Abstract
Abilities to successfully quit smoking display substantial evidence for heritability in classic and molecular genetic studies. Genome-wide association (GWA) studies have demonstrated single-nucleotide polymorphisms (SNPs) and haplotypes that distinguish successful quitters from individuals who were unable to quit smoking in clinical trial participants and in community samples. Many of the subjects in these clinical trial samples were aided by nicotine replacement therapy (NRT). We now report novel GWA results from participants in a clinical trial that sought dose/response relationships for “precessation” NRT. In this trial, 369 European-American smokers were randomized to 21 or 42 mg NRT, initiated 2 wks before target quit dates. Ten-week continuous smoking abstinence was assessed on the basis of self-reports and carbon monoxide levels. SNP genotyping used Affymetrix 6.0 arrays. GWA results for smoking cessation success provided no P value that reached “genome-wide” significance. Compared with chance, these results do identify (a) more clustering of nominally positive results within small genomic regions, (b) more overlap between these genomic regions and those identified in six prior successful smoking cessation GWA studies and (c) sets of genes that fall into gene ontology categories that appear to be biologically relevant. The 1,000 SNPs with the strongest associations form a plausible Bayesian network; no such network is formed by randomly selected sets of SNPs. The data provide independent support, based on individual genotyping, for many loci previously nominated on the basis of data from genotyping in pooled DNA samples. These results provide further support for the idea that aid for smoking cessation may be personalized on the basis of genetic predictors of outcome.
INTRODUCTION
Cigarette smoking is a significant cause of premature death and disease (1). Although abstinence reduces risks to smokers, success rates after attempts to quit smoking remain modest. One year after unaided attempts to quit smoking, abstinence rates are <5%. Even with pharmacologic aids that increase success, long-term abstinence rates are <25% (2). Twin studies document substantial heritability for smokers’ abilities to successfully abstain from smoking, suggesting substantial genetic components to individual differences in abilities to quit (3,4).
We recently reported genome-wide association (GWA) studies for success in quitting smoking in six independent samples of carefully monitored individuals who attempted to quit smoking in clinical trials or in community quitters, using carefully validated DNA pooling approaches (5–8). No result from any of these studies achieves “genome-wide” significance. However, the molecular genetic results from these independent samples display substantial convergence with each other (that is, the nominally positive results from each of these samples cluster in small chromosomal regions to extents much greater than expected by chance, and the same small chromosomal regions are identified by the clustered, nominally positive results from different samples to greater extents than those expected by chance) (5,9–13).
We report GWA studies of smoking cessation success in individually genotyped European-American participants in a smoking cessation trial that examined effects of 21 versus 42 mg/24 h precessation nicotine replacement therapy (NRT) (14). Although the sample size is modest for GWA, we nevertheless described the highly significant overlap between the chromosomal regions identified in this work and those identified by nominally significant associations with successfully quitting in other studies of smoking cessation. We identify specific gene ontology classes into which candidate “quit success” genes (that are identified in these analyses) fall more often than expected by chance. We describe a Bayesian network into which the quit success–associated SNPs fall.
MATERIALS AND METHODS
Subjects
Adult smokers who expressed desires to quit were recruited and screened at one of four North Carolina centers. Participants provided written informed consent; reported smoking an average of ≥10 cigarettes/day that each yielded ≥0.5 mg nicotine; displayed end-expired air carbon monoxide (CO) ≥10 ppm; failed to display any exclusionary features on history, physical examination or laboratory evaluations; and were compensated up to $140. Smokers were subdivided into low- and high-dependence subgroups (Fagerström Test for Nicotine Dependence [FTND] scores ≤6 or >6, respectively), and individuals in each of these subgroups were randomly assigned to 21 mg/24 h or 42 mg/24 h nicotine patch doses. During seven study sessions, brief supportive counseling was provided, clinical trial materials were dispensed and dependent measures were assessed. Dependent measures included measured end-expired air CO and reports of smoking, withdrawal symptoms and adverse effects including nausea and/or emesis.
Each participant wore two skin patches daily for 6 wks, beginning 2 wks before the target quit date. One 21-mg active patch (GlaxoSmithKline, Research Triangle Park, NC, USA) was applied in the morning. At noon, either another 21-mg patch (42 mg/day) or a placebo patch (Rejuvenation Labs, Cadillac, MI, USA) (21 mg/day) was applied. NRT doses were gradually reduced beginning 4 or 6 wks after the quit date for the 42 and 21 mg/24 h groups, respectively. Participants with sleep disturbances removed patches at bedtime and applied new ones upon awakening. Subjects experiencing other symptoms of nicotine toxicity reduced doses until symptoms abated according to the following sequence: reduce morning patch from 21 to 14 to 7 to 0 mg/day and then discontinue the afternoon patch. All participants were provided with denicotinized cigarettes (<0.05 mg nicotine yield; Vector Tobacco, Mebane, NC, USA) to smoke during the 2-wk precessation period.
The primary outcome—continuous abstinence from the target quit date through the end of treatment (10 wks)—was assessed on the basis of self-reports of continuous abstinence that were confirmed by end-expired CO levels ≤10 ppm. An intent-to-treat criterion was used. Participants who withdrew from the study or were lost to follow-up were classified as nonabstinent.
Genotyping and Assignment of Genetic Background Groups
DNA was extracted from blood, quantitated and genotyped by using Affymetrix 6.0 microarrays according to the manufacturer’s instructions. Genotypes for each individual passed Affymetrix quality control metrics with a contrast quality control threshold >0.4 and provided calls for >97% of SNP genotypes. Imputation using PLINK (15) with a confidence threshold >0.95 determined most missing genotype calls. We assessed data from 905,273 SNPs, of which 868,154 were autosomal, 36,862 were located on X and 257 were located on Y.
Genetic background was assigned for each individual on the basis of principal component analyses of data from all SNPs and was confirmed by self-report in almost all cases (14). Data from the 369 participants of European- American descent who were identified in this way are analyzed herein.
Analyses
Differences between allele frequencies in successful quitters versus unsuccessful quitters were compared by using the χ2 test. We performed preplanned primary “nontemplate” GWA analyses similar to those we have previously described (16). We identified SNPs that (a) display χ2 values with P < 0.01 “nominally positive” significance compared with data from individuals who were successful versus unsuccessful in quitting smoking and (b) cluster in small chromosomal regions, so that at least four of these nominally positive SNPs lie within 25 kb of at least one other positive SNP. A number of these clustered, nominally positive SNPs identify genes; many also lie between currently annotated genes.
To seek additional support for the chromosomal regions identified by these clusters of nominally positive SNPs, we sought additional association signals in these same regions from clustered, nominally positive SNPs identified in relevant independent GWA studies: (a) Uhl et al.: 1,000,000 SNP studies of smokers who quit versus those who continued to smoke in the “patch in practice” study of NRT in UK smokers (8,17); (b) Uhl et al.: 1,000,000 SNP GWA studies of smokers who quit versus those who continued to smoke in a clinical trial of denicotinized cigarettes (7); (c) Drgon et al.: 500,000 SNP GWA studies of smokers who quit versus those who continued to smoke in community settings (6); (d–f) each of three samples from Uhl et al.: 500–600,000 SNP GWA studies of smokers who were successful versus unsuccessful in quitting in clinical trial settings (5); and (g) Bierut et al.: 38,000 SNP GWA studies of nondependent (FTND) versus dependent (FTND) smokers (11). To provide insight into some of the genes likely to harbor variants that contribute to individual differences in ability to quit, we identify genes that are identified by clustered, nominally positive SNPs from the current sample and at least two other quit success or nicotine dependence samples.
We compare observed results for smoking cessation success to those expected by chance using 10,000 Monte Carlo simulation trials, as described (18). For each trial, a randomly selected set of SNPs from the current data set was assessed to see if it provided results equal to or greater than the results that we actually observed. The number of Monte Carlo trials for which the randomly selected SNPs displayed (at least) the same features as the observed results was then tallied to generate an empirical P value. These simulations thus corrected for the number of repeated comparisons made in these analyses, an important consideration in evaluating these GWA data sets. We also performed permutation analyses using PLINK to provide a secondary assessment of significance.
To assess the power of our current approach for smoking cessation success, we used current sample sizes and standard deviations, the program PS v2.1.31 (19,20) and α = 0.05. To provide controls for the possibility that differences between quitters and nonquitters observed herein were due to occult ethnic/racial allele frequency differences or noisy assays, we assessed the overlap between the results obtained here and the SNPs that displayed the largest (a) allele frequency differences between African-American versus European-American control individuals and (b) the largest assay “noise.”
Bayesian networks are probabilistic graphical models that represent a set of variables as nodes and their conditional interdependencies as edges. These networks thus provide data-driven probabilistic classifications that can identify ways in which results from sets of SNPs provide a reasonable network, which SNPs provide the most direct relationship to quit success and which SNPs provide a more indirect relationship to quit success. We thus used BayesWare (Markov Chain Monte Carlo methods; BayesWare™, http://www.bayesware.com) to seek networks for sets of the 25, 50, 100, 200, 500 and 1,000 SNPs that displayed the strongest evidence for association with quit success from the current data, or from sets of 25, 50, 100, 200 and 500 SNPs that came from lists in which there were random relationships between the P values and SNPs. The numbers of SNPs included in the networks formed were tabulated for each set of SNPs from true and permuted control data sets. The network based on 1,000 true SNPs was used for subsequent analyses that sought relationships between SNPs and quit success and between SNPs in the inner versus outer circles of this Bayesian network.
Gene ontology analyses were performed in BioBase™. The gene names in lists of genes identified by clustered, nominally significant results were matched to BioBase gene annotations. Functional enrichment analyses were performed by using “biological process” gene ontology (GO) terms as defined in the BioBase knowledge base. Functional enrichment was tested by using hypergeometric tests. To provide a control, random gene lists of the same size were assembled from the list of all genes using a Perl script (Drgon et al., unpublished data); GO analysis was then performed on these random gene lists. The hypergeometric test P value distributions of the randomized gene lists analyses were compared with the P value distributions obtained from GO analysis of the bona fide lists.
