Abstract
S-nitrosylation by nitric oxide (NO) is a major mode of signaling to cellular proteins1, including prominent nuclear proteins such as HDAC22 and PARP13. The high reactivity of the NO group with protein thiols implies the existence of selective targeting mechanisms. Specificity of NO signaling is often achieved by the binding of NO synthase (NOS) to target proteins, either directly4 or through scaffolding proteins such as PSD-955 and CAPON6. As the three principal isoforms of NOS - neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS) - are primarily non-nuclear, the mechanisms by which nuclear proteins are selectively nitrosylated have been elusive. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is physiologically nitrosylated at its Cys150 residue, conferring upon it the ability to bind to Siah1, which possesses a nuclear localization signal and conveys nitrosylated GAPDH (SNO-GAPDH) to the nucleus7. We now show that SNO-GAPDH physiologically transnitrosylates nuclear proteins, including the deacetylating enzyme SIRT1, histone deacetylase-2 (HDAC2), and DNA-activated protein kinase (DNA-PK). Our findings reveal a novel mechanism for targeted nitrosylation of nuclear proteins and suggest that protein-protein transfer of NO groups may be a general mechanism in cellular signal transduction.
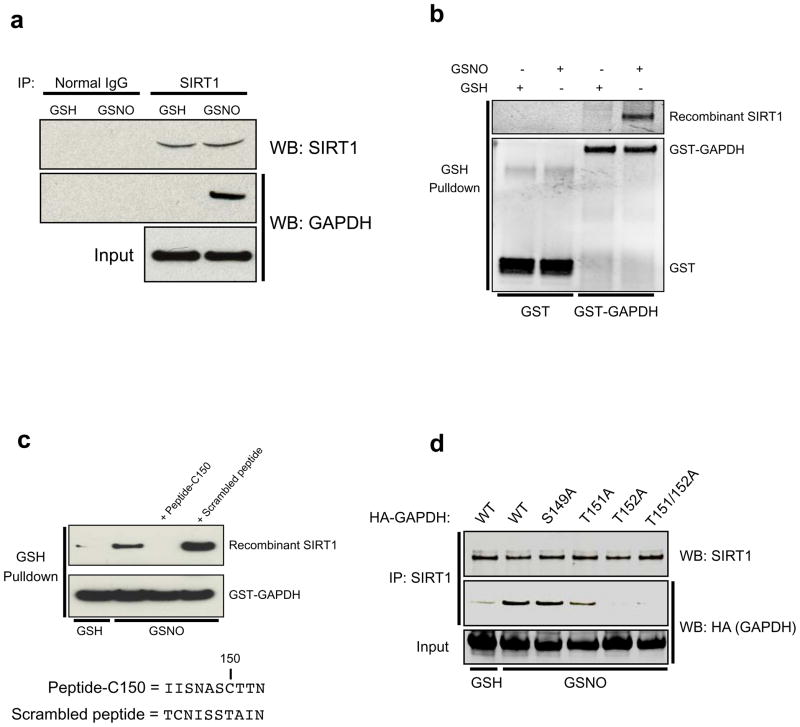
Previous work in our laboratory has shown that nuclear GAPDH regulates protein acetylation by binding to and activating the acetyltransferases p300 and CBP8. We wondered whether GAPDH might also regulate the reverse process of protein deacetylation by interaction with deacetylases. A two-hybrid-based protein-interaction map of the Drosophila melanogaster proteome created by Giot et al.9 revealed a potential physical interaction between GAPDH and the Drosophila protein Sir2, a prominent NAD-activated nuclear protein deacetylase. This led us to examine whether GAPDH physically interacts with SIRT1, a close mammalian homolog of Drosophila Sir210. In intact cells, SIRT1 robustly binds GAPDH, but only upon treatment with the NO donor S-nitrosoglutathione (GSNO) (Fig. 1a). These findings do not merely reflect GSNO eliciting movement of GAPDH to the nucleus, as nitrosylation of purified GAPDH is required for its interaction with SIRT1 in vitro (Fig. 1b). Mutation of the nitrosylated Cys150 residue of GAPDH abolishes binding (Supplementary Information, Fig. S1a). Moreover, GAPDH-C150 appears to be within the binding site for SIRT1, as a ten amino acid peptide which spans Cys150 (Peptide-C150), but not a scrambled peptide, selectively prevents the binding of SNO-GAPDH to SIRT1 in vitro (Fig. 1c). Further characterization of the interaction domain reveals a critical role for the single amino acid Thr152 of GAPDH, as mutation of this residue abolishes the physical interaction with SIRT1 (Fig. 1d).
Figure 1. SNO-GAPDH interacts with SIRT1 near its nitrosylated Cys150 residue.
(a) Endogenous co-immunoprecipitation of SIRT1 and GAPDH in HEK293 cells treated with NO donor. Cells were treated with 200 μM GSH or GSNO for 16 hr prior to lysis. (b) Nitrosylated GAPDH (SNO-GAPDH) binds directly to SIRT1 in vitro. GST or GST-GAPDH was pre-treated with 100 μM GSH or GSNO for 30 min at 37°C. After desalting, recombinant SIRT1 was added and binding assessed by a GSH-agarose pulldown assay. (c) A small peptide corresponding to the region of GAPDH that spans Cys150 (Peptide-C150) blocks the interaction between SNO-GAPDH and SIRT1. The assay was performed as in b. (d) Mutation of Thr152 of GAPDH abolishes binding to SIRT1. Twenty-four hours after transfection with wild-type HA-GAPDH or the indicated point mutants, HEK293 cells were treated with 200 μM GSH or GSNO for 16 hr. Cell lysates were immunoprecipitated with anti-SIRT1 antibody and analyzed by western blotting with anti-HA antibody. HA-GAPDH, HA-tagged GAPDH.
