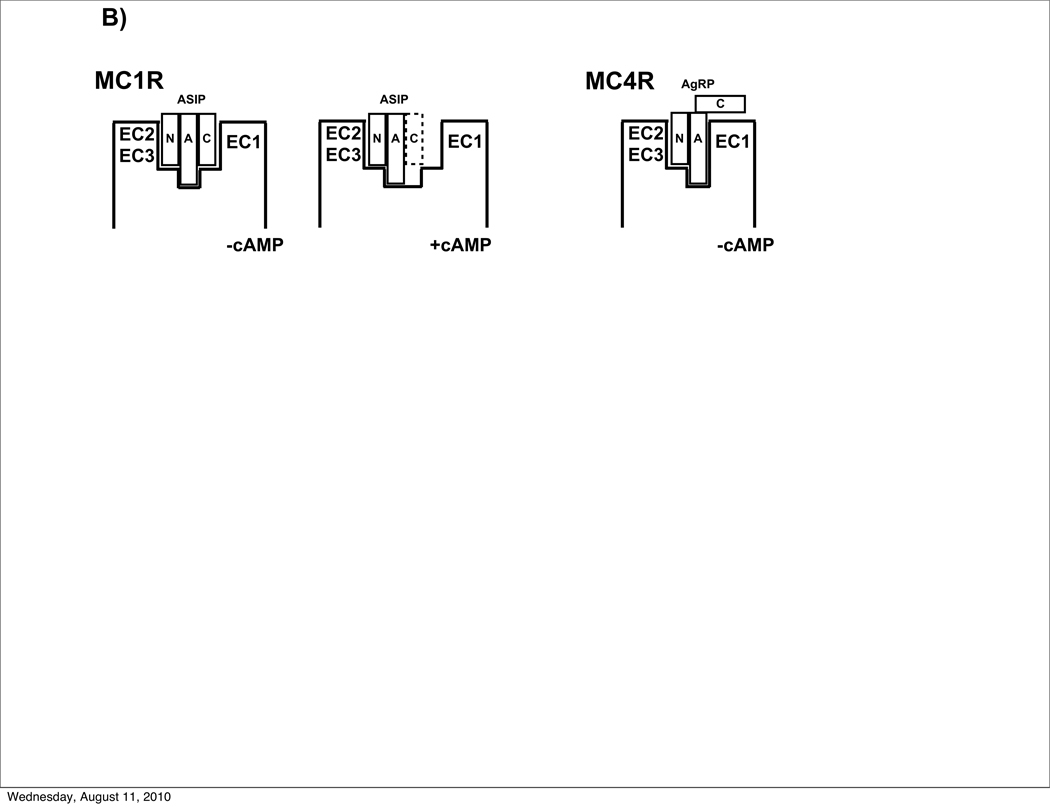
A) Model of ASIP (red) docked to the extracellular domain of MC1R (blue with cyan extracellular loops), developed by Chai et al.39 Data presented here demonstrate the importance of the ASIP C-terminal loop in MC1R recognition. We propose that the C-terminal loop interacts with EC1 through hydrophobic contacts (non-polar side chains are in white; Ala residues are not shown). In addition, R109 from the MC1R EC1 is positioned to form a salt bridge with ASIP D109, providing further stabilization. Coordinates, kindly provided by H. Mosberg and I. Pogozheva, are available at http://mosberglab.phar.umich.edu/resources/index.php. B) Schematic of ASIP-MC1R vs AgRP-MC4R interactions. Whereas AgRP requires only its N-terminal (N) and active (A) loops, ASIP also requires its C-terminal loop (C), to favor the non-signaling receptor state. With an altered (dashed lines) or missing ASIP C-terminal loop, ASIP functions as an agonist.