RESULTS
Unsuccessful Versus Successful Quitters
When comparing data from European-American trial participants who were unsuccessful with successful quitters, there is significant clustering of nominally positive SNPs in small chromosomal regions. Thus, there are 5,898 “nominally positive” SNPs with nominal P < 0.01. A total of 2,147 of these SNPs lie in 338 clusters, each containing at least four nominally positive SNPs separated from each other by ≤25 kb. We would expect eight such clusters by chance (Monte Carlo P < 0.0001). A total of 176 of the regions identified by these clustered, nominally positive SNPs contain a total of 206 genes (Table 1). None of 10,000 permutation tests in which individuals were randomly assigned to be “pseudo abstinent” or “pseudo nonabstinent” ever identified as many SNPs that achieved nominally significant results and that clustered in small chromosomal regions as found in the actual data set (thus P < 0.0001).
Table 1.
Genomic regions that contain clustered, nominally positive SNPs for success in smoking cessation.
| Chromosome | bp: Start | bp: End | No. SNPs | Gene(s) | Pmin SNP | Pmin |
|---|---|---|---|---|---|---|
| 1 | 4,514,682 | 4,527,839 | 5 | rs241275 | 5.10E-04 | |
| 1 | 6,632,197 | 6,693,727 | 9 | DNAJC11 | rs7549198 | 9.61E-04 |
| 1 | 10,231,218 | 10,268,359 | 5 | KIF1B | rs17034615 | 4.40E-04 |
| 1 | 23,620,353 | 23,638,820 | 6 | DDEFL1 and TCEA3 | rs1077514 | 6.38E-03 |
| 1 | 34,455,349 | 34,457,938 | 4 | C1orf94 | rs10158529 | 1.09E-03 |
| 1 | 37,211,157 | 37,308,655 | 8 | GRIK3 | rs12077898 | 7.80E-05 |
| 1 | 57,609,677 | 57,646,636 | 9 | DAB1 | rs2405994 | 8.50E-05 |
| 1 | 67,964,539 | 67,970,259 | 5 | GNG12 | rs2803462 | 1.93E-03 |
| 1 | 88,442,597 | 88,446,200 | 4 | rs1336577 | 2.01E-03 | |
| 1 | 89,531,807 | 89,553,114 | 5 | rs4658084 | 4.67E-03 | |
| 1 | 89,873,914 | 89,908,021 | 7 | LRRC8C | rs10801757 | 2.17E-03 |
| 1 | 96,268,987 | 96,289,166 | 4 | rs161107 | 1.65E-04 | |
| 1 | 111,376,530 | 111,506,446 | 16 | CEPT1 and TMEM77 | rs7551294 | 1.39E-04 |
| 1 | 114,328,943 | 114,369,687 | 5 | HIPK1 and OLFML3 | rs3006998 | 4.01E-03 |
| 1 | 154,610,316 | 154,623,104 | 4 | RHBG | rs942679 | 2.95E-03 |
| 1 | 166,614,722 | 166,624,383 | 7 | MIRN557 | rs2268546 | 8.65E-04 |
| 1 | 170,094,146 | 170,139,395 | 7 | DNM3 | rs6660011 | 3.70E-05 |
| 1 | 172,111,855 | 172,158,440 | 14 | SERPINC1 | rs6663875 | 9.37E-04 |
| 1 | 175,222,602 | 175,247,214 | 6 | ASTN | rs228002 | 5.90E-03 |
| 1 | 193,993,579 | 194,086,960 | 19 | rs2942926 | 1.79E-03 | |
| 1 | 196,958,987 | 197,044,377 | 10 | PTPRC | rs6696533 | 8.26E-04 |
| 1 | 220,660,638 | 220,663,363 | 7 | rs11591051 | 4.15E-04 | |
| 1 | 227,859,399 | 227,870,277 | 5 | KIAA0133 | rs879265 | 2.47E-03 |
| 1 | 245,318,105 | 245,338,921 | 4 | ZNF669 | rs6426218 | 5.47E-04 |
| 2 | 16,804,278 | 16,827,433 | 4 | rs1035308 | 1.43E-04 | |
| 2 | 18,819,735 | 18,844,740 | 4 | rs6531118 | 1.55E-03 | |
| 2 | 21,042,552 | 21,057,986 | 5 | rs6544366 | 3.23E-03 | |
| 2 | 24,800,554 | 24,850,219 | 4 | NCOA1 | rs11682130 | 1.47E-03 |
| 2 | 38,048,618 | 38,105,376 | 11 | FAM82A | rs1348748 | 9.95E-04 |
| 2 | 43,128,594 | 43,141,133 | 4 | rs4953720 | 4.41E-03 | |
| 2 | 45,387,134 | 45,403,164 | 4 | rs12473388 | 1.08E-03 | |
| 2 | 46,271,312 | 46,310,086 | 8 | PRKCE | rs2218549 | 2.89E-03 |
| 2 | 47,337,642 | 47,344,246 | 4 | rs6755555 | 2.29E-03 | |
| 2 | 67,972,791 | 68,001,847 | 4 | rs2047816 | 2.26E-03 | |
| 2 | 79,973,446 | 79,998,431 | 6 | CTNNA2 | rs1434098 | 5.96E-05 |
| 2 | 85,376,787 | 85,422,321 | 7 | TCF7L1 and TGOLN2 | rs1061782 | 6.19E-05 |
| 2 | 108,178,476 | 108,251,772 | 8 | SULT1C3 | rs12712018 | 7.01E-04 |
| 2 | 123,072,621 | 123,097,366 | 4 | rs13427932 | 4.17E-03 | |
| 2 | 127,092,461 | 127,095,999 | 4 | rs6760443 | 8.73E-03 | |
| 2 | 130,043,114 | 130,130,607 | 16 | rs3109254 | 9.19E-05 | |
| 2 | 136,523,244 | 136,535,189 | 5 | rs11693502 | 7.14E-04 | |
| 2 | 137,369,412 | 137,413,690 | 13 | rs567483 | 2.74E-03 | |
| 2 | 139,920,877 | 139,994,049 | 7 | rs10200212 | 4.38E-03 | |
| 2 | 148,004,334 | 148,011,288 | 4 | rs12691758 | 6.93E-03 | |
| 2 | 173,519,672 | 173,586,587 | 10 | RAPGEF4 | rs3754753 | 2.69E-04 |
| 2 | 183,108,430 | 183,112,207 | 4 | rs1430154 | 1.25E-03 | |
| 2 | 183,154,780 | 183,173,866 | 4 | rs1527878 | 1.81E-04 | |
| 2 | 206,296,221 | 206,302,386 | 5 | NRP2 | rs868196 | 3.58E-03 |
| 2 | 222,633,911 | 222,677,368 | 11 | rs348995 | 2.59E-03 | |
| 2 | 224,266,128 | 224,288,164 | 6 | rs1992191 | 6.10E-04 | |
| 2 | 229,320,230 | 229,349,994 | 4 | rs7589424 | 1.14E-04 | |
| 2 | 229,515,119 | 229,536,702 | 4 | rs7593561 | 6.00E-04 | |
| 3 | 14,674,247 | 14,690,891 | 4 | C3orf19 | rs2276754 | 2.49E-03 |
| 3 | 15,386,896 | 15,439,322 | 6 | METTL6 | rs6442522 | 6.31E-04 |
| 3 | 15,469,006 | 15,477,134 | 6 | COLQ | rs12633820 | 4.61E-03 |
| 3 | 20,739,135 | 20,755,593 | 4 | rs4610242 | 1.83E-04 | |
| 3 | 29,306,863 | 29,326,141 | 4 | RBMS3 | rs1025644 | 7.19E-05 |
| 3 | 32,190,375 | 32,194,730 | 4 | GPD1L | rs6784980 | 4.43E-03 |
| 3 | 59,994,590 | 60,038,259 | 8 | FHIT | rs212059 | 9.10E-04 |
| 3 | 61,209,872 | 61,228,928 | 5 | FHIT | rs815718 | 3.58E-04 |
| 3 | 65,279,856 | 65,286,293 | 4 | rs1479959 | 1.60E-03 | |
| 3 | 72,417,549 | 72,453,713 | 4 | rs4677135 | 3.64E-05 | |
| 3 | 106,744,838 | 106,779,003 | 4 | ALCAM | rs526297 | 4.33E-03 |
| 3 | 110,125,036 | 110,153,292 | 4 | GUCA1C | rs2593962 | 1.27E-03 |
| 3 | 111,630,473 | 111,640,366 | 4 | rs12632602 | 6.97E-03 | |
| 3 | 111,669,241 | 111,693,318 | 8 | rs11715989 | 2.02E-03 | |
| 3 | 120,865,217 | 120,894,965 | 6 | COX17 | rs13091305 | 2.03E-03 |
| 3 | 121,481,748 | 121,485,834 | 4 | rs4146299 | 2.66E-04 | |
| 3 | 129,575,855 | 129,610,333 | 6 | EEFSEC | rs7373685 | 3.55E-03 |
| 3 | 138,330,528 | 138,421,502 | 13 | rs1461512 | 1.23E-04 | |
| 3 | 144,583,316 | 144,615,415 | 4 | SLC9A9 | rs11707857 | 3.95E-03 |
| 3 | 145,284,332 | 145,302,343 | 6 | rs1530479 | 1.46E-03 | |
| 3 | 145,735,062 | 145,784,349 | 4 | rs12631899 | 5.29E-03 | |
| 3 | 149,689,756 | 149,704,154 | 4 | rs35942196 | 2.00E-03 | |
| 3 | 154,252,230 | 154,297,605 | 8 | rs515099 | 3.05E-03 | |
| 3 | 169,420,213 | 169,432,275 | 4 | rs2067678 | 4.65E-03 | |
| 3 | 180,872,350 | 180,943,519 | 11 | USP13 | rs4854948 | 5.98E-05 |
| 3 | 184,522,643 | 184,559,448 | 8 | MCF2L2 | rs9882117 | 2.23E-05 |
| 3 | 188,467,212 | 188,480,342 | 4 | MASP1 | rs710471 | 6.01E-04 |