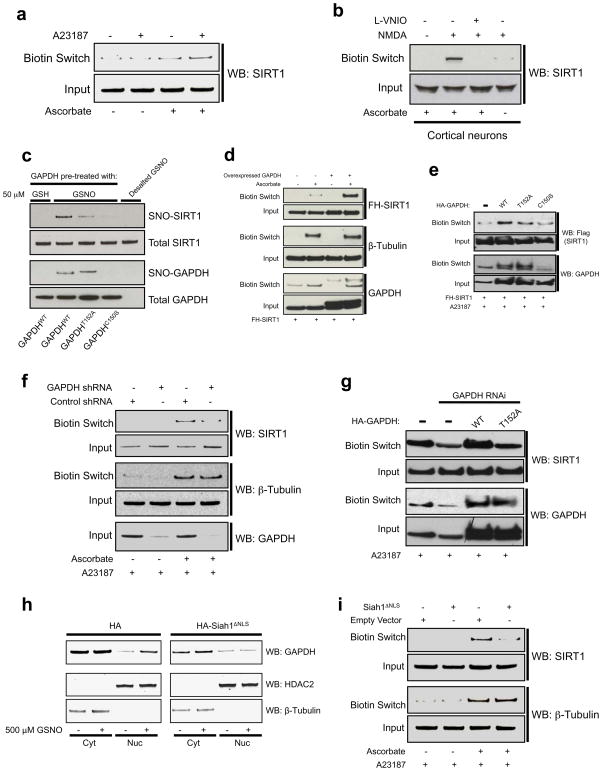
Given the role of NO in the GAPDH-SIRT1 interaction, we wondered whether SIRT1 is itself nitrosylated. To test this, we utilized the biotin switch assay11. SIRT1 is nitrosylated in cells treated with GSNO (Supplementary Information, Fig. S1b) as well as when exposed in vitro to GSNO (Supplementary Information, Fig. S1c), confirming observations of Kaneki et al12, 13. To examine whether endogenous SIRT1 is nitrosylated by an endogenous source of NO, we performed a biotin switch from HEK293 cells stably expressing nNOS (293-nNOS) (Fig. 2a). SIRT1 is robustly nitrosylated within two hours of treatment with the calcium ionophore A23187 (5 μM). SIRT1 is also nitrosylated in mouse embryonic cortical neurons following treatment with the glutamate derivative N-methyl-D-aspartate (NMDA) (Fig. 2b). Pre-treatment of these cells with the specific nNOS inhibitor Vinyl-L-NIO (L-VNIO, 100 μM) abolishes NMDA-induced SIRT1 nitrosylation.
Figure 2. Nuclear SNO-GAPDH mediates nitrosylation of SIRT1 via transnitrosylation.
(a) Endogenous SIRT1 is nitrosylated in 293-nNOS cells treated with the calcium ionophore A23187 (5 μM, 2 hr). (b) SIRT1 is nitrosylated in cortical neurons treated with NMDA. Nitrosylation is abolished by pre-treatment with the nNOS inhibitor L-VNIO (100 μM, 2 hr). (c) In vitro transnitrosylation assay. Recombinant SIRT1 was incubated with recombinant GAPDH (wild-type, T152A, or C150S, approx. 0.75 μM) that had been pre-treated with 50 μM GSH or GSNO and desalted to remove excess small molecules. A biotin switch assay was then performed. (d) Overexpression of GAPDH augments SIRT1 nitrosylation in 293-nNOS cells. (e) Mutation of Thr152 or Cys150 abrogates the effect of GAPDH overexpression. (f) Depletion of GAPDH by RNAi in 293-nNOS cells leads to a loss of nitrosylation of endogenous SIRT1. (g) The effect of GAPDH knockdown on SIRT1 nitrosylation is rescued by wild-type GAPDH but not GAPDH-T152A. (h) Siah1ΔNLS prevents NO-induced nuclear translocation of GAPDH. 293T cells were transfected with the indicated plasmid and treated with or without 500 μM GSNO for 3 hr followed by nuclear fractionation. (i) Siah1ΔNLS abolishes nitrosylation of endogenous SIRT1 in 293-nNOS cells. FH-SIRT1, Flag-HA-tagged SIRT1. Where indicated, cells were treated with 5 μM A23187 for 2 hr. Results from this figure are quantified in Supplementary Information, Fig. S2.
Mutational analysis identified Cys387 and Cys390, residues that are critical to SIRT1 function14, as the sites of SIRT1 nitrosylation (Supplementary Information, Fig. S1d). Since single mutation of either of these residues abolishes the biotin switch signal, it is unclear whether both residues are nitrosylated or, alternatively, allosteric interactions result in the mutation of one residue preventing the nitrosylation of the other. Since these two cysteines participate in coordination of a zinc ion, one could ask whether their mutation abolishes nitrosylation by eliciting protein misfolding. However, mutation of Cys363 and Cys366, which participate with Cys387 and Cys390 in zinc ion coordination, has no effect on SIRT1 nitrosylation.