Figure 6B shows a general scheme that brings together our findings here and molecular details from modeling studies. At MC4R, AgRP interacts primarily through its N-terminal and active loops, which bind to EC2/3 and the transmembrane pocket, respectively, thus stabilizing the inactive state and suppressing cAMP production. The AgRP C-terminal loop makes only a weak contact with the receptor. In contrast, suppression of cAMP production by MC1R requires ASIP binding through all three of its loops. Without the C-terminal loop, the ASIP knottin domain, defined by its N-terminal and active loops, binds to a receptor conformation that stimulates release of intracellular cAMP.
The strict requirements for ASIP mediated antagonism at MC1R may have important functional and evolutionary implications. It is likely that ancestral vertebrates possessed a single melanocortin receptor, and a set of cognate ligands, that served to simultaneously regulate skin pigmentation, body temperature and metabolism.28 With differentiation of MCR subtypes later in vertebrate evolution, the receptor-specific functional responses became fine-tuned to their respective biological requirements. MC4R is found in a number of internal tissues, with its most well characterized function in the brain where it regulates feeding and metabolism. Here, specificity of spatial action depends on synaptic contacts where there is a tightly controlled and precisely regulated microenvironment with ligand and receptor typically expressed in specialized pre-synaptic and post-synaptic regions, respectively. By contrast, MC1R is expressed primarily by melanocytes in the skin, and its major ligand, ASIP, has a radius of action that must reach several cell diameters but no further, since ASIP is normally released from dermal papilla cells at the base of hair follicles, and affects MC1R function in melanocytes of overlying but not adjacent follicles. Thus, one possible explanation for the more elaborate binding mechanisms that exist between ASIP and MC1R compared to AgRP (or ASIP) and MC4R is that spatial requirements for MC1R action do not make use of a specialized anatomic structures, and instead depend more on molecular features that have evolved in both ligand and receptor. Moreover, melanocytes are exposed to a wide range of hormones, growth factors and other small signaling proteins.42 The remarkable ASIP-MC1R specificity may filter out spurious signaling, especially from peptide and proteins possessing positively charged loop segments.
The strict requirements for MC1R antagonism might suggest that ASIP’s action is absolutely unique, but a recent investigation of canine coat color identified β-defensins as a surprising new class of MC1R ligands.43 β-Defensins are produced by the innate immune system and possess well characterized antimicrobial activity. A current hypothesis is that β-defensin 3 (BDEF103 in dogs) acts as a neutral antagonist blocking ASIP, thus driving black coat pigmentation, a question that can be addressed by additional structure-function studies analogous to the ones described here.
Understanding more about the biochemistry of MC1R signaling could have important health consequences. Normal variation in human skin color is thought to represent a fine balance between the beneficial and detrimental effects of UV irradiation on vitamin D absorption and skin damage, respectively. MC1R agonists stimulate DNA repair, thus complementing strategies for reducing the risk of skin damage and melanoma.27 Our work indicating that all three MC1R exoloops are involved in antagonist function suggests that fragment based drug development, based on linking together low affinity binding elements at the individual loops,44 might serve as a useful platform for the development of synthetic MC1R specific antagonists or agonists.