| 3 | 188,885,209 | 188,900,089 | 4 | RTP2 | rs10937316 | 5.91E-03 |
| 3 | 189,170,534 | 189,221,370 | 9 | rs1348637 | 2.75E-05 | |
| 3 | 190,539,426 | 190,548,763 | 4 | rs13059863 | 3.64E-04 | |
| 3 | 190,648,097 | 190,715,335 | 15 | rs2633448 | 1.09E-03 | |
| 3 | 190,815,442 | 190,827,550 | 4 | TP73L | rs4398409 | 2.20E-03 |
| 3 | 191,257,452 | 191,290,326 | 9 | LEPREL1 | rs9879082 | 1.20E-03 |
| 3 | 191,960,031 | 191,960,376 | 4 | rs9858906 | 5.22E-03 | |
| 3 | 193,616,813 | 193,633,511 | 5 | FGF12 | rs6444640 | 7.21E-04 |
| 3 | 198,680,894 | 198,694,948 | 4 | rs1897298 | 7.42E-03 | |
| 4 | 5,680,199 | 5,690,519 | 4 | EVC2 | rs13133528 | 9.58E-04 |
| 4 | 23,835,312 | 23,885,637 | 10 | rs10023214 | 3.23E-05 | |
| 4 | 54,097,128 | 54,112,431 | 4 | LNX1 | rs11723168 | 1.58E-03 |
| 4 | 64,201,517 | 64,229,222 | 10 | rs1961776 | 1.39E-03 | |
| 4 | 70,481,520 | 70,549,400 | 13 | UGT2A1 | rs7662309 | 1.91E-04 |
| 4 | 96,613,817 | 96,647,190 | 6 | UNC5C | rs7697199 | 5.73E-04 |
| 4 | 106,794,718 | 106,813,023 | 5 | rs17036090 | 7.95E-04 | |
| 4 | 106,921,128 | 106,950,112 | 4 | GSTCD | rs11732298 | 5.93E-04 |
| 4 | 126,643,891 | 126,694,831 | 11 | rs13112740 | 6.38E-04 | |
| 4 | 148,647,977 | 148,659,051 | 4 | EDNRA and GTF2F2L | rs7674137 | 1.41E-03 |
| 4 | 180,561,608 | 180,602,617 | 4 | rs17090633 | 2.39E-05 | |
| 4 | 180,638,793 | 180,675,229 | 10 | rs2681357 | 1.95E-04 | |
| 4 | 180,817,055 | 180,846,530 | 5 | rs17067909 | 6.16E-04 | |
| 4 | 181,395,374 | 181,434,417 | 5 | rs10007307 | 2.24E-03 | |
| 4 | 184,506,382 | 184,515,862 | 5 | rs4862161 | 5.46E-04 | |
| 5 | 3,127,414 | 3,132,233 | 6 | rs10475190 | 6.72E-05 | |
| 5 | 3,225,052 | 3,266,546 | 7 | rs1215667 | 4.03E-04 | |
| 5 | 24,128,887 | 24,146,110 | 6 | rs17444609 | 3.31E-03 | |
| 5 | 30,609,959 | 30,638,252 | 4 | rs4091500 | 2.59E-03 | |
| 5 | 31,427,481 | 31,456,427 | 6 | RNASEN | rs2330693 | 3.59E-04 |
| 5 | 33,797,323 | 33,828,213 | 4 | ADAMTS12 | rs10062147 | 1.34E-03 |
| 5 | 51,906,987 | 51,933,802 | 4 | rs350431 | 9.88E-04 | |
| 5 | 56,136,753 | 56,154,632 | 5 | MAP3K1 | rs1423621 | 6.54E-03 |
| 5 | 58,334,117 | 58,349,350 | 4 | PDE4D | rs7727206 | 7.03E-03 |
| 5 | 66,773,558 | 66,827,933 | 6 | rs747919 | 9.91E-04 | |
| 5 | 108,544,843 | 108,586,419 | 8 | FER | rs11240992 | 1.80E-04 |
| 5 | 108,619,178 | 108,619,907 | 4 | rs1363212 | 7.04E-04 | |
| 5 | 115,313,413 | 115,329,965 | 6 | FLJ90650 | rs4920898 | 1.79E-03 |
| 5 | 131,752,849 | 131,771,364 | 8 | SLC22A5 | rs6596075 | 8.90E-04 |
| 5 | 148,737,396 | 148,748,096 | 4 | IL17B | rs353275 | 2.60E-03 |
| 5 | 155,340,456 | 155,354,167 | 4 | rs6866134 | 5.53E-03 | |
| 5 | 155,681,772 | 155,746,581 | 17 | SGCD | rs7722398 | 6.05E-04 |
| 5 | 168,238,733 | 168,254,799 | 6 | SLIT3 | rs11742567 | 2.41E-04 |
| 5 | 169,729,345 | 169,754,582 | 5 | KCNMB1 and KCNIP1 | rs7726856 | 3.78E-03 |
| 5 | 171,754,276 | 171,773,330 | 5 | SH3PXD2B | rs13356223 | 7.01E-03 |
| 5 | 172,891,689 | 172,951,609 | 12 | rs735059 | 1.37E-04 | |
| 6 | 1,234,462 | 1,296,928 | 8 | FOXQ1 | rs12201633 | 1.62E-04 |
| 6 | 1,385,456 | 1,400,822 | 5 | rs9328053 | 3.15E-04 | |
| 6 | 2,607,546 | 2,627,058 | 5 | rs6939996 | 3.59E-04 | |
| 6 | 77,557,859 | 77,650,512 | 10 | rs13219726 | 2.03E-04 | |
| 6 | 82,378,692 | 82,412,699 | 5 | rs10943827 | 6.04E-04 | |
| 6 | 86,222,521 | 86,253,709 | 5 | NT5E | rs4373339 | 4.54E-03 |
| 6 | 97,549,867 | 97,576,111 | 6 | KIAA1900 | rs6924307 | 1.56E-03 |
| 6 | 101,914,228 | 101,951,050 | 7 | GRIK2 | rs1832411 | 3.19E-03 |
| 6 | 106,107,523 | 106,120,691 | 4 | rs4946673 | 7.21E-04 | |
| 6 | 112,454,425 | 112,464,056 | 4 | rs4947157 | 1.44E-03 | |
| 6 | 115,333,820 | 115,390,578 | 4 | rs4945528 | 1.78E-03 | |
| 6 | 132,881,158 | 132,901,302 | 6 | STX7 | rs2840839 | 1.08E-03 |
| 6 | 149,165,112 | 149,209,898 | 6 | UST | rs9498164 | 8.60E-04 |
| 6 | 161,599,178 | 161,612,276 | 5 | AGPAT4 | rs747866 | 1.75E-03 |
| 6 | 162,757,512 | 162,770,899 | 6 | PARK2 | rs9295187 | 5.03E-03 |
| 6 | 166,432,591 | 166,437,713 | 5 | rs1445277 | 3.99E-03 | |
| 7 | 4,303,003 | 4,305,939 | 4 | rs10232703 | 1.24E-03 | |
| 7 | 13,661,977 | 13,668,606 | 6 | rs10260350 | 8.62E-03 | |
| 7 | 14,697,226 | 14,721,583 | 4 | DGKB | rs6947566 | 4.02E-03 |
| 7 | 17,978,277 | 18,049,332 | 10 | PRPS1L1 | rs4236293 | 6.58E-04 |
| 7 | 42,497,140 | 42,554,082 | 13 | rs1991769 | 1.45E-04 | |
| 7 | 42,634,401 | 42,693,143 | 7 | rs2583879 | 1.68E-03 | |
| 7 | 49,183,816 | 49,227,470 | 10 | rs6963695 | 7.37E-04 | |
| 7 | 77,878,213 | 77,888,531 | 6 | MAGI2 | rs6967983 | 1.66E-03 |
| 7 | 79,652,863 | 79,688,271 | 5 | GNAI1 | rs6973616 | 2.89E-03 |
| 7 | 83,543,807 | 83,573,774 | 12 | SEMA3A | rs17298417 | 2.22E-03 |
| 7 | 112,553,318 | 112,589,711 | 11 | rs10252483 | 4.84E-05 | |
| 7 | 123,087,091 | 123,137,160 | 7 | WASL | rs1005567 | 2.05E-03 |
| 7 | 141,952,539 | 141,996,982 | 4 | TRB@ and TRBV17-15 | rs12703485 | 2.02E-03 |
| 7 | 149,982,834 | 150,023,276 | 7 | GIMAP2 | rs6965369 | 1.12E-03 |
| 7 | 150,061,644 | 150,077,753 | 8 | GIMAP3P and GIMAP5 | rs6972271 | 1.43E-04 |
| 7 | 155,991,372 | 156,020,645 | 5 | rs1543989 | 7.32E-04 | |
| 8 | 3,032,637 | 3,051,975 | 6 | CSMD1 | rs12545450 | 9.18E-04 |
| 8 | 8,972,080 | 9,013,363 | 8 | RNU7P4 | rs11775551 | 5.07E-04 |
| 8 | 13,209,117 | 13,220,673 | 5 | DLC1 | rs13271362 | 4.68E-03 |
| 8 | 16,579,753 | 16,599,436 | 5 | rs4922125 | 2.56E-03 | |
| 8 | 18,724,146 | 18,757,428 | 12 | PSD3 | rs6992325 | 6.82E-05 |
| 8 | 18,862,099 | 18,898,958 | 6 | PSD3 | rs1426918 | 1.15E-03 |
| 8 | 26,881,502 | 26,895,504 | 4 | rs13280864 | 5.55E-03 | |
| 8 | 40,359,266 | 40,379,708 | 9 | rs11776669 | 2.08E-03 | |
| 8 | 59,580,896 | 59,609,008 | 5 | rs1582824 | 4.78E-03 | |
| 8 | 83,983,766 | 84,024,130 | 9 | rs1449827 | 2.05E-03 | |
| 8 | 85,313,939 | 85,345,627 | 4 | rs13261650 | 2.10E-04 | |
| 8 | 85,492,450 | 85,530,449 | 7 | rs317954 | 1.02E-04 | |
| 8 | 118,454,399 | 118,479,415 | 4 | rs2635123 | 2.93E-03 | |
| 8 | 131,903,175 | 131,948,250 | 4 | ADCY8 | rs7843541 | 7.12E-04 |
| 8 | 134,090,570 | 134,109,272 | 4 | TG | rs10110664 | 1.03E-03 |
| 8 | 134,582,532 | 134,610,513 | 5 | ST3GAL1 | rs10100754 | 1.02E-03 |
| 8 | 135,306,059 | 135,343,379 | 9 | rs12542306 | 6.36E-04 | |
| 8 | 135,573,080 | 135,605,416 | 4 | ZNF406 | rs7010252 | 1.71E-04 |
| 8 | 139,722,571 | 139,745,264 | 6 | COL22A1 | rs13271565 | 1.88E-03 |