Since SNO-GAPDH translocates to the nucleus and binds SIRT1 near its nitrosylated Cys150 residue, we hypothesized that SNO-GAPDH is the physiologic source of NO for SIRT1 nitrosylation via a protein-protein transnitrosylation reaction. To determine whether SNO-GAPDH can transfer its NO group to SIRT1, we performed an in vitro transnitrosylation assay (Fig. 2c; Supplementary Information, Fig. S2a). Purified GAPDH (wild-type, T152A, or C150S) was exposed to GSNO (50 μM) to induce nitrosylation, with glutathione (GSH) treatment serving as a control. After desalting to remove excess NO donor, the proteins were incubated with purified SIRT1, followed by a biotin switch assay to determine the extent of SIRT1 nitrosylation. Incubation with wild-type GAPDH, but not GAPDH-C150S, leads to nitrosylation of SIRT1, reflecting transfer of the NO group from the Cys150 residue of SNO-GAPDH to SIRT1. Furthermore, a binding interaction between the two proteins is required, as mutation of GAPDH-T152 abrogates transnitrosylation of SIRT1. Peptide-C150, which blocks GAPDH-SIRT1 binding, also prevents SIRT1 nitrosylation by SNO-GAPDH (Supplementary Information, Fig. S3a). Additionally, as further evidence for the specificity of the reaction, nitrosylated albumin does not nitrosylate SIRT1 in vitro (Supplementary Information, Fig. S3b).
Interestingly, SNO-GAPDH appears to be much more efficient than GSNO at nitrosylating SIRT1, as approximately 0.75 μM total GAPDH leads to substantial SIRT1 nitrosylation in vitro, with the concentration of SNO-GAPDH being less than that of total GAPDH. By contrast, in vitro nitrosylation with GSNO requires a minimum concentration of 10 μM (Supplementary Information, Fig. S1c).
To directly examine an influence of GAPDH upon SIRT1 nitrosylation in cells, we overexpressed SIRT1 in 293-nNOS cells with or without co-overexpression of GAPDH (Fig. 2d; Supplementary Information, Fig. S2b). Overexpression of GAPDH leads to substantially elevated levels of SIRT1 nitrosylation. By contrast, β-tubulin is nitrosylated to the same extent in the absence or presence of overexpressed GAPDH. GAPDH-SIRT1 binding is required for this effect, as overexpression of GAPDH-T152A fails to augment SIRT1 nitrosylation to the same extent as wild-type (Fig. 2e; Supplementary Information, Fig. S2c). Mutation of Cys150 also abrogates the effect of GAPDH on nitrosylation of SIRT1. Importantly, GAPDH-mediated nitrosylation of SIRT1 does not require NOS, as overexpression of GAPDH substantially augments SIRT1 nitrosylation in 293 cells treated with GSNO (Supplementary Information, Fig. S3c). Furthermore, mutation of Thr152 of GAPDH has no appreciable effect on its glycolytic activity (Supplementary Information, Fig. S3d). The biotin switch signal from the nitrosylation-null SIRT1-C387/390S mutant is unaffected by GAPDH overexpression (Supplementary Information, Fig. S3e).
To ascertain whether basal levels of GAPDH physiologically regulate nitrosylation of endogenous SIRT1, we depleted GAPDH from 293-nNOS cells by RNA interference, which leads to a loss of SIRT1 nitrosylation with no alteration in β-tubulin nitrosylation (Fig. 2f; Supplementary Information, Fig. S2d). The effect of GAPDH knockdown is rescued by expression of wild-type GAPDH but not GAPDH-T152A (Fig. 2g; Supplementary Information, Fig. S2e). GAPDH depletion also abolishes nitrosylation of exogenously expressed SIRT1 (Supplementary Information, Fig. S3f).
These experiments establish that GAPDH-SIRT1 binding is required for SIRT1 nitrosylation but do not prove that nuclear GAPDH specifically mediates the nitrosylation. Accordingly, we devised an approach to prevent nuclear translocation of GAPDH. GAPDH is translocated to the nucleus by binding to Siah1, which possesses a nuclear localization signal. We employed Siah1 lacking a nuclear localization signal (Siah1ΔNLS), which acts in a dominant-negative manner to prevent nuclear translocation of GAPDH (Fig. 2h). Siah1ΔNLS abolishes nitrosylation of endogenous SIRT1 but has no effect on β-tubulin nitrosylation (Fig. 2i; Supplementary Information, Fig. S2f). Siah1ΔNLS also abolishes nitrosylation of exogenously expressed SIRT1 (Supplementary Information, Fig. S3g).
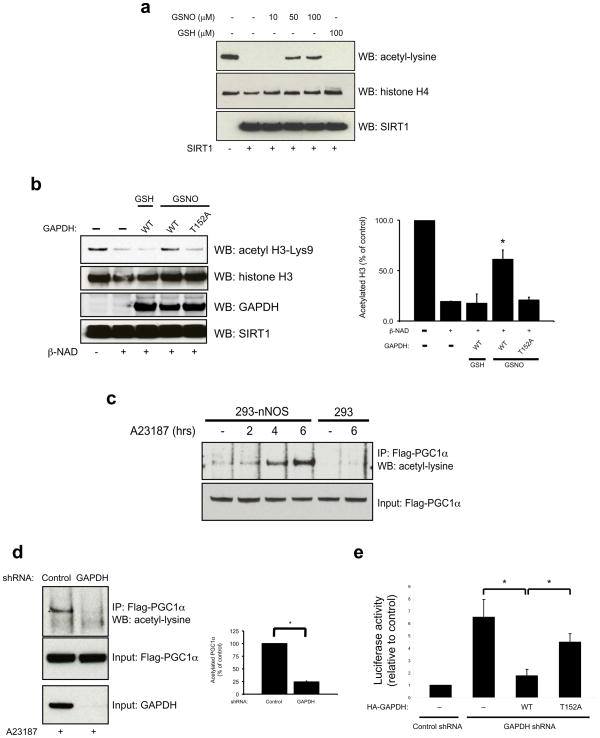
If SNO-GAPDH physiologically nitrosylates SIRT1, it should also regulate influences of SIRT1 nitrosylation upon the enzyme’s catalytic activity. To test this, we utilized an in vitro histone deacetylation assay. GSNO treatment inhibits SIRT1 catalytic activity (Fig. 3a), confirming observations of Kaneki et al12, 13 that nitrosylation inhibits the enzymatic function of SIRT1. SNO-GAPDH also inhibits SIRT1 activity, an effect that is abolished by mutation of Thr152 (Fig. 3b) or Cys150 (Supplementary Information, Fig. S3h) of GAPDH.