Materials and Method
Peptide Synthesis and Folding
All peptides were synthesized using Fmoc chemistry beginning with a rink amide resin. Appropriately protected amino acids were added in stepwise fashion and the only variation was the incorporation of cysteines. In order to avoid enantiomerization, preactivated Cys was incorporated base-free. Upon completion of the synthesis of each peptide, the final Fmoc group was removed and the amino terminus acetylated. Peptides were cleaved from resin with 95% TFA, 2% TIS, 2% EDT, 1% phenol leaving the final peptide with an amidated C-terminus and acetylated N-terminus. After cleavage, each peptide was precipitated out of the TFA solution by addition of ethyl ether. Mixtures were then centrifuged to separate the precipitated peptide, and the pellet was redissolved in a 50:50 (volume:volume) acetonitrile-water solution, diluted to 95% water and purified using reverse phase HPLC. The resulting fractions were analyzed using electrospray mass spectrometry. Peptides were folded in an oxidizing buffer (1.6 M GuHCL, 0.1 M THAM, 4.8 mM reduced glutathione, 0.48 mM oxidized glutathione and 10% DMSO (v/v) at pH 8). Folding was monitored using reverse phase HPLC and was allowed to continue until one peak dominated over other partially folded and unfolded species (time ranged from 6 hours - 72 hours). The fraction corresponding to the folded peptide was subjected to another round of reverse phase HPLC purification. Analytical HPLC and electrospray mass spectrometry were conducted on a small sample of each folded peptide to confirm the presence of a single species of correct molecular weight (within 1 amu), accounting for the loss of two atomic masses for each disulfide bond. Amino acid analysis (UC Davis Molecular Structure Facility) was used to determine subsequent peptide concentrations in aqueous solution. These concentrations were then used to calibrate the UV-Vis absorbance and from which molar extinction coefficient was obtained, giving an accuracy of 10% to 15% for all peptide solutions 45. Peptide sequences are listed in table 1.19; 20; 24; 25
All constructs containing the ASIP active loop were based on the ASIP-YY sequence, which contains the mutations Q115Y and S124Y (human sequence) to ensure complete folding.19 Despite the stability of ASIP-YY, minimal constructs with ASIP’s N-terminal loop, but lacking a C-terminal loop, failed to fold to completion. Considering the known Xaa-Pro isomerization and multiple conformations in the PPAPA sequence (residues 102 – 106 in human ASIP) within the N-terminal loop, 19 an additional double Pro→Ala mutation was introduced to give PAAAA. Previous pharmacological studies showed that the PAAAA sequence facilitates efficient folding without significantly altering ASIP’s binding or inhibition at MC1R or MC4R.19 To probe for possible conformational or fold perturbation relative to the wildtype, full NMR structures were determined for both C-terminal and mini-ASIP (ASIP(93–126)) with P103A and P105A mutations (PDB codes 2KZA and 2L1J, respectively; see Supplement). Each protein gives a single low energy conformation, confirming inhibition of Xaa-Pro isomerization, with disulfide connectivities, β-sheet structure and hydrogen bonding patterns consistent with wildtype C-terminal ASIP. Backbone RMSD values comparing these two constructs to their respective polypeptide segments in wildtype, C-terminal ASIP found deviations of approximately 1.0 Å. (Structures, NMR statistics and NOE patterns are available in the Supplement.)
Binding Assays
All binding assays were performed on intact HEK 293T cells that were transiently transfected with human melanocortin receptor expression constructs. Transfection with calcium phosphate was achieved by adding 10 µg of each melanocortin receptor expression construct to a 10 cm dish of 293T cells and monitored by comparing to parallel transfections with a GFP-tagged MC4R construct. The media was changed after 12–16 hours, and after 24 hours the cells were treated with trypsin, then washed in media, and resuspended in binding buffer. After resuspension, the mixture was partitioned into a 96-well Acrowell filtration plate (Pall). A DELFIA lanthanide-based detection system was used to monitor displacement.43 The assays were setup with 1.8–3.0 nM of Eu-NDP-MSH and varying concentrations of the chimeric peptide. After incubating for 2 hours at 37 °C, the binding buffer was filtered by centrifugation and the wells were washed three times with 200 µL of ice-cold buffer. Next, 150 µL of enhancement solution (Perkin-Elmer) was added to each well and incubated for 45 minutes. Time-resolved fluorescence was measured using a FluoStar Optima plate reader (BMG Labtech). The cell concentration (20,000–70,000 per well) and Eu-NDP-MSH concentration (1.8–3.0 nM) were estimated from pilot experiments. After estimation of Bmax and Ki, concentrations were adjusted to maintain a dynamic range and avoid depletion of free ligand by more than 10%. Nonspecific binding was measured using the same procedure, but in the presence of varying concentrations of unlabelled NDP-MSH. Data were fit with a sigmoidal dose response curve with variable slope and analyzed using Graphpad and reported as Ki values (Graphpad Software, San Diego, CA).