| 8 | 140,660,752 | 140,683,139 | 4 | rs2111571 | 1.74E-04 | |
| 9 | 2,295,133 | 2,320,907 | 6 | rs17407787 | 6.69E-04 | |
| 9 | 8,941,749 | 8,979,686 | 4 | PTPRD | rs10977426 | 1.83E-03 |
| 9 | 10,541,203 | 10,572,586 | 6 | rs1322281 | 9.02E-04 | |
| 9 | 11,281,395 | 11,329,109 | 4 | rs2171661 | 3.10E-04 | |
| 9 | 11,356,549 | 11,425,129 | 9 | rs10959753 | 3.89E-04 | |
| 9 | 11,455,508 | 11,491,589 | 4 | rs10959836 | 1.33E-04 | |
| 9 | 11,673,979 | 11,737,876 | 11 | rs372412 | 4.65E-05 | |
| 9 | 11,764,821 | 11,792,691 | 6 | rs12377084 | 6.90E-04 | |
| 9 | 13,944,734 | 13,960,315 | 4 | rs17192702 | 1.47E-03 | |
| 9 | 14,161,645 | 14,190,005 | 7 | NFIB | rs12377502 | 9.96E-04 |
| 9 | 20,872,525 | 20,886,938 | 5 | KIAA1797 | rs10738569 | 3.89E-03 |
| 9 | 21,836,285 | 21,884,495 | 6 | MTAP | rs7850937 | 4.05E-03 |
| 9 | 24,895,556 | 24,966,888 | 6 | rs4514074 | 3.41E-04 | |
| 9 | 27,739,775 | 27,782,012 | 5 | rs10812663 | 3.09E-04 | |
| 9 | 32,922,194 | 32,989,419 | 6 | APTX | rs10813916 | 5.19E-04 |
| 9 | 70,652,673 | 70,685,840 | 4 | PIP5K1B | rs11143995 | 1.81E-03 |
| 9 | 77,455,592 | 77,461,845 | 4 | rs4745430 | 5.21E-04 | |
| 9 | 85,456,055 | 85,518,478 | 8 | UBQLN1 | rs10746721 | 2.86E-03 |
| 9 | 100,370,139 | 100,378,998 | 4 | GABBR2 | rs2779524 | 6.42E-03 |
| 9 | 109,843,796 | 109,876,206 | 6 | rs10481656 | 1.94E-03 | |
| 9 | 110,451,542 | 110,499,911 | 5 | rs12350675 | 1.44E-03 | |
| 9 | 115,927,888 | 115,961,345 | 5 | COL27A1 | rs2002284 | 1.77E-03 |
| 9 | 116,490,065 | 116,516,444 | 5 | rs10123202 | 2.68E-03 | |
| 9 | 119,676,622 | 119,710,655 | 7 | rs4836705 | 1.10E-03 | |
| 9 | 130,590,543 | 130,622,446 | 6 | TBC1D13, C9orf114, and ENDOG | rs2977998 | 2.83E-03 |
| 10 | 530,161 | 537,658 | 4 | DIP2C | rs885593 | 3.47E-03 |
| 10 | 609,316 | 626,509 | 4 | DIP2C | rs12245224 | 8.59E-05 |
| 10 | 2,107,537 | 2,136,617 | 4 | rs964291 | 4.56E-03 | |
| 10 | 8,551,393 | 8,572,641 | 6 | rs10795631 | 1.14E-03 | |
| 10 | 15,375,814 | 15,441,543 | 11 | C10orf38 | rs10906883 | 3.28E-04 |
| 10 | 44,746,622 | 44,772,461 | 4 | RASSF4 | rs6593452 | 8.61E-04 |
| 10 | 59,011,566 | 59,034,855 | 4 | rs12778784 | 1.78E-03 | |
| 10 | 60,128,925 | 60,134,399 | 4 | BICC1 | rs11006230 | 4.11E-03 |
| 10 | 72,378,282 | 72,407,490 | 4 | rs827287 | 2.87E-04 | |
| 10 | 72,463,858 | 72,480,077 | 5 | rs12261506 | 1.62E-03 | |
| 10 | 82,518,813 | 82,553,424 | 5 | rs1863044 | 1.06E-04 | |
| 10 | 115,419,114 | 115,503,088 | 13 | CASP7 and C10orf81 | rs7085113 | 8.74E-04 |
| 10 | 123,053,164 | 123,085,176 | 4 | rs11199898 | 2.11E-03 | |
| 11 | 4,365,143 | 4,391,233 | 5 | TRIM21 | rs1426378 | 2.06E-03 |
| 11 | 17,542,957 | 17,557,795 | 4 | OTOG | rs757985 | 1.83E-04 |
| 11 | 17,900,240 | 17,932,697 | 8 | SERGEF | rs11603299 | 3.05E-03 |
| 11 | 30,708,854 | 30,731,101 | 11 | rs628029 | 7.43E-04 | |
| 11 | 35,294,776 | 35,334,645 | 7 | SLC1A2 | rs4756221 | 3.53E-03 |
| 11 | 40,418,273 | 40,436,826 | 6 | rs11035841 | 3.56E-03 | |
| 11 | 40,465,084 | 40,512,119 | 4 | rs979531 | 5.18E-03 | |
| 11 | 64,026,514 | 64,066,226 | 9 | rs6421690 | 1.94E-03 | |
| 11 | 71,935,566 | 71,947,033 | 4 | rs4943927 | 2.54E-03 | |
| 11 | 85,536,065 | 85,583,777 | 4 | rs12786057 | 2.29E-04 | |
| 11 | 91,737,172 | 91,777,262 | 5 | FAT3 | rs11019944 | 6.61E-03 |
| 11 | 113,321,587 | 113,348,687 | 4 | HTR3B | rs17116164 | 3.07E-03 |
| 11 | 114,126,679 | 114,146,011 | 5 | rs4145058 | 5.27E-03 | |
| 11 | 117,459,594 | 117,514,489 | 10 | TMPRSS4 and SCN4B | rs10790240 | 2.49E-03 |
| 11 | 121,667,158 | 121,693,657 | 4 | rs485139 | 3.85E-03 | |
| 11 | 125,461,319 | 125,532,083 | 6 | rs668171 | 3.89E-04 | |
| 12 | 10,012,614 | 10,050,191 | 9 | CLEC12A, CLEC1B, and CLEC12B | rs479499 | 1.45E-03 |
| 12 | 23,632,891 | 23,661,597 | 4 | SOX5 | rs17383893 | 6.67E-03 |
| 12 | 23,941,586 | 23,968,615 | 6 | SOX5 | rs7970953 | 9.15E-04 |
| 12 | 43,268,257 | 43,287,053 | 5 | NELL2 | rs10506250 | 5.34E-04 |
| 12 | 54,252,624 | 54,262,230 | 4 | OR2AP1 | rs2371189 | 6.81E-05 |
| 12 | 54,317,540 | 54,339,134 | 6 | OR10AE3P, PSMB3P, and OR10P1 | rs10876844 | 9.08E-04 |
| 12 | 83,408,629 | 83,450,589 | 10 | rs1031681 | 2.52E-03 | |
| 12 | 86,192,156 | 86,204,576 | 5 | rs17577874 | 5.81E-03 | |
| 12 | 87,365,359 | 87,371,132 | 4 | rs10858738 | 2.11E-03 | |
| 12 | 93,106,608 | 93,149,172 | 5 | PLXNC1 | rs7307255 | 3.59E-03 |
| 12 | 96,863,518 | 96,876,686 | 5 | rs11109296 | 2.01E-03 | |
| 12 | 100,517,711 | 100,531,564 | 8 | MYBPC1 | rs11830848 | 6.08E-04 |
| 12 | 102,435,173 | 102,466,844 | 6 | rs4540923 | 3.43E-04 | |
| 12 | 107,684,502 | 107,707,330 | 5 | SSH1 | rs744043 | 4.03E-03 |
| 12 | 130,029,070 | 130,044,192 | 4 | GPR133 | rs11061274 | 3.89E-03 |
| 12 | 130,440,792 | 130,468,902 | 7 | rs7135162 | 2.24E-04 | |
| 13 | 23,537,175 | 23,552,362 | 8 | rs12872637 | 7.95E-04 | |
| 13 | 29,334,889 | 29,356,737 | 5 | rs7321345 | 3.15E-04 | |
| 13 | 35,009,742 | 35,042,078 | 9 | NBEA | rs9544663 | 2.21E-04 |
| 13 | 39,636,316 | 39,658,938 | 4 | rs2039623 | 7.70E-04 | |
| 13 | 39,697,109 | 39,749,380 | 12 | rs10492680 | 8.11E-04 | |
| 13 | 43,735,145 | 43,791,548 | 10 | rs2031996 | 4.95E-04 | |
| 13 | 47,139,485 | 47,154,561 | 4 | rs1172397 | 1.13E-03 | |
| 13 | 58,512,555 | 58,540,818 | 5 | rs4600350 | 8.61E-04 | |
| 13 | 75,251,157 | 75,282,563 | 5 | LMO7 | rs17065046 | 1.42E-03 |
| 13 | 89,698,915 | 89,734,895 | 5 | rs16944259 | 1.08E-03 | |
| 13 | 89,897,985 | 89,939,352 | 6 | rs9522984 | 1.66E-03 | |
| 13 | 92,323,593 | 92,395,330 | 11 | rs7328931 | 4.09E-04 | |
| 13 | 93,396,157 | 93,497,436 | 17 | GPC6 | rs8000417 | 1.75E-04 |
| 13 | 95,989,215 | 96,008,274 | 9 | HS6ST3 | rs7323727 | 2.38E-03 |
| 13 | 97,955,399 | 97,988,906 | 4 | STK24 | rs17471066 | 5.34E-04 |
| 14 | 24,453,213 | 24,477,023 | 4 | STXBP6 | rs12232232 | 5.68E-03 |
| 14 | 32,965,535 | 32,990,025 | 4 | NPAS3 | rs10129955 | 3.76E-03 |
| 14 | 33,068,792 | 33,082,637 | 12 | NPAS3 | rs10134389 | 3.20E-04 |
| 14 | 36,253,365 | 36,272,464 | 4 | SLC25A21 | rs17105125 | 1.69E-03 |
| 14 | 50,894,900 | 50,900,116 | 4 | rs8019638 | 4.06E-04 | |
| 14 | 71,757,205 | 71,849,388 | 15 | RGS6 | rs2283394 | 2.26E-03 |
| 14 | 71,877,312 | 71,953,627 | 17 | RGS6 | rs7159300 | 4.28E-06 |
| 14 | 84,233,182 | 84,262,668 | 4 | rs4904196 | 9.40E-04 | |
| 14 | 95,518,120 | 95,526,439 | 4 | rs17093634 | 7.89E-04 | |
| 14 | 95,602,656 | 95,625,170 | 5 | C14orf132 | rs3208738 | 1.73E-03 |
| 14 | 95,673,756 | 95,705,352 | 4 | rs10144552 | 2.01E-03 | |