Figure 3. SNO-GAPDH mediates inhibition of SIRT1 enzymatic activity by nitric oxide.
(a) GSNO treatment inhibits SIRT1 catalytic activity in an in vitro histone deacetylation assay. SIRT1 was pre-treated with the indicated concentration of GSH or GSNO and desalted prior to the assay. (b) Pre-incubation with SNO-GAPDH inhibits SIRT1 catalytic activity in vitro. This effect is lost with mutation of GAPDH-T152. The assay was performed as in a. *P<0.05, n=3, mean±s.e.m., one-way ANOVA. (c) Activation of nNOS leads to increased acetylation of PGC1α. Twenty-four hours after transfection with Flag-PGC1α, HEK293 or 293-nNOS cells were treated with A23187 (5 μM) for the indicated times prior to lysis. (d) Depletion of GAPDH in 293-nNOS cells by RNAi leads to decreased acetylation of PGC1α. Cells were treated with A23187 (5 μM) for 6 hr prior to lysis. *P<0.01, n=3, mean ± s.e.m., student’s t-test. (e) GAPDH regulates PGC1α/HNF4α transcriptional activity via nitrosylation of SIRT1. Depletion of GAPDH in 293-nNOS cells transfected with plasmids encoding HNF4α and an HNF4α binding site luciferase leads to increased luciferase activity. This effect is rescued by wild-type GAPDH but not GAPDH-T152A. Cells were treated with 5 μM A23187 for 3 hr. *P<0.05, n=3, mean ± s.e.m., student’s t-test. Flag-PGC1α, Flag-tagged PGC1α.
We examined regulation of SIRT1 by SNO-GAPDH in intact cells. GAPDH augments the acetylation by p300/CBP of numerous substrates8. PGC1α, which is deacetylated exclusively by SIRT115, is one of the few, if not the only, SIRT1 substrates that is not acetylated by p300/CBP16 and so is an optimal substrate for experiments investigating the functional effects of GAPDH transnitrosylation of SIRT1. Treatment of 293-nNOS cells with the calcium ionophore A23187 (5 μM) leads to substantially elevated levels of acetylated PGC1α, reflecting inhibition of SIRT1 enzyme activity (Fig. 3c). The effect of A23187 reflects Ca2+/CaM-activation of nNOS, as A23187 has no effect on PGC1α acetylation in 293 cells. Depletion of GAPDH in 293-nNOS cells by RNA interference reduces levels of acetylated PGC1α, consistent with preventing the SNO-GAPDH mediated transnitrosylation and inhibition of SIRT1 (Fig. 3d). Conversely, overexpression of GAPDH in 293-nNOS cells markedly increases PGC1α acetylation (Supplementary Information, Fig. S3i). Nuclear localization of GAPDH is required for this effect, as GAPDH-K225A, which does not enter the nucleus7, fails to inhibit PGC1α deacetylation.
PGC1α is a transcriptional co-activator that induces gluconeogenic gene expression through its interaction with the transcription factor HNF4α17. Acetylation of PGC1α prevents its coactivation of HNF4α15. To further explore the physiological effects of GAPDH-mediated nitrosylation of SIRT1, we utilized a luciferase reporter assay to measure PGC1α/HNF4α transcriptional activity. As predicted, depletion of GAPDH leads to a substantial increase in PGC1α/HNF4α transcriptional activity, an effect that is rescued by expression of wild-type GAPDH but not GAPDH-T152A (Fig 3e).