cAMP Production
cAMP production assays were conducted using the TRM 432 kit (Amersham Pharmacia Biotech) on HEK-293 cells, stably transfected with the human melanocortin receptors (MC1R and MC4R).46 After removal of the cell culture medium, the cells were incubated for 30 minutes at 37 °C with 0.5 mL of Earle’s balanced salt solution (EBSS), 10−3 M isobutlymethylxanthine (IBMX), NDP-MSH (10−10–10−6 M) and the peptide of interest (10−7M). The reaction is quenched by adding 500 µL/well of ice-cold 100% ethanol and then centrifuged for 10 minutes at 1900 × g. After centrifugation, the supernatant was evaporated with pre-purified nitrogen gas, while kept at 55°C using a water bath. cAMP generation was measured according to the kit protocols. All experiments were performed in triplicate and data analyzed using Graphpad Prism (Graphpad Software, San Diego, CA) or KaleidaGraph (Synergy Software).47; 48 Direct agonist and inverse agonist function, probed in the absence of NDP-MSH, followed the same procedure.
Supplementary Material
Acknowledgements
The authors are grateful to Professor Henry I. Mosberg and Dr. Irina Pogozheva, University of Michigan, for providing ASIP-MC1R model coordinates and for insightful comments on interpretation of our data, and to Professor Carrie Haskell-Luevano for helpful discussion.
Supported by NIH Grant and ARRA supplement DK064265 (G. L. M.).
Abbreviations
- α-MSH
α-melanocyte stimulating hormone
- ACTH
adrenocortotropic hormone
- AgRP
agouti-related protein
- ASIP
agouti signaling protein
- AMU
atomic mass unit
- cAMP
cyclic adenosine monophosphate
- DMSO
dimethyl sulfoxide
- EC50
half maximal effective concentration
- EDT
ethane dithiol
- Eu
europium
- Fmoc
fluorenylmethyloxycarbonyl
- GuHCL
guanidine hydrochloride
- HPLC
high-performance liquid chromatography
- IC50
half maximal inhibitory concentration
- ICK
inhibitor cystine knot
- MCR
melanocortin receptor
- MC1R/MC3R/MC4R
melanocortin receptor 1/3/4
- NDP-MSH
[Nle4, D-Phe7]alpha-MSH
- NMR
nuclear magnetic resonance
- TIS
triisopropylsilane
- TFA
trifluoroacetic acid
- THAM
tris(hydroxymethyl)aminomethane;
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dinulescu DM, Cone RD. Agouti and agouti-related protein: analogies and contrasts. J Biol Chem. 2000;275:6695–6698. doi: 10.1074/jbc.275.10.6695. [DOI] [PubMed] [Google Scholar]
- 2.Wikberg JES, Muceniece R, Mandrika I, Prusis P, Lindblom J, Post C, Skottner A. New aspects on the melanocortins and their receptors. Pharmacological Research. 2000;42:393–420. doi: 10.1006/phrs.2000.0725. [DOI] [PubMed] [Google Scholar]
- 3.Gantz I, Fong TM. The melanocortin system. Am J Physiol Endocrinol Metab. 2003;284:E468–E474. doi: 10.1152/ajpendo.00434.2002. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Malek ZA. Melanocortin receptors: their functions and regulation by physiological agonists and antagonists. Cell Mol Life Sci. 2001;58:434–441. doi: 10.1007/PL00000868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mountjoy KG. Functions for pro-opiomelanocortin-derived peptides in obesity and diabetes. Biochem J. 2010;428:305–324. doi: 10.1042/BJ20091957. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard LE, White A. Neuropeptide processing and its impact on melanocortin pathways. Endocrinology. 2007;148:4201–4207. doi: 10.1210/en.2006-1686. [DOI] [PubMed] [Google Scholar]