| 14 | 97,249,110 | 97,257,396 | 5 | rs4905614 | 3.64E-04 | |
| 14 | 97,287,217 | 97,294,334 | 4 | rs11160403 | 1.74E-04 | |
| 15 | 23,924,404 | 23,944,630 | 4 | rs17669037 | 2.63E-03 | |
| 15 | 51,558,394 | 51,611,521 | 9 | WDR72 | rs1995318 | 3.39E-05 |
| 15 | 52,031,769 | 52,034,625 | 5 | rs1478190 | 3.54E-04 | |
| 15 | 58,117,071 | 58,142,791 | 4 | rs1425935 | 2.64E-03 | |
| 15 | 68,562,262 | 68,579,051 | 7 | rs7161778 | 1.67E-03 | |
| 15 | 78,442,521 | 78,453,010 | 5 | rs11072909 | 6.12E-03 | |
| 15 | 93,340,362 | 93,368,675 | 4 | rs8036547 | 1.41E-03 | |
| 16 | 4,461,750 | 4,482,118 | 4 | HMOX2 | rs17137051 | 3.76E-04 |
| 16 | 6,646,077 | 6,659,894 | 6 | A2BP1 | rs1029967 | 1.52E-03 |
| 16 | 11,253,200 | 11,296,549 | 4 | SOCS1 | rs193778 | 6.38E-04 |
| 16 | 15,777,026 | 15,787,511 | 6 | MYH11 | rs8048077 | 1.91E-03 |
| 16 | 24,007,245 | 24,024,978 | 4 | PRKCB1 | rs2470688 | 2.95E-03 |
| 16 | 26,116,243 | 26,150,812 | 6 | rs763980 | 5.97E-05 | |
| 16 | 49,031,049 | 49,060,345 | 4 | rs1592538 | 3.70E-03 | |
| 16 | 53,536,288 | 53,547,149 | 4 | rs8054521 | 5.77E-03 | |
| 16 | 54,055,550 | 54,067,401 | 4 | MMP2 | rs12924764 | 5.38E-03 |
| 16 | 62,400,870 | 62,475,972 | 12 | rs322575 | 5.08E-04 | |
| 16 | 82,329,363 | 82,384,267 | 7 | CDH13 | rs690836 | 3.57E-03 |
| 17 | 10,621,615 | 10,628,617 | 4 | rs9897496 | 3.37E-04 | |
| 17 | 12,053,119 | 12,060,510 | 6 | rs9910495 | 1.23E-03 | |
| 17 | 19,117,656 | 19,175,068 | 8 | EPN2 | rs3785778 | 7.51E-04 |
| 17 | 61,942,803 | 61,966,341 | 9 | PRKCA | rs16959526 | 7.42E-04 |
| 17 | 73,899,797 | 73,911,147 | 7 | PGS1 | rs12944051 | 4.72E-03 |
| 17 | 76,176,198 | 76,219,409 | 4 | KIAA1303 | rs11653499 | 7.29E-03 |
| 17 | 76,321,112 | 76,357,049 | 4 | KIAA1303 | rs9899782 | 1.50E-03 |
| 18 | 5,503,917 | 5,517,864 | 4 | EPB41L3 | rs1618055 | 2.33E-03 |
| 18 | 24,500,885 | 24,521,689 | 4 | rs16945100 | 3.75E-03 | |
| 18 | 34,074,362 | 34,089,743 | 7 | rs8083420 | 2.22E-03 | |
| 18 | 41,488,464 | 41,498,356 | 10 | SLC14A2 | rs9304318 | 4.65E-03 |
| 18 | 63,727,755 | 63,734,371 | 4 | rs12455531 | 7.02E-03 | |
| 18 | 66,882,313 | 66,917,659 | 4 | rs17179440 | 4.97E-04 | |
| 19 | 15,685,071 | 15,718,958 | 4 | rs12975815 | 1.57E-03 | |
| 19 | 46,867,000 | 46,883,137 | 4 | CEACAM7 | rs7251886 | 8.66E-03 |
| 19 | 59,541,275 | 59,565,647 | 4 | LAIR1 and LILRA4 | rs2004431 | 2.05E-04 |
| 20 | 22,256,842 | 22,274,938 | 6 | rs1012800 | 2.54E-04 | |
| 20 | 36,099,124 | 36,172,404 | 8 | C20orf77 | rs6022796 | 4.79E-03 |
| 20 | 46,208,923 | 46,235,831 | 17 | rs151050 | 2.08E-04 | |
| 20 | 58,755,693 | 58,772,191 | 6 | rs6071344 | 3.18E-04 | |
| 21 | 18,415,630 | 18,426,726 | 5 | rs2150385 | 4.67E-04 | |
| 21 | 19,469,347 | 19,587,446 | 19 | SLC6A6P | rs8134931 | 7.07E-04 |
| 21 | 23,287,429 | 23,334,156 | 10 | rs244230 | 8.12E-04 | |
| 21 | 24,030,348 | 24,092,754 | 15 | rs1157277 | 2.31E-04 | |
| 21 | 24,303,999 | 24,355,766 | 10 | rs8134281 | 6.61E-04 | |
| 22 | 24,752,980 | 24,778,643 | 7 | MYO18B | rs6004901 | 2.43E-03 |
| 22 | 31,662,712 | 31,713,419 | 4 | SYN3 | rs17779789 | 2.61E-03 |
| 22 | 40,052,810 | 40,077,663 | 4 | ZC3H7B | rs3817999 | 3.90E-03 |
| 22 | 42,418,082 | 42,454,835 | 4 | FLJ23588 | rs1894489 | 2.67E-03 |
| 22 | 47,156,058 | 47,181,983 | 5 | rs130785 | 7.21E-03 | |
| 23 | 5,289,710 | 5,324,870 | 9 | rs12011665 | 1.37E-03 | |
| 23 | 5,521,502 | 5,547,657 | 4 | rs34291001 | 3.46E-03 | |
| 23 | 7,351,237 | 7,371,322 | 6 | rs17269009 | 2.63E-03 | |
| 23 | 7,559,572 | 7,571,315 | 4 | rs6639914 | 2.59E-03 | |
| 23 | 14,021,870 | 14,066,309 | 8 | rs5935694 | 3.91E-03 | |
| 23 | 15,858,474 | 15,924,353 | 10 | rs705857 | 1.00E-04 | |
| 23 | 26,704,945 | 26,730,141 | 4 | rs4898189 | 1.67E-03 | |
| 23 | 65,661,456 | 65,675,958 | 4 | rs6624988 | 5.23E-03 | |
| 23 | 68,704,238 | 68,751,820 | 6 | EDA | rs4844179 | 2.41E-03 |
| 23 | 83,765,563 | 83,775,407 | 4 | rs830240 | 3.81E-03 | |
| 23 | 83,802,507 | 83,866,535 | 12 | rs707677 | 3.85E-03 | |
| 23 | 86,001,607 | 86,017,818 | 4 | rs1936029 | 4.13E-03 | |
| 23 | 86,669,781 | 86,714,253 | 16 | KLHL4 | rs6617426 | 1.10E-03 |
| 23 | 111,364,987 | 111,397,689 | 5 | ZCCHC16 | rs17307753 | 7.49E-03 |
| 23 | 120,290,236 | 120,347,792 | 16 | rs7054144 | 1.00E-03 | |
| 23 | 144,107,310 | 144,120,517 | 6 | rs9792699 | 5.30E-03 |
Columns list the chromosome and bp coordinates for the beginning and end of the genomic region identified by clustered, nominally positive SNPs from the current study; number of clustered, nominally positive SNPs that lie in clusters within the region in the current sample; the gene(s) (if any) that lie within this chromosomal region; the SNP that displays the nominally smallest P value in the cluster; and the P value displayed by that SNP. Note that several genes are identified by more than one cluster of nominally positive SNPs. Genes identified by clusters of nominally positive SNPs for which ≥ 25% are among the SNPs for which assay variance is largest for Affymetrix 6.0 arrays in other studies are identified in boldfaced italics.
Power for Quit Success Comparisons
We calculated the power of these samples for detection of 5%, 7.5% and 10% differences in allele frequency. We used the mean 0.24 minor allele frequency that we found for nominally positive SNPs in these samples. The power to detect these differences was 0.15, 0.28 and 0.43, respectively.
Overlap with Data from Previous Quit Success Samples
These data for clustered, nominally positive SNPs from the current data set provide significant chromosomal overlap with genes that have been identified by other relevant data sets, largely those derived from validated pooled genotyping approaches (Table 2). These approaches identify the same genes that are identified by nominally positive results in other studies to extents much greater than what we would expect by chance. The overlaps between the clustered, nominally positive SNPs from the current sample and the clustered, nominally positive SNPs from at least two other samples of successful versus unsuccessful quitters and/or nicotine dependence identify 59 genes. Whereas the empirical P values associated with most of these genes do not withstand stringent Bonferroni corrections for multiple testing, several of these gene-wise P values do yield P values <0.0008 and thus survive this correction for multiple testing (21) (Table 2).
Table 2.