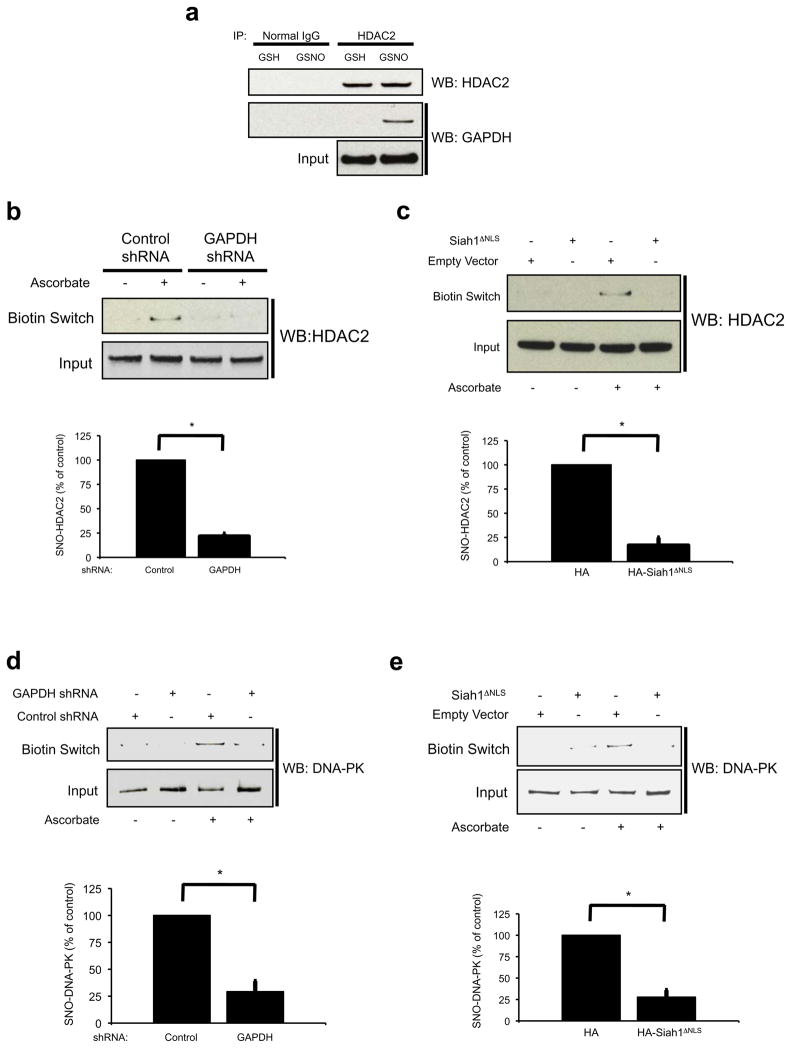
These experiments establish that nitrosylation of nuclear SIRT1 and inhibition of its catalytic activity are physiologically mediated by nuclear SNO-GAPDH via transnitrosylation. We wondered whether this process is limited to SIRT1 or reflects a more general mechanism for nitrosylating nuclear proteins. We first examined HDAC2, a nuclear protein shown by Riccio and associates2 to be physiologically nitrosylated. GAPDH binds to HDAC2 in intact cells treated with GSNO (Fig. 4a). We confirm nitrosylation of endogenous HDAC2 and show that depletion of GAPDH by RNA interference abolishes this nitrosylation (Fig. 4b). HDAC2 nitrosylation requires nuclear localization of GAPDH, as the nitrosylation is abolished in cells expressing Siah1ΔNLS (Fig. 4c). DNA-PK was also recently shown to be nitrosylated18, and we identified it in a screen for nuclear proteins whose nitrosylation is enhanced by GAPDH overexpression (Supplementary Information, Fig. S4). As with SIRT1 and HDAC2, nitrosylation of endogenous DNA-PK is abolished by depletion of GAPDH by RNA interference (Fig. 4d), as well as by expression of the Siah1ΔNLS dominant-negative construct (Fig. 4e).
Figure 4. Identification of HDAC2 and DNA-PK as nuclear targets of SNO-GAPDH mediated transnitrosylation.
(a) Endogenous co-immunoprecipitation of HDAC2 and GAPDH in HEK293 cells treated with NO donor. Cells were treated with 200 μM GSH or GSNO for 16 hr prior to lysis. (b) and (c) GAPDH knockdown (b) and Siah1ΔNLS expression (c) lead to a loss of HDAC2 nitrosylation in 293- nNOS cells. (d) and (e) Similar results were obtained for DNA-PK. *P<0.05, n=3, mean±s.e.m., student’s t-test.
The present study has established that nuclear proteins such as SIRT1, HDAC2, and DNA-PK are physiologically transnitrosylated by SNO-GAPDH. Previously, Stamler and colleagues reported the transnitrosylative transfer of NO from hemoglobin (Hb) to the anion exchanger AE1 at the plasma membrane of red blood cells, implicating the process in the systemic transport and release of NO bioactivity19. Not only was Hb required for AE1 nitrosylation, but transfer of NO from Hb to AE1 was necessary for NO-induced hypoxic vasodilation. Subsequently, Marletta and associates20,21 described movement of NO from Cys73 of thioredoxin-1 to caspase-3, while Stamler and colleagues22 reported that thioredoxin-1 and −2 denitrosylate caspase-3 via a conserved Cys32 residue. Very recently, Lipton and colleagues23 noted that caspase-3 can transfer NO to the antiapoptotic protein XIAP. Our findings firmly establish a role for protein-protein transnitrosylation in intracellular signaling by NO. Specifically, our identification of GAPDH-mediated nitrosylation of proteins following nuclear translocation reveals a mechanism for the selective propagation of nitrosylation-dependent signals between subcellular loci, yielding new insight into the compartmentalization of NO bioactivity.
There are some reports in the literature of nuclear localization of NOS. Nuclear eNOS and iNOS have been described in rodent brown adipocytes24, and nuclear translocation of eNOS has been observed in estradiol treated endothelial cells25, 26. The physiologic relevance of these reports is unclear, as most studies have found eNOS excluded from the nucleus, cycling instead between soluble and membrane-bound cytosolic compartments27. Sessa and colleagues have found major reductions in eNOS catalytic activity when it is targeted to the nucleus via fusion to an exogenous nuclear localization signal28. To our knowledge, there are no reports of nuclear nNOS. In the present study, we have found that GAPDH mediates SIRT1 nitrosylation independently of NOS, as overexpression of GAPDH augments SIRT1 nitrosylation following treatment with NO donor in cells lacking NOS expression (Fig. S3c).