- 7.Ollmann MM, Lamoreux ML, Wilson BD, Barsh GS. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998;12:316–330. doi: 10.1101/gad.12.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 9.Cone RD. Studies on the physiological functions of the melanocortin system. Endocrine Reviews. 2006;27:736–749. doi: 10.1210/er.2006-0034. [DOI] [PubMed] [Google Scholar]
- 10.Millhauser GL, McNulty JC, Jackson PJ, Thompson DA, Barsh GS, Gantz I. Loops and links: structural insights into the remarkable function of the agouti-related protein. Annals of the New York Academy of Sciences. 2003;994:27–35. doi: 10.1111/j.1749-6632.2003.tb03159.x. [DOI] [PubMed] [Google Scholar]
- 11.Adan RAH, Kas MJH. Inverse agonism gains weight. Trends Pharmacol Sci. 2003;24:315–321. doi: 10.1016/S0165-6147(03)00130-5. [DOI] [PubMed] [Google Scholar]
- 12.Chai BX, Neubig RR, Millhauser GL, Thompson DA, Jackson PJ, Barsh GS, Dickinson CJ, Li JY, Lai YM, Gantz I. Inverse agonist activity of agouti and agouti-related protein. Peptides. 2003;24:603–609. doi: 10.1016/s0196-9781(03)00104-9. [DOI] [PubMed] [Google Scholar]
- 13.Le Pape E, Wakamatsu K, Ito S, Wolber R, Hearing VJ. Regulation of eumelanin/pheomelanin synthesis and visible pigmentation in melanocytes by ligands of the melanocortin 1 receptor. Pigment Cell & Melanoma Research. 2008;21:477–486. doi: 10.1111/j.1755-148X.2008.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- 15.Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 16.Kaelin CB, Candille SI, Yu B, Jackson PJ, Thompson DA, Nix MA, Binkley J, Millhauser GL, Barsh GS. New ligands for melanocortin receptors. Int J Obes Relat Metab Disord. 2008;32 Suppl 7:S19–S27. doi: 10.1038/ijo.2008.234. [DOI] [PubMed] [Google Scholar]
- 17.Yen TT, Gill AM, Frigeri LG, Barsh GS, Wolff GL. Obesity, diabetes, and neoplasia in yellow A(vy)/- mice: ectopic expression of the agouti gene. FASEB J. 1994;8:479–488. doi: 10.1096/fasebj.8.8.8181666. [DOI] [PubMed] [Google Scholar]
- 18.Shutter JR, Graham M, Kinsey AC, Scully S, Lüthy R, Stark KL. Hypothalamic expression of ART, a novel gene related to agouti, is up-regulated in obese and diabetic mutant mice. Genes & Development. 1997;11:593–602. doi: 10.1101/gad.11.5.593. [DOI] [PubMed] [Google Scholar]
- 19.McNulty JC, Jackson PJ, Thompson DA, Chai BX, Gantz I, Barsh GS, Dawson PE, Millhauser GL. Structures of the agouti signaling protein. J Mol Biol. 2005;346:1059–1070. doi: 10.1016/j.jmb.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 20.Jackson PJ, McNulty JC, Yang YK, Thompson DA, Chai BX, Gantz I, Barsh GS, Millhauser GL. Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity. Biochemistry. 2002;41:7565–7572. doi: 10.1021/bi012000x. [DOI] [PubMed] [Google Scholar]
- 21.McNulty JC, Thompson DA, Bolin KA, Wilken J, Barsh GS, Millhauser GL. High-resolution NMR structure of the chemically-synthesized melanocortin receptor binding domain AGRP(87–132) of the agouti-related protein. Biochemistry. 2001;40:15520–15527. doi: 10.1021/bi0117192. [DOI] [PubMed] [Google Scholar]