Genes that contain clustered, nominally positive SNPs from the current study and clustered, nominally positive SNPs from at least two additional 500,000, 600,000 or 1,000,000 SNP GWA studies of smoking cessation success in pooled DNA samples from subjects of European genetic backgrounds.
| Gene | Chromo-some | bp: Start | bp: End | Current | PIP | V | H | L | R | B | Bi | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIF1B | 1 | 10193418 | 10364242 | 5 | 16 | 8 | 0.0005 | |||||
| DAB1 | 1 | 57236167 | 58488799 | 9 | 93 | 7 | 2 | 3 | 0.0021 | |||
| DNM3 | 1 | 170077261 | 170648480 | 7 | 17 | 3 | 1 | 0.0095 | ||||
| ASTN | 1 | 175096826 | 175400647 | 6 | 12 | 2 | 1 | 0.0082 | ||||
| CTNNA2 | 2 | 79593634 | 80729416 | 6 | 57 | 2 | 2 | 7 | 0.0066 | |||
| TCF7L1 | 2 | 85214245 | 85391016 | 5 | 1 | 2 | 0.0122 | |||||
| RAPGEF4 | 2 | 173308853 | 173625861 | 10 | 6 | 1 | 0.0052 | |||||
| RBMS3 | 3 | 29297947 | 30021624 | 4 | 11 | 2 | 8 | 0.0068 | ||||
| FHIT | 3 | 59710076 | 61212164 | 11 | 105 | 7 | 2 | 2 | 0.0033 | |||
| EEFSEC | 3 | 129355003 | 129610179 | 6 | 17 | 2 | 0.0077 | |||||
| SLC9A9 | 3 | 144466754 | 145049979 | 4 | 33 | 2 | 5 | 0.0099 | ||||
| TP73L | 3 | 190831910 | 191097759 | 3 | 4 | 1 | 1 | 0.0145 | ||||
| LEPREL1 | 3 | 191157316 | 191321412 | 9 | 11 | 4 | 2 | 0.0010 | ||||
| FGF12 | 3 | 193342413 | 193928066 | 5 | 5 | 3 | 2 | 0.0175 | ||||
| RNASEN | 5 | 31436926 | 31567925 | 6 | 1 | 1 | 0.0094 | |||||
| PDE4D | 5 | 58302468 | 58918032 | 4 | 15 | 1 | 0.0428 | |||||
| SLC22A5 | 5 | 131733343 | 131759205 | 6 | 8 | 1 | 0.0039 | |||||
| SLIT3 | 5 | 168025857 | 168660554 | 6 | 24 | 5 | 3 | 3 | 1 | 0.0012 | ||
| KCNIP1 | 5 | 169713459 | 170096214 | 5 | 23 | 3 | 0.0088 | |||||
| KIAA1900 | 6 | 97479324 | 97694980 | 6 | 6 | 1 | 0.0125 | |||||
| GRIK2 | 6 | 101953675 | 102623474 | 2 | 16 | 2 | 2 | 1 | 0.0139 | |||
| UST | 6 | 149110157 | 149439818 | 6 | 41 | 4 | 0.0039 | |||||
| PARK2 | 6 | 161689661 | 163068793 | 6 | 91 | 6 | 2 | 8 | 0.0044 | |||
| DGKB | 7 | 14153770 | 14847413 | 4 | 38 | 1 | 0.0262 | |||||
| MAGI2 | 7 | 77484310 | 78920826 | 6 | 51 | 2 | 1 | 0.0314 | ||||
| SEMA3A | 7 | 83428426 | 83661848 | 12 | 2 | 1 | 0.0024 | |||||
| CSMD1 | 8 | 2782789 | 4839736 | 6 | 191 | 4 | 10 | 10 | 5 | 12 | 0.0015 | |
| DLC1 | 8 | 12985243 | 13416766 | 5 | 17 | 2 | 0.0169 | |||||
| PSD3 | 8 | 18432343 | 18915476 | 18 | 23 | 1 | 0.0013 | |||||
| TG | 8 | 133948387 | 134216325 | 4 | 11 | 3 | 0.0123 | |||||
| ST3GAL1 | 8 | 134540312 | 134653344 | 5 | 9 | 2 | 1 | 0.0042 | ||||
| ZNF406 | 8 | 135559213 | 135794463 | 4 | 21 | 3 | 0.0072 | |||||
| COL22A1 | 8 | 139669660 | 139995418 | 6 | 21 | 1 | 0.0119 | |||||
| PTPRD | 9 | 8307268 | 9008735 | 4 | 42 | 2 | 8 | 2 | 0.0026 | |||
| KIAA1797 | 9 | 20648309 | 20985954 | 5 | 5 | 2 | 0.0214 | |||||
| PIP5K1B | 9 | 70510436 | 70813912 | 4 | 51 | 1 | 0.0030 | |||||
| GABBR2 | 9 | 100090187 | 100511300 | 4 | 19 | 5 | 0.0101 | |||||
| DIP2C | 10 | 311432 | 725606 | 8 | 7 | 3 | 1 | 0.0043 | ||||
| BICC1 | 10 | 59942910 | 60258851 | 4 | 8 | 2 | 0.0244 | |||||
| NRAP | 10 | 115338573 | 115413795 | 1 | 11 | 1 | 1 | 0.0057 | ||||
| CASP7 | 10 | 115428925 | 115480654 | 11 | 10 | 1 | 2 | 0.0004 | ||||
| SLC1A2 | 11 | 35229329 | 35397372 | 7 | 34 | 3 | 3 | 0.0004 | ||||
| SOX5 | 12 | 23576498 | 24606647 | 10 | 48 | 2 | 2 | 0.0053 | ||||
| MYBPC1 | 12 | 100512878 | 100603789 | 8 | 4 | 1 | 0.0047 | |||||
| GPR133 | 12 | 130004790 | 130189786 | 4 | 26 | 1 | 0.0090 | |||||
| NBEA | 13 | 34414456 | 35144873 | 9 | 5 | 1 | 0.0206 | |||||
| LMO7 | 13 | 75092571 | 75332003 | 5 | 23 | 2 | 0.0076 | |||||
| GPC5 | 13 | 90848930 | 92317491 | 1 | 25 | 2 | 1 | 0.1015 | ||||
| GPC6 | 13 | 92677096 | 93853948 | 17 | 40 | 1 | 4 | 0.0013 | ||||
| STK24 | 13 | 97902414 | 98027350 | 4 | 22 | 1 | 0.0075 | |||||
| NPAS3 | 14 | 32478200 | 33340702 | 16 | 70 | 5 | 1 | <0.0001 | ||||
| RGS6 | 14 | 71469586 | 72100407 | 32 | 15 | 1 | 4 | <0.0001 | ||||
| WDR72 | 15 | 51594652 | 51839151 | 7 | 14 | 2 | 2 | 0.0021 | ||||
| HMOX2 | 16 | 4466447 | 4500349 | 4 | 1 | 2 | 0.0113 | |||||
| A2BP1 | 16 | 6009133 | 7702500 | 6 | 181 | 3 | 14 | 12 | 13 | <0.0001 | ||
| CDH13 | 16 | 81218079 | 82387702 | 7 | 160 | 5 | 8 | 3 | 7 | 2 | <0.0001 | |
| PRKCA | 17 | 61729388 | 62237324 | 9 | 21 | 4 | 1 | 0.0036 | ||||
| SLC14A2 | 18 | 41448764 | 41517070 | 10 | 11 | 4 | 0.0002 | |||||
| MYO18B | 22 | 24468120 | 24757007 | 5 | 32 | 5 | 3 | 1 | 0.0015 |
Columns list the gene symbol, chromosome and bp coordinates for the beginning and end of the gene, and numbers of nominally positive, clustered SNPs that fall within the gene from the current study. PIP: 1,000,000 SNP GWA from “patch in practice samples” (8); V: 1,000,000 SNP GWA from a trial of the efficacy of denicotinized cigarettes: Vector samples (7), 500,000 SNP GWA of samples of community quitters and continuing smokers; H: Hamer samples (6); L: Lerman; B: Brown; R: Rose from smoking cessation samples 1–3 from Uhl et al. (5). Bi: Data from comparison of 38,000 SNPs identified in comparisons to smokers with and without FTND dependence from Bierut et al. (11). Monte Carlo P values note the number of times in 10,000 simulation trials that results this strong or stronger are obtained by randomly sampling the same numbers of SNPs from the same data sets. Boldfaced entries denote the genes in which at least three samples identify the same region within the gene.
Control for occult stratification was based on examining the overlap between the 2,147 clustered, nominally positive SNPs from the present quit success analyses with the 2.5% of the SNPs for which the racial/ethnic differences in control individuals from prior data sets were largest. We identified 48 SNPs with these properties; 50 would have been expected by chance. Controls for noisy SNPs found that 70 of the clustered, nominally positive SNPs overlapped with the set of SNPs that provided the largest variance in other assessments of these SNPs using Affymetrix 6.0 arrays, while 50 would be expected by chance. We identify the clusters that contain SNPs that provide greater assay variance in Table 1.
Bayesian Network Analysis
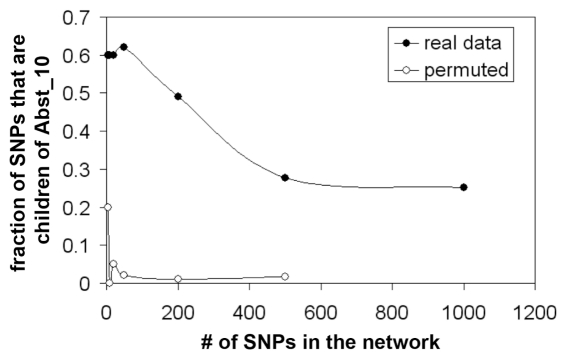
Bayesian networks incorporated many of the SNPs that provided the strongest 25, 50, 100, 200, 500 or 1,000 P values for the true quit success data when analyzed by using BayesWare (Figure 1) (http://bayesware.com [22–25]). By contrast, only a few SNPs were included in the corresponding analyses of data from permutated control sets of SNPs in which there were random relationships between SNPs and the set of P values obtained from the bona fide data (Figure 1). Figure 2 provides a graphic representation of the Bayesian network for data from the 1,000 SNPs with the strongest P values. Interestingly, the relationship between the SNPs for which data directly predicts abstinence in this data set (for example, those in the inner circle forming the “Markov Blanket” of the outcome node) and the SNPs located in the outer circle can be explained by the linkage disequilibrium between the SNPs (data not shown). This relationship would be expected if the network was detecting true biological relationships, but not if the network was detecting noise. However, there were relatively few interrelationships between these “inner circle” SNPs (data not shown), suggesting that linkage disequilibrium was not responsible for much of the influence of these SNPs on quit success.
Figure 1.
Generation of Bayesian network for prediction of abstinence. SNPs are first sorted based on nominal P value, and SNPs with the 5 to 1,000 lowest P values are used. Networks are generated from real data using the Markov Chain Monte Carlo methods using the BayesWare factor.
Figure 2.
Bayesian network including the 1,000 SNPs with the strongest P values in the current study. The SNPs in the inner circle (Markov Blanket of the outcome node) provide the most direct, strongest relationship to abstinence at week 10 of this trial. Many of the relationships between SNPs in the first ring with those in the second, third and fourth rings can be explained by linkage disequilibrium.
The 5,898 SNPs, for which alleles are identified by these results as directly predicting abstinence, display P values that range from 0.0000028 to 0.01 in the primary data set. A total of 960 of these SNPs also display nominally significant association with quit success in at least one other previously reported quit success data set, whereas 32 of these SNPs display such nominally significant associations in at least two prior samples.