We have shown that transnitrosylation by GAPDH inhibits SIRT1 enzymatic activity, leading to decreased transcriptional activity of PGC1α, a well established SIRT1 substrate. Given the importance of SIRT1 regulation of PGC1 α in metabolic pathways29, and particularly in the metabolic dysfunction associated with obesity30,31, the effect of GAPDH on the SIRT1-PGC1 α axis may have clinical relevance. Chronic inflammation is associated with the metabolic alterations seen in type 2 diabetes32, and NO is a major mediator of inflammation33. Thus, GAPDH-mediated inhibition of SIRT1 may afford a link between inflammation and metabolic dysfunction.
Because of its abundance in all cells and its well characterized nuclear translocation following nitrosylation, GAPDH is ideally suited to donate NO to nuclear proteins. In addition to proteins that primarily exist in the nucleus, transcription factors act in the nucleus but often cycle between the nucleus and cytosol, and some have been shown to be physiologically nitrosylated, e.g. NFkB34 and HIF1α35. It seems reasonable to speculate that some of these proteins may also be targets of transnitrosylation by SNO-GAPDH. Thus, SNO-GAPDH may regulate diverse nuclear activities. We have also detected substantial levels of SNO-GAPDH in cytosolic extracts (data not shown), suggesting that GAPDH may transnitrosylate cytosolic targets. Besides its conventional catalytic activity, GAPDH has been implicated in other cellular events such as transcriptional regulation36, DNA repair37, nuclear export of tRNAs38, and regulation of mRNA stability39.
In summary, our findings establish a novel signaling pathway whereby nuclear proteins can be physiologically nitrosylated, with GAPDH providing nuclear access for NO. Binding of GAPDH to target proteins provides selectivity in the nitrosylation process. Earlier formulations of NO as freely diffusing within cells failed to explain specificity of signaling. The binding of NOS isoforms directly to targets4 or via scaffolding proteins such as CAPON6 permits selectivity for cytosolic proteins. Transnitrosylation via GAPDH extends such specificity to nuclear proteins with substantial functional implications. Protein-protein transnitrosylation may be a general mechanism for selectively propagating the NO signal within defined cellular pathways.
Methods
Reagents
Unless otherwise stated, all biochemical reagents were purchased from Sigma and all cell culture reagents from Gibco. L-VNIO was from Alexis Biochemicals. Peptide C-150 (IISNASCTTN) and a scrambled peptide (TCNISSTAIN) were synthesized by GenScript. GAPDH and Siah1ΔNLS mammalian expression constructs were produced as described7. Flag-HA-tagged SIRT1 and Flag-tagged PGC1α were a kind gift from P. Puigserver. HNF4α expression construct was purchased from Open Biosystems, and HNF4α luciferase reporter assay kit was from SABiosciences. Antibodies were obtained from the following companies and used at the stated dilutions: anti-GAPDH monoclonal antibody (Calbiochem, WB 1:5,000); human SIRT1 was immunoprecipitated with polyclonal antibody from Abcam (1–2 μl/IP) and detected by western blot with polyclonal antibody from Biomol (1:1,000); mouse-derived SIRT1 was detected by western blot with polyclonal mouse specific antibody from Cell Signaling Technology (1:1,000); anti-β-Tubulin monoclonal antibody (Upstate, WB 1:5,000); anti-HA monoclonal antibody (Covance, WB 1:5,000); anti-biotin monoclonal antibody (Sigma, WB 1:2,000); anti-HDAC2 (WB 1:1,000), anti-acetylated-lysine polyclonal (WB 1:2,000), anti-acetylated H3-K9 (WB 1:5,000), anti-histone H3 (WB 1:1,000), anti-histone H4 (WB 1:500), and anti-DNA-PK (WB 1:1,000) antibodies were from Cell Signaling Technology; HRP-conjugated secondary antibodies were from GE Healthcare or eBioscience (TrueBlot system).
Cell culture
HEK293 and 293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) plus 10% FBS, 1 mM L-glutamine, and Pen-Strep at 37°C with a 5% CO2 atmosphere in a humidified incubator. 293-nNOS cells were maintained as above but with Pen-Strep-Neo. All transfections were performed with PolyFect reagent (Qiagen) according to the manufacturer’s protocol. Mouse embryonic cortical neurons were maintained in Neurobasal medium including B27 supplement, and experiments were performed at 7 days in vitro (DIV 7). To activate NMDA receptors, neurons were treated with Mg2+-free Earle’s balanced salt solution containing 300 μM NMDA and 5 μM glycine for 10 min. The NMDA-containing solution was then removed, fresh Neurobasal medium with B27 supplement was added, and neurons were incubated at 37°C for another 16 hours before being subjected to the biotin switch assay.
RNA interference
Two methods for RNA interference were used in this study. In the first, control and GAPDH RNA interference were performed using, respectively, Non-silencing- and GAPDH-GIPZ lentiviral shRNAmir viral particles from Open Biosystems according to the manufacturer’s protocol. After infection, 1 mM sodium pyruvate (Sigma) was added to the culture medium, along with 10 μg/ml puromycin (selection marker, Sigma).
In the second method, used in Fig. 2g, small oligonucleotides from Dharmacon RNA Technologies were used as described previously7. For rescue experiment, transfection with rescue plasmids occurred 24 hours after siRNA treatment. Cells were then incubated at 37°C for another 24 hours before lysis. The rescue plasmids were not RNAi resistant; rather, rescue was achieved (and confirmed via western blot) by the high level of expression of the plasmids.
Western blotting
Western blots were visualized using either the SuperSignal West ECL system from Pierce followed by film exposure or the LI-COR Odyssey infrared imaging system.
