Abstract
We have expressed human p53 cDNA in the yeast Saccharomyces cerevisiae and shown that the level of production and the length of the p53 protein depends on the presence of untranslated mRNA regions (UTRs). The expression of the ORF alone leads to a p53 protein of correct size (53 kDa) that accumulates to high levels, concomitantly with the presence of a small amount of a p40 protein (40 kDa). However, when either the entire 5′-UTR and a part of the 3′- or 5′-UTR alone is used, this leads to the production of small amounts of the 40 kDa truncated form only. The p40 protein corresponds to a truncated form of p53 at the C-terminal extremity since it reacts only with a monoclonal antibody recognising the N-terminal epitope. This effect on the amount and length of p53 protein had no correlation at the mRNA level, suggesting that translational control probably occurs through the 5′-UTR. We propose a model of structural interaction between this UTR and a part of the ORF mRNA for the regulation of p53 expression in this heterologous context.
INTRODUCTION
The tumour suppressor gene p53 is mutated or deficient in 50% of all human cancers (1–3). The disruption of p53 function is associated with genetic instability and tumourigenesis (4). The p53 protein plays a central role in cell cycle check-point in response to DNA damage, triggering G1 arrest or apoptosis. The p53 protein is a nuclear transcription factor whose target genes are involved in cell cycle control, such as p21 CIP1/WAF1, which inhibits cyclin-dependent pRB kinase (5,6); bax and bcl2 genes whose products enhance and repress the apoptotic pathway, respectively (7,8); Gadd45 gene, which may be involved in DNA repair (9); and Mdm2 gene, which negatively regulates p53 synthesis by mediating its proteolysis (10).
The p53 protein is active as a tetramer and the 393 amino acid monomer is constituted by an acidic N-terminal region containing a transactivator domain, a central DNA binding domain and a C-terminus which includes an oligomerisation region, a nuclear localisation signal and a site for DNA damage recognition (for reviews see 3,11). Most of the p53 missense mutations in tumours are located in the DNA binding domain with four mutational hot spots affecting residues R175, R248, R249 and R273 (12–14). In wild-type cells the p53 level is very low compared to tumour cells where the mutant p53 is accumulated due to an increase in its half-life from a few minutes to several hours (15). Following DNA damage, the wild-type p53 level increases rapidly and seems to be an important event for the initiation of G1 arrest or apoptosis (11).
Several lines of enquiry show that p53 expression is regulated at least in part at the translational level (16–19). It has been shown that the murine p53 protein has the ability to bind to the 5′-UTR and inhibit its own mRNA translation in an in vitro assay system (20,21). On the other hand, the p53 3′-UTR can repress translation of the corresponding mRNA and of heterologous transcripts in cell-free extracts (22,23). The authors have demonstrated that a negative regulatory element in the distal end of the p53 mRNA (containing an Alu-like sequence) is involved in this translational inhibition in vivo. Possible interactions between RNA-binding factors and the p53 mRNA UTR might contribute to the regulation of human p53 mRNA translation (23).
Human p53 has been expressed in heterologous fungal systems such as the yeast Saccharomyces cerevisiae (24) and Schizosaccharomyces pombe (25). A growth arrest response is observed only in this latter yeast, while in the former such expression led to the development of the ‘fassay’, a functional assay to screen and detect p53 mutations in cell lines, blood and tumours (26).
In this paper, we report the effect of the 5′-UTR and part of the 3′-UTR on human p53 expression in yeast S.cerevisiae. We show that the expression of p53 ORF alone leads to a recombinant protein of 53 kDa accumulating in large amounts after galactose induction of the Gal10/cyc1 promoter, and a limited accumulation of a shorter protein (40 kDa). However, if we express the p53 ORF with the above mentioned portions of UTRs, we obtain only limited amounts of the p40 protein, which corresponds to a truncated form of p53 since it is recognised specifically with a p53 monoclonal antibody directed against the N-terminal but not against the C-terminal epitope. The difference between the p53 protein level in the two recombinant yeast strains was not correlated with differences at the mRNA level since we have shown that the p53 mRNA abundance is quite similar in the two strains (yp53 and yΔp53). In view of this result, we suggest that human p53 expression is controlled at a translational level via the UTRs in this heterologous context. To identify which UTR was involved, we deleted the 5′- or 3′-UTR of p53 and showed that the presence of the 3′-UTR does not affect p53 synthesis since we obtain high levels of the 53 kDa protein. However, when only the 5′-UTR was present, the p40 protein was expressed at a low level, suggesting that the negative control was exerted by the 134 bp of the 5′-UTR. We propose a structural model to explain our results on the translational regulation of human p53 expression in yeast S.cerevisiae.
MATERIALS AND METHODS
Strains and media
The yeast strain W303-1B (α leu2 ura3 trp1 his3 ade2 canR) was used for transformation by LiCl procedure as described (27).
The recombinant strains (yp53, yΔp53, y5′Δp53 and y3′Δp53) were cultivated on minimum (0.67% yeast nitrogen base) or rich (1% bactopeptone, 1% yeast extract) media supplemented with 0.5% glucose as a carbon source and 2% galactose as an inducer.
The Escherichia coli strain was DH5α (supE44Δlac U169, hsdR17, recA1, endA1, gyrA96, thi-1, relA1). Transformation was carried out as described (28).
Plasmids
The YepDP8-1 (provided by D.Pompon, C.G.M., CNRS-Gif sur Yvette, France) was used as a vector to express human p53 cDNA, under the control of the Gal10/cyc1 yeast promoter. This vector contains the 2µ replication origin and the ura3 marker. The wild-type human p53 cDNA was isolated from pBSKII/p53 (provided by Prof. Ozturk, Bilkent University, Ankara, Turkey) as a fragment of 1.478 kb by BamHI–KpnI. After purification from Nusieve agarose gel (TEBU), the p53 cDNA fragment was inserted into BamHI–KpnI sites of YepDP8-1 and the recombinant plasmid was introduced in yeast W303-1B. To construct the p53 cDNA deleted in both 5′- and 3′-UTR portions or in only one of each UTR, we used two oligonucleotides covering the ATG and TGA codons. PCR was done on 100 ng of pBSKII/p53 as follows: 30 s at 94°C, 30 s at 55°C and 2 min at 72°C for 30 cycles. The primers used were: sense primer, 5′-GGGATCCCATGGAGGAGCCGCAGTCA; antisense primer, 5′-GGTACCGAGTGAGTCAGTCTGAGTC. The PCR products were analysed on a 1.2% agarose gel and the fragments were purified on Nusieve agarose gel, cloned in pMosblue-T vector (Amersham) and finally inserted into BamHI–KpnI sites of the YepDP8-1, downstream of the Gal10/cyc1 promoter.
RNA extraction and northern blot analysis
Total RNA was isolated from yeast cells collected from 100 ml of strains yp53, yΔp53, y5′Δp53 and y3′Δp53 cultured on rich medium containing 0.5% glucose, until OD = 3, then supplemented with 2% galactose to induce the Gal10/cyc1 promoter, for 12 h. Cells (1 g) were broken with 1 g of alumina in 10 ml TE (10 mM Tris–HCl pH 8, 1 mM EDTA) buffer. After two phenol–chloroform extractions, nucleic acids were precipitated overnight at –20°C with 2 vol ethanol, and RNA were collected after precipitation by an equal volume of 8 M LiCl at 4°C. Thirty micrograms of total RNA were fractionated by electrophoresis on a formaldehyde–agarose gel and transferred to nylon membranes (Amersham). Filters were hybridised with p53 cDNA 32P-labelled using a Rediprime kit (Amersham). Hybridisations were done in 50% formamide, 1% Denhardt, 1% SDS, 6× SSC (150 mM NaCl, 15 mM sodium citrate) at 42°C overnight followed by two 10 min washes at room temperature with 2× SSC, 0.1% SDS and two 15 min washes at 42°C in 2× SSC, 0.1% SDS.
Protein extraction and western blot
The recombinant strains yp53, yΔp53, y5′Δp53 and y3′Δp53 were cultivated on rich medium containing 0.5% glucose. After 24 h, 2% galactose was added and the cultures were maintained at 30°C for 12 h. Cells were collected and broken by glass beads in 10 mM Tris–HCl pH 7.5, 1 mM EDTA pH 8, 150 mM NaCl, 1% Nonidet P-40 in the presence of protease inhibitors (1 mM PMSF, 2 µg/ml leupeptine and 1 mg/ml pepstatine). After centrifugation at 12 000 r.p.m. (Eppendorf) for 15 min, the supernatant was collected and protein concentration determined by Bio-Rad assay. Thirty micrograms of total protein were separated by 10% SDS–PAGE and electrotransferred to nitrocellulose membranes (Amersham). Immunoblotting was carried out with one of two primary monoclonal anti-p53 antibodies: (i) HR53C1 (from Prof. Ozturk), which recognises N-terminal epitope (121–130 amino acids) and was used at 1:100 v/v dilution or (ii) Ab1 (Amersham), which recognises the C-terminal epitope of the p53 (371–380 amino acids) and was used at 1:100 v/v dilution.
The secondary antibody used was an anti-mouse conjugated to alkaline phosphatase, revealed by NBT and BCIP as recommended by the supplier (Bio-Rad).
RESULTS
Expression of human p53 in yeast S.cerevisiae and its effect on cell growth
We have expressed the wild-type human p53 cDNA using the yeast multicopy vector YepDP8-1 under the control of an inducible promoter Gal10/cyc1. The wild-type p53 cDNA contains, in addition to the 1190 bp of the ORF, the entire 5′-UTR (134 bases) and the beginning of the 3′-UTR (154 bases).