Functional Genomic Analyses
A number of genes identified by clusters of nominally significant SNPs in this work fall into several functional classes identified by gene ontology. Functional enrichment analysis (BioBase) that compares the representation of functional classes with all human genes identified significant overrepresentation, when corrected for false discovery rate (FDR), of genes involved in the following: molecular functions, the membrane/plasma membrane, synapses and synaptic transmission of nerve impulses, cell communication, radial glia-guided migration of Purkinje cells, protein binding, neuron projections, protein kinase C activity, cell–cell signaling and communication, cell migration in hindbrain, negative regulation of response to stimulus, localization of the cell, hindbrain radial glia-guided cell migration, cell motion, axon guidance, binding, cell junctions, hind-brain development, signal transduction, nucleoside monophosphate and cAMP metabolic process, G-protein complexes and glutamate receptor activity.
DISCUSSION
The current results provide independent support, from individually genotyped GWA, for data derived from prior studies of smoking cessation success in clinical trial and community settings that used validated methods for pooled genotyping. The substantial overlaps between the autosomal data obtained with individual genotyping and those obtained previously in pooled DNA samples provide mutual validation for the current and previous data sets. The current results provide additional support for polygenic contributions to individual differences in the ability to quit smoking.
These observations can be discussed in light of the strengths and limitations of the current data set. The data display several strengths: (a) the successful and unsuccessful subjects were recruited at the same time from the same study centers, providing significant assurance that contributions of underlying stratification to the results obtained herein have been minimized; (b) both the careful clinical and biochemical monitoring of these participants support the accuracy of smoking cessation assessments; (c) nominally positive results from this work cluster into small chromosomal regions to extents greater than expected by chance; (d) many more of the positive results from this work than we would expect by chance identify the same chromosomal regions that were identified by other studies of smoking cessation and/or vulnerability to develop nicotine dependence in smokers; (e) in these same subjects, a single genotype score per subject that was based on data from the study by Uhl et al. (5) predicted quit success via interactions with nicotine dose and FTND dependence significantly better than at random (P = 0.015 [14]); (f) the true results from this trial, but not permuted results, form a plausible Bayesian network; and (g) the genes identified by these results provide overrepresentation of plausible groups of biological mechanisms in functional enrichment analyses (BioBase).
There are also limitations of these analyses. First, the sample is of modest size from the perspective of GWA, although it is relatively large from the perspective of a clinical trial. This modest sample size provides modest power. This modest power led us to forego analyses of subgroups, such as comparisons between subjects treated with 21 versus 42 mg nicotine. It reduces our confidence in the genes that are identified in this work but not in prior studies and in the negative data concerning genes that have been reproducibly identified in prior studies but not in the current work. Second, individuals in this trial were recruited so that an equal number of participants with FTND scores ≤6 and >6 were randomized to 42 or 21 mg NRT. We combined individuals treated with both doses in the current analysis to increase power, since overall effects of dose on quit success rates were not significant (although effects can be noted in subsets of subjects). Third, we identify no large effects of any SNP assessed here. Data for individual SNPs are less robust than data for clusters of nominally positive SNPs or sets of these clustered SNPs. The data from individual SNPs from this trial, for example, fail to achieve significance in permutation analyses (data not shown). Fourth, more than one-quarter of the SNPs that form seven of the clusters identified in Table 1 are found among the sets of SNPs for which assay variation is large in prior studies using these same Affymetrix 6.0 reagents. Although no cluster is identified solely on the basis of SNPs with these properties, we label these clusters in Table 1 to provide additional cautions in interpreting these results. Fifth, we have not used SNPs, samples or treatments that are identical to those used in prior smoking cessation GWA studies. Each of these issues has limited our enthusiasm about use of SNP-by-SNP meta-analyses, although these metaanalyses might be appropriate when larger data sets are assessed (26–29). Sixth, because some of the chromosomal clusters contain genes with related functions, by selecting all of the genes in a cluster for BioBase analyses, some selection bias may be introduced.
Clustering of SNPs whose allele frequencies display nominally significant differences between successful quitters and those who were not successful provides a major preplanned signal that lies at the core of the analyses used herein. We would anticipate the observed highly significant clustering of SNPs that display nominally positive results in this and several additional independent samples if many of these positive SNPs lay near and were in linkage disequilibrium with functional allelic variants that distinguished subjects who were more able to quit smoking from those who were less able to quit. We would not anticipate this degree of clustering if the results were due to chance. The Monte Carlo P values noted here are thus likely to receive contributions from both the extent of linkage disequilibrium among the clustered, nominally positive SNPs and the extent of linkage disequilibrium between these SNPs and the functional haplotype(s) that lead to associations with quit success. These Monte Carlo P values thus weigh against two null hypotheses: (a) that all of the results are random “noise” (Monte Carlo P values for clustering data from the current study alone) and (b) that the results are caused by stochastic differences in haplo-type frequencies between the successful versus unsuccessful quitters (Monte Carlo P value for clustering data from the current versus prior quit success GWA studies).
The current work has thus identified a set of SNPs that, based on Bayesian network analyses and overlap with prior data sets, are likely to identify a network of SNPs and genes with true biological relationships. Indeed, the genes identified in the current and prior smoking cessation studies are overrepresented in specific GO categories (Table 3). Most of these genes are expressed in the brain, as we might expect for addiction-related traits. Many can be related to neurotransmission processes, as we again might expect for such traits. Although the large number of genes identified in this work precludes detailed discussions of each gene, it is especially interesting to note the substantial representation of “cell adhesion”–related genes among those likely to contain allelic variants that associate with the ability to quit smoking. These genes include DAB1, ASTN, CTNNA2, FHIT, SLIT3, MAGI2, SEMA3A, CSMD1, PTPRD, GPC5, GPC6 and CDH13 (30). It is also interesting that the GO results point to several kinds of biological processes of importance for development of and function of selected brain circuits. We could speculate that variations in such genes could influence brain development, alter basal or preexisting behavioral traits and thus indirectly influence smoking cessation (31).
Table 3.
Gene ontology classes identified by genes from Table 2.
| GO identifier | Group size | Gene symbol(s) | GO term | No. of Hits |
P | |
|---|---|---|---|---|---|---|
| Obsb | Expb | |||||
| GO:0021942 | 2 | CTNNA2, DAB1 | Radial glia guided migration of Purkinje cell | 2 | 1 | 0.004625 |
| GO:0021535 | 4 | CTNNA2, DAB1 | Cell migration in hindbrain | 2 | 1 | 0.005527 |
| GO:0043005 | 194 | CDH13, CTNNA2, DNM3, GABBR2, PARK2, SLC1A2 | Neuron projection | 6 | 1 | 0.005752 |
| GO:0007268 | 304 | CTNNA2, GABBR2, GRIK2, KCNIP1, KIF1B, PARK2, SLC1A2 | Synaptic transmission | 7 | 1 | 0.006198 |
| GO:0007399 | 912 | CTNNA2, DAB1, DLC1, DNM3, FGF12, PARK2, SEMA3A, SLC1A2, SLIT3, SOX5 | Nervous system development | 11 | 3 | 0.006541 |
| GO:0021932 | 3 | CTNNA2, DAB1 | Hindbrain radial glia guided cell migration | 2 | 1 | 0.006923 |
| GO:0007267 | 609 | CTNNA2, FGF12, GABBR2, GRIK2, KCNIP1, KIF1B, PARK2, SLC1A2, TP63 | Cell–cell signaling | 9 | 2 | 0.007025 |
| GO:0030054 | 490 | CTNNA2, DLC1, GABBR2, GRIK2, LMO7, MAGI2, NRAP, PSD3 | Cell junction | 8 | 2 | 0.007113 |
| GO:0007417 | 351 | CTNNA2, DAB1, DLC1, PARK2, SLC1A2, SLIT3, SOX5 | Central nervous system development | 7 | 2 | 0.007605 |
| GO:0042805 | 5 | LMO7, NRAP | Actinin binding | 2 | 1 | 0.007661 |
| GO:0019226 | 349 | CTNNA2, GABBR2, GRIK2, KCNIP1, KIF1B, PARK2, SLC1A2 | Transmission of nerve impulse | 7 | 2 | 0.008389 |
| GO:0051674 | 520 | ASTN1, CDH13, CTNNA2, DAB1, DLC1, PRKCA, SEMA3A, SLIT3 | Localization of cell | 8 | 2 | 0.009009 |
| GO:0043395 | 8 | GPC5, GPC6 | Heparan sulfate proteoglycan binding | 2 | 1 | 0.009135 |
| GO:0006928 | 520 | ASTN1, CDH13, CTNNA2, DAB1, DLC1, PRKCA, SEMA3A, SLIT3 | Cell motion | 8 | 2 | 0.009759 |
| GO:0010259 | 10 | SLC1A2, TP63 | Multicellular organismal aging | 2 | 1 | 0.010234 |
| GO:0043394 | 10 | GPC5, GPC6 | Proteoglycan binding | 2 | 1 | 0.010773 |
| GO:0044456 | 197 | DNM3, GABBR2, GRIK2, PSD3, SLC1A2 | Synapse part | 5 | 1 | 0.011012 |
| GO:0045202 | 294 | DNM3, GABBR2, GRIK2, MAGI2, PSD3, SLC1A2 | Synapse | 6 | 1 | 0.011148 |
| GO:0010646 | 866 | CDH13, DLC1, GRIK2, PARK2, PRKCA, PSD3, RAPGEF4, RGS6, TCF7L1, TP63 | Regulation of cell communication | 10 | 3 | 0.011326 |
| GO:0007154 | 4,037 | CDH13, CTNNA2, DAB1, DGKB, DKFZp434B1272, DLC1, FGF12, GABBR2, GRIK2, KCNIP1 | Cell communication | 25 | 14 | 0.011507 |
| GO:0005515 | 7,814 | A2BP1, ASTN1, CASP7, CDH13, CTNNA2, DAB1, DGKB, DIP2C, DLC1, DNM36 | Protein binding | 38 | 26 | 0.01181 |
| GO:0043616 | 12 | CDH13, TP63 | Keratinocyte proliferation | 2 | 1 | 0.012998 |
| GO:0001964 | 12 | CTNNA2, PARK2 | Startle response | 2 | 1 | 0.013588 |
| GO:0030902 | 52 | CTNNA2, DAB1, DLC1 | Hindbrain development | 3 | 1 | 0.01365 |
| GO:0016477 | 336 | ASTN1, CDH13, CTNNA2, DAB1, DLC1, PRKCA | Cell migration | 6 | 2 | 0.01407 |