- 22.Tota MR, Smith TS, Mao C, MacNeil T, Mosley RT, Van der Ploeg LHT, Fong TM. Molecular interaction of Agouti protein and Agouti-related protein with human melanocortin receptors. Biochemistry. 1999;38:897–904. doi: 10.1021/bi9815602. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer LL, Veal JM, Mountjoy KG, Wilkison WO. Melanocortin receptor binding determinants in the agouti protein. Biochemistry. 1998;37:991–997. doi: 10.1021/bi971913h. [DOI] [PubMed] [Google Scholar]
- 24.Bolin KA, Anderson DJ, Trulson JA, Thompson DA, Wilken J, Kent SB, Gantz I, Millhauser GL. NMR structure of a minimized human agouti related protein prepared by total chemical synthesis. FEBS Lett. 1999;451:125–131. doi: 10.1016/s0014-5793(99)00553-0. [DOI] [PubMed] [Google Scholar]
- 25.Jackson PJ, Yu B, Hunrichs B, Thompson DA, Chai BX, Gantz I, Millhauser GL. Chimeras of the agouti-related protein: insights into agonist and antagonist selectivity of melanocortin receptors. Peptides. 2005;26:1978–1987. doi: 10.1016/j.peptides.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 26.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Malek ZA, Kadekaro AL, Kavanagh RJ, Todorovic A, Koikov LN, McNulty JC, Jackson PJ, Millhauser GL, Schwemberger S, Babcock G, Haskell-Luevano C, Knittel JJ. Melanoma prevention strategy based on using tetrapeptide alpha-MSH analogs that protect human melanocytes from UV-induced DNA damage and cytotoxicity. FASEB J. 2006;20:1561–1563. doi: 10.1096/fj.05-5655fje. [DOI] [PubMed] [Google Scholar]
- 28.Jackson PJ, Douglas NR, Chai BX, Binkley J, Sidow A, Barsh GS, Millhauser GL. Structural and molecular evolutionary analysis of Agouti and Agouti-related proteins. Chem Biol. 2006;13:1297–1305. doi: 10.1016/j.chembiol.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creemers JWM, Pritchard LE, Gyte A, Le Rouzic P, Meulemans S, Wardlaw SL, Zhu X, Steiner DF, Davies N, Armstrong D, Lawrence CB, Luckman SM, Schmitz CA, Davies RA, Brennand JC, White A. Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)83–132: interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology. 2006;147:1621–1631. doi: 10.1210/en.2005-1373. [DOI] [PubMed] [Google Scholar]
- 30.Arolas JL, Aviles FX, Chang JY, Ventura S. Folding of small disulfide-rich proteins: clarifying the puzzle. Trends Biochem Sci. 2006;31:292–301. doi: 10.1016/j.tibs.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39:43–60. doi: 10.1016/s0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 32.Ho G, MacKenzie RG. Functional characterization of mutations in melanocortin-4 receptor associated with human obesity. J Biol Chem. 1999;274:35816–35822. doi: 10.1074/jbc.274.50.35816. [DOI] [PubMed] [Google Scholar]
- 33.Chhajlani V, Wikberg JE. Molecular cloning and expression of the human melanocyte stimulating hormone receptor cDNA. FEBS Lett. 1992;309:417–420. doi: 10.1016/0014-5793(92)80820-7. [DOI] [PubMed] [Google Scholar]
- 34.Haskell-Luevano C, Monck EK, Wan YP, Schentrup AM. The agouti-related protein decapeptide (Yc[CRFFNAFC]Y) possesses agonist activity at the murine melanocortin-1 receptor. Peptides. 2000;21:683–689. doi: 10.1016/s0196-9781(00)00194-7. [DOI] [PubMed] [Google Scholar]
- 35.Siegrist W, Drozdz R, Cotti R, Willard DH, Wilkison WO, Eberle AN. Interactions of alpha-melanotropin and agouti on B16 melanoma cells: evidence for inverse agonism of agouti. J Recept Signal Transduct Res. 1997;17:75–98. doi: 10.3109/10799899709036595. [DOI] [PubMed] [Google Scholar]