Immunoprecipitation
For SIRT1-GAPDH co-immunoprecipitation, cells were lysed in binding buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.1% Chaps, 100 μM deferoxamine, 1 mM EDTA, 0.1 mg/ml BSA) and homogenized by passing through a 26-gage needle. Cell extracts were subjected to immunoprecipitation with anti-SIRT1 or anti-HDAC2 antibody and 30 μl TrueBlot anti-Rabbit Ig IP beads for either 3 hr or overnight at 4°C. The beads were washed five times with binding buffer.
For PGC1α immunoprecipitation, cells were lysed by sonication in lysis buffer (50 mM Tris pH 8.0, 100 mM NaCl, 2 mM EGTA, 0.4% Triton-X100, 10 mM nicotinamide, 10 μM TSA, 100 μM deferoxamine), and extracts were subjected to immunoprecipitation with anti-flag antibody and Protein G agarose beads (Calbiochem) for a minimum of 3 hr. at 4°C. Beads were washed four times with lysis buffer.
After washes, beads were resuspended in 30 μl SDS sample buffer. The samples were resolved by SDS-PAGE and analyzed by western blotting.
Purification of recombinant proteins
Constructs were subcloned into the pGEX-6p-2 vector (GE Healthcare) and transformed into BL21-Gold competent bacterial cells (Stratagene). Protein purification was performed according to Procedure 2.2.1 as described40. Proteins were either eluted with 10mM glutathione or cleaved from GST using PreScission Protease (GE Healthcare) according to the manufacturer’s protocol. Proteins were then dialyzed into HEN buffer (250mM HEPES pH 7.7, 1mM EDTA, 0.1mM neocuproine).
In vitro binding assay
Equimolar amounts of GST or GST-GAPDH were incubated with 100 μM GSH or GSNO for 30 min. at 37°C, desalted with protein desalting spin columns (Pierce), and then added to 1 ml of binding buffer along with recombinant SIRT1 (100–300 ng). Where indicated, 50 μM Peptide-C150 or scrambled peptide was also added. Samples were rotated for 15 min. at room temperature, 20 μl of 50% glutathione-sepharose slurry (Amersham Biosciences) were added, and samples were rotated for one additional hour at 4°C. Samples were then washed five times with binding buffer. The beads were resuspended in SDS sample buffer, and proteins were resolved by SDS-PAGE and analyzed by western blotting.
Nuclear fractionation
Cells were lysed for 10 min. on ice in buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.05% NP40, pH 7.9) and then centrifuged at 1,000g for 10 min. at 4°C. The supernatant was removed and saved as the cytosolic fraction, and the pellet was resuspended in buffer B (5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 25% glycerol (v/v), pH 7.9) and lysed by sonication. After another 30 min. incubation on ice, the samples were cleared by centrifugation at 16,000g at 4°C, and the supernatant was removed and saved as the nuclear fraction.
Biotin switch assay
The assay was performed as described41, with minor modifications. Briefly, cells were first lysed for 10 min. on ice in NP40/HEN (HEN buffer adjusted to contain 0.4% NP40) and then centrifuged at 1,000g for 10 min. at 4°C. The pellet was then resuspended in RIPA/HEN (HEN buffer adjusted to contain 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) and cleared by centrifugation at 16,000g at 4°C. The supernatant was then used for the assay. Additionally, labeling was performed in the dark for 2–3 hours at room temperature with 5 mM sodium ascorbate and 0.4 mM biotin-HPDP (Pierce), and pulldown was performed overnight with high-capacity neutravidin agarose (Pierce).
For the in vitro transnitrosylation assay, GAPDH was first treated with 50 μM GSH or GSNO for 30 min. at 37°C. The GAPDH samples were then desalted and incubated with SIRT1 for an additional 30 min. at 37°C. Where indicated, 5 μM Peptide-C150 or scrambled peptide was pre-incubated with SIRT1 for 15 min. at room temperature prior to the addition of GAPDH. The biotin switch assay was then performed as described32, and biotinylated proteins were detected using anti-biotin monoclonal antibody.
In vitro SIRT1 activity assay
Core histones were either purchased from Upstate and chemically acetylated as described42 or purchased from Roche and used as is, as the Roche histones were found to be basally acetylated. Acetylated histones were incubated in deacetylation buffer (25 mM Tris pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2) in the presence or absence of recombinant SIRT1 and 0.5 mM β-NAD for 1 hr. at 30°C. Reactions were resolved by SDS-PAGE and analyzed by western blotting. For the effect of nitrosylation on SIRT1 activity, SIRT1 was first incubated with GSH or GSNO for 30 min. at 37°C, desalted, and then added to the other reagents. For the effect of GAPDH on SIRT1 activity, GAPDH was treated with 50 μM GSH or GSNO for 30 min. at 37°C, desalted, and incubated with SIRT1 for 20 min. at room temperature before addition of the other reagents.
Luciferase reporter assay
293-nNOS cells infected with Non-silencing-or GAPDH-GIPZ lentiviral shRNAmir viral particles were transfected with HNF4α, an HNF4α binding site luciferase, and either empty vector, wild-type GAPDH, or GAPDH-T152A. Rescue was achieved via the high level of expression of the rescue plasmids. Twenty-four hours after transfection, cells were treated with A23187 (5 μM) for 3 hrs. Luciferase activity was then measured using the Promega Dual-Luciferase Reporter Assay Kit.
GAPDH glycolytic activity assay
This assay was performed as described7.
Statistical analysis
P values were calculated by student’s t-test or one-way ANOVA using Microsoft Excel.
Supplementary Material
Acknowledgments
We are grateful to M. Koldobskiy, B. Selvakumar, S.F. Kim, P. Kim, K. Werner, and all members of the Snyder laboratory for insight and discussion. We thank P. Puigserver for SIRT1 and PGC1α plasmids. We thank B. Ziegler for organizing the manuscript. This work was supported by USPHS grant DA-000266 and Research Scientist Award DA-00074 to SHS.