| GO:0005912 | 139 | CTNNA2, DLC1, LMO7, NRAP | Adherens junction | 4 | 1 | 0.016601 |
| GO:0040011 | 495 | ASTN1, CDH13, CTNNA2, DAB1, DLC1, PRKCA, SEMA3A | Locomotion | 7 | 2 | 0.017088 |
| GO:0050927 | 15 | CDH13, PRKCA | Positive regulation of positive chemotaxis | 2 | 1 | 0.017505 |
| GO:0045296 | 15 | CDH13, CTNNA2 | Cadherin binding | 2 | 1 | 0.018178 |
| GO:0048870 | 370 | ASTN1, CDH13, CTNNA2, DAB1, DLC1, PRKCA | Cell motility | 6 | 2 | 0.018515 |
| GO:0050926 | 15 | CDH13, PRKCA | Regulation of positive chemotaxis | 2 | 1 | 0.018905 |
| GO:0042995 | 514 | CDH13, CTNNA2, DNM3, GABBR2, PARK2, SLC1A2, SLC22A5 | Cell projection | 7 | 2 | 0.019186 |
| GO:0007626 | 257 | CDH13, NPAS3, PARK2, PRKCA, SEMA3A | Locomotory behavior | 5 | 1 | 0.020432 |
| GO:0003674 | 15,439 | A2BP1, ASTN1, BICC1, CASP7, CDH13, COL22A1, CTNNA2, DAB1, DGKB, DIP2C | Cell adhesion | 57 | 51 | 0.02068 |
| GO:0050918 | 18 | CDH13, PRKCA | Positive chemotaxis | 2 | 1 | 0.020739 |
| GO:0005626 | 685 | HMOX2, MAGI2, PDE4D, PRKCA, PSD3, RAPGEF4, SLC14A2, SLC1A2 | Insoluble fraction | 8 | 3 | 0.020892 |
| GO:0070161 | 157 | CTNNA2, DLC1, LMO7, NRAP | Anchoring junction | 4 | 1 | 0.021548 |
| GO:0051179 | 3,205 | A2BP1, ASTN1, CDH13, CTNNA2, DAB1, DLC1, DNM3, GRIK2, KCNIP1, KIF1B | Localization | 20 | 11 | 0.024066 |
| GO:0021575 | 21 | DAB1, DLC1 | Hindbrain morphogenesis | 2 | 1 | 0.025229 |
| GO:0007610 | 417 | CDH13, NPAS3, PARK2, PRKCA, SEMA3A, SLC1A2 | Behavior | 6 | 2 | 0.026079 |
| GO:0016337 | 284 | ASTN1, CDH13, CTNNA2, DAB1, LMO7 | Cell–cell adhesion | 5 | 1 | 0.02663 |
| GO:0043197 | 23 | DNM3, SLC1A2 | Dendritic spine | 2 | 1 | 0.027998 |
| GO:0044459 | 2,123 | CDH13, CTNNA2, DLC1, GABBR2, GPC5, GPC6, GRIK2, LMO7, MAGI2, NRAP | Plasma membrane part | 15 | 7 | 0.028616 |
| GO:0022610 | 768 | ASTN1, CDH13, COL22A1, CTNNA2, DAB1, DLC1, LMO7, MYBPC1 | Biological adhesion | 8 | 3 | 0.028908 |
| GO:0005913 | 24 | LMO7, NRAP | Cell–cell adherens junction | 2 | 1 | 0.029029 |
| GO:0007155 | 767 | ASTN1, CDH13, COL22A1, CTNNA2, DAB1, DLC1, LMO7, MYBPC1 | Cell adhesion | 8 | 3 | 0.029264 |
| GO:0040017 | 25 | CDH13, PRKCA | Positive regulation of locomotion | 2 | 1 | 0.029388 |
| GO:0050920 | 26 | CDH13, PRKCA | Regulation of chemotaxis | 2 | 1 | 0.029785 |
Genes identified by data from the current study and at least two additional 500,000, 600,000 or 1,000,000 SNP GWA studies of smoking cessation success were subjected to BioBase functional enrichment analyses. Columns list the GO identifier, number of genes supporting the GO class, list of the first several genes that support the class, definition of the GO term, number of genes observed, number of genes expected by chance and FDR-corrected P value. The 48 GO terms with the lowest FDR-corrected P values are listed.
Obs, observed; Exp, expected by chance.
The current data add appreciably to the increasingly robust sets of studies that document molecular genetic contributions to the ability to quit smoking. The present results add to the support for personalized approaches to smoking cessation treatment that come from recent analyses of single genotype–based scores for each of these subjects (14). In this work, abstinence varied on the basis of individual and/or interactive effects of genotype score, nicotine dose and baseline level of nicotine dependence in predicting the degree to which participants were able to reduce smoking during a two-week precessation treatment with NRT. We need to continue to work to apply an integrated sum of SNPs in the context of appropriate clinical information (http://www.genome.gov/27529204) to match individuals with the best type and/or intensity of therapy to maximize benefits and minimize side effects in smoking cessation. One current stepped-care approach based on these aggregate data might entail the following: (a) initial use of NRT, with assignment of nicotine dose based on dependence level and quit success genotype scores, (b) identification of individuals who do not reduce CO sufficiently during initial NRT and (c) prompt reassignment of such non–CO reducers to alternative therapies, such as bupropion or varenecline.
More precise information about genetic influences on the ability to quit smoking from these and prior data sets will aid us in constructing improved “quit success” genotype scores. In subsequent studies, for example, we can test whether the quit success scores in which data from SNPs are selected and weighted by P values (14) perform better or worse than quit success scores in which data from SNPs are selected and weighted on the basis of participation in Bayesian networks, such as those documented here. It is conceivable that such scores may also help us to assess the genetic determinants of generalized abilities to change other health-related behaviors. For both dependent individuals and individuals with other health problems that can be modified through behavior change, these data might thus add to an increasingly rich basis for improved understanding and for development of personalized treatment strategies.
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH)– Intramural Research Program, National Institute on Drug Abuse, Department of Health and Human Services (GRU); a grant to Duke University (JE Rose, principal investigator) from Philip Morris USA (Richmond, VA, USA). The funders had no role in the planning or execution of the study, data analysis or publication of results. We are grateful for help from Joseph E. Herskovic, PhD, Eric C. Westman, MD, Qing-Rong Liu, PhD, and Donna Walther, MS. The underlying clinical trial was registered with clinicaltrials. gov (ID# NCT00734617). This study used BioBase (http://biobase-international.com), installed on the Helix System at the Center for Information Technology (CIT), National Institutes of Health, Bethesda, Maryland (http://helix.nih.gov).
Footnotes
DISCLOSURE
GR Uhl and JE Rose are listed as inventors for a patent application filed by Duke University that specifies sets of genomic markers that distinguish successful quitters from unsuccessful quitters in data from other clinical trials. MF Ramoni has financial interest in BayesWare LLC.
Online address: http://www.molmed.org
REFERENCES
- 1.Adhikari B, Kahende J, Malarcher A, Pechacek T, Tong V. Smoking-attributable mortality, years of potential life lost, and productivity losses: United States, 2000–2004. MMVR Weekly. 2008;57:1226–28. [PubMed] [Google Scholar]
- 2.Zhu S, Melcer T, Sun J, Rosbrook B, Pierce JP. Smoking cessation with and without assistance: a population-based analysis. Am J Prev Med. 2000;18:305–11. doi: 10.1016/s0749-3797(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 3.Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9:64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- 4.Lessov CN, et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34:865–79. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- 5.Uhl GR, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch. Gen. Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drgon T, et al. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Mol Med. 2009;15:21–27. doi: 10.2119/molmed.2008.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drgon T, Johnson C, Walther D, Albino AP, Rose JE, Uhl GR. Genome-wide association for smoking cessation success: participants in a trial with adjunctive denicotinized cigarettes. Mol Med. 2009;15:268–74. doi: 10.2119/molmed.2009.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uhl GR, et al. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics. 11:357–67. doi: 10.2217/pgs.09.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhl GR, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch. Gen. Psychiatry. 2008;65:345–55. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 10.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR. Personalized smoking cessation: interactions between nicotine dose, dependence and quit success genotype score. Mol Med. 2010;16:247–53. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhl GR, et al. Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify “connectivity constellation” and drug target genes with pleiotropic effects. Ann N Y Acad Sci. 2008;1141:318–81. doi: 10.1196/annals.1441.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aveyard P, et al. Weekly versus basic smoking cessation support in primary care: a randomised controlled trial. Thorax. 2007;62:898–903. doi: 10.1136/thx.2006.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141B:918–25. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- 19.Dupont WD, Plummer WD., Jr Power and sample size calculations: a review and computer program. Control Clin Trials. 1990;11:116–28. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 20.Dupont WD, Plummer WD., Jr Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19:589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 21.Gordon A, Glazko G, Qiu X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Ann Appl Stat. 2007;1:179–90. [Google Scholar]
- 22.Ramoni RB, Saccone NL, Hatsukami DK, Bierut LJ, Ramoni MF. A testable prognostic model of nicotine dependence. J Neurogenet. 2009;23:283–92. doi: 10.1080/01677060802572911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himes BE, Dai Y, Kohane IS, Weiss ST, Ramoni MF. Prediction of chronic obstructive pulmonary disease (COPD) in asthma patients using electronic medical records. J Am Med Inform Assoc. 2009;16:371–79. doi: 10.1197/jamia.M2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himes BE, et al. Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics. 2009;10:1393–1412. doi: 10.2217/pgs.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramoni RB, Himes BE, Sale MM, Furie KL, Ramoni MF. Predictive genomics of cardioembolic stroke. Stroke. 2009;40:S67–70. doi: 10.1161/STROKEAHA.108.533273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakaoka H, Inoue I. Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner’s curse. J Hum Genet. 2009;54:615–23. doi: 10.1038/jhg.2009.95. [DOI] [PubMed] [Google Scholar]
- 27.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–47. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorgeirsson TE, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li CY, Liu QR, Zhang PW, Li XM, Wei L, Uhl GR. OKCAM: an ontology-based, human-centered knowledgebase for cell adhesion molecules. Nucleic Acids Res. 2009;37:D251–60. doi: 10.1093/nar/gkn568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiroi N, Scott D. Constitutional mechanisms of vulnerability and resilience to nicotine dependence. Mol. Psychiatry. 2009;14:653–67. doi: 10.1038/mp.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]