- 36.Haskell-Luevano C, Cone RD, Monck EK, Wan YP. Structure activity studies of the melanocortin-4 receptor by in vitro mutagenesis: identification of agouti-related protein (AGRP), melanocortin agonist and synthetic peptide antagonist interaction determinants. Biochemistry. 2001;40:6164–6179. doi: 10.1021/bi010025q. [DOI] [PubMed] [Google Scholar]
- 37.Yang YK, Dickinson CJ, Zeng Q, Li JY, Thompson DA, Gantz I. Contribution of melanocortin receptor exoloops to Agouti-related protein binding. J Biol Chem. 1999;274:14100–14106. doi: 10.1074/jbc.274.20.14100. [DOI] [PubMed] [Google Scholar]
- 38.Wilczynski A, Wang XS, Joseph CG, Xiang Z, Bauzo RM, Scott JW, Sorensen NB, Shaw AM, Millard WJ, Richards NG, Haskell-Luevano C. Identification of putative agouti-related protein(87–132)-melanocortin-4 receptor interactions by homology molecular modeling and validation using chimeric peptide ligands. J Med Chem. 2004;47:2194–2207. doi: 10.1021/jm0303608. [DOI] [PubMed] [Google Scholar]
- 39.Chai B-X, Pogozheva ID, Lai Y-M, Li J-Y, Neubig RR, Mosberg HI, Gantz I. Receptor-antagonist interactions in the complexes of agouti and agouti-related protein with human melanocortin 1 and 4 receptors. Biochemistry. 2005;44:3418–3431. doi: 10.1021/bi0478704. [DOI] [PubMed] [Google Scholar]
- 40.Haitina T, Ringholm A, Kelly J, Mundy NI, Schioth HB. High diversity in functional properties of melanocortin 1 receptor (MC1R) in divergent primate species is more strongly associated with phylogeny than coat color. Mol Biol Evol. 2007;24:2001–2008. doi: 10.1093/molbev/msm134. [DOI] [PubMed] [Google Scholar]
- 41.Mundy NI, Kelly J. Evolution of a pigmentation gene, the melanocortin-1 receptor, in primates. Am J Phys Anthropol. 2003;121:67–80. doi: 10.1002/ajpa.10169. [DOI] [PubMed] [Google Scholar]
- 42.Slominski A, Wortsman J, Plonka PM, Schallreuter KU, Paus R, Tobin DJ. Hair follicle pigmentation. J Invest Dermatol. 2005;124:13–21. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Millhauser GL, Barsh GS. A -defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annual review of biophysics and biomolecular structure. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 45.Yu B, Millhauser GL. Chemical disulfide mapping identifies an inhibitor cystine knot in the agouti signaling protein. FEBS Lett. 2007;581:5561–5565. doi: 10.1016/j.febslet.2007.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y-K, Dickinson C, Haskell-Luevano C, Gantz I. Molecular basis for the interaction of [Nle4,D-Phe7]melanocyte stimulating hormone with the human melanocortin-1 receptor. J Biol Chem. 1997;272:23000–23010. doi: 10.1074/jbc.272.37.23000. [DOI] [PubMed] [Google Scholar]
- 47.Yang YK, Thompson DA, Dickinson CJ, Wilken J, Barsh GS, Kent SB, Gantz I. Characterization of Agouti-related protein binding to melanocortin receptors. Mol Endocrinol. 1999;13:148–155. doi: 10.1210/mend.13.1.0223. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Celik A, Georgeson KE, Harmon CM, Yang YK. Molecular basis of melanocortin-4 receptor for AGRP inverse agonism. Regul Pept. 2006;136:40–49. doi: 10.1016/j.regpep.2006.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.