Footnotes
Author contributions
MDK designed and performed most of the experiments, analyzed the data, prepared the figures, helped write the manuscript, and contributed to project design. NS performed experiments investigating the effects of GAPDH mutants on SIRT1 nitrosylation in intact cells. He also performed the GAPDH glycolytic activity assay and the luciferase reporter assay. He analyzed the data and prepared the figures for these experiments. MRH identified the physical interaction between GAPDH and SIRT1. KRJ identified S-nitrosylation of SIRT1, helped with SIRT1 assay design, and prepared constructs. JVKN performed some in vitro binding and enzyme activity assays. AMS performed site-directed mutagenesis and prepared plasmids. LL helped perform some experiments. LDH generated neuronal cultures. SHS designed and supervised the project, wrote the manuscript, and provided financial support.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nature Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 3.Yu Z, Kuncewicz T, Dubinsky WP, Kone BC. Nitric oxide-dependent negative feedback of PARP-1 trans-activation of the inducible nitric-oxide synthase gene. J Biol Chem. 2006;281:9101–9109. doi: 10.1074/jbc.M511049200. [DOI] [PubMed] [Google Scholar]
- 4.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 5.Lipton SA, et al. Cysteine regulation of protein function as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- 6.Fang M, et al. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 7.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 8.Sen N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nature Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 10.Haigis MC, Guarente LP. Mammalian sirtuins emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 11.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nature Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 12.Chang K, Shimizu N, Fukushima Y, Martyn J, Kaneki M. iNOS Inactivates Sirt1, a Key Regulator of Stress Resistance and Metabolism, by S-Nitrosylation. 2006 Annual Meeting American Society of Anesthesiologists conference abstract; 2006. p. A1067. [Google Scholar]
- 13.Chang K, Shimizu N, Fukushima Y, Martyn J, Kaneki M. Inducible Nitric Oxide Synthase-Mediated p53 Activation and Apoptosis in Muscle after Burn Injury. 2007 Annual Meeting American Society of Anesthesiologists conference abstract; 2007. p. A1856. [Google Scholar]
- 14.Chen L, et al. Dual role of Zn2+ in maintaining structural integrity and suppressing deacetylase activity of SIRT1. J Inorg Biochem. 2010;104:180–185. doi: 10.1016/j.jinorgbio.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 16.Lerin C, et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Rhee J, et al. Regulation of hepatic fasting response PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci USA. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester MT, et al. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nature Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nature Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano A, et al. Evidence for a functional nitric oxide synthase system in brown adipocyte nucleus. FEBS Lett. 2002;514:135–140. doi: 10.1016/s0014-5793(02)02245-7. [DOI] [PubMed] [Google Scholar]
- 25.Gobeil F, et al. Nitric oxide signaling via nuclearized endothelial nitric-oxide synthase modulates expression of the immediate early genes iNOS and mPGES-1. J Biol Chem. 2006;281:16058–16067. doi: 10.1074/jbc.M602219200. [DOI] [PubMed] [Google Scholar]
- 26.Grasselli A, et al. Estrogen receptor-alpha and endothelial nitric oxide synthase nuclear complex regulates transcription of human telomerase. Circ Res. 2008;103:34–42. doi: 10.1161/CIRCRESAHA.107.169037. [DOI] [PubMed] [Google Scholar]
- 27.Dudzinski DM, Michel T. Life history of eNOS: Partners and pathways. Cardiovascular Research. 2007;75:247–260. doi: 10.1016/j.cardiores.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagnandan D, Sessa WC, Fulton D. Intracellular location regulates calcium-calmodulin-dependent activation of organelle-restricted eNOS. Am J Physiol Cell Physiol. 2005;289:C1024–33. doi: 10.1152/ajpcell.00162.2005. [DOI] [PubMed] [Google Scholar]
- 29.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt HHHW, Walter U. NO at work. Cell. 1994;78:919–925. doi: 10.1016/0092-8674(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 34.Marshall HE, Stamler JS. Inhibition of NF-κB by S-nitrosylation. Biochemistry. 2001;40:1688–1693. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- 35.Yasinska IM, Sumbayev VV. S-nitrosation of Cys-800 of HIF-1alpha protein activates its interaction with p300 and stimulates its transcriptional activity. FEBS Lett. 2003;549:105–109. doi: 10.1016/s0014-5793(03)00807-x. [DOI] [PubMed] [Google Scholar]
- 36.Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 37.Meyer-Siegler K, Rahman-Mansur N, Wurzer JC, Sirover MA. Proliferative dependent regulation of the glyceraldehyde-3-phosphate dehydrogenase/uracil DNA glycosylase gene in human cells. Carcinogenesis. 1992;13:2127–2132. doi: 10.1093/carcin/13.11.2127. [DOI] [PubMed] [Google Scholar]
- 38.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehydes-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, et al. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol Cancer Res. 2008;6:1375–1384. doi: 10.1158/1541-7786.MCR-07-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.North BJ, Schwer B, Ahuja N, Marshall B, Verdin E. Preparation of enzymatically active recombinant class III protein deacetylases. Methods. 2005;36:338–345. doi: 10.1016/j.ymeth.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Science STKE. 2001;86:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 42.Parsons XH, Garcia SN, Pillus L, Kadonaga JT. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc Natl Acad Sci USA. 2003;100:1609–1614. doi: 10.1073/pnas.0434064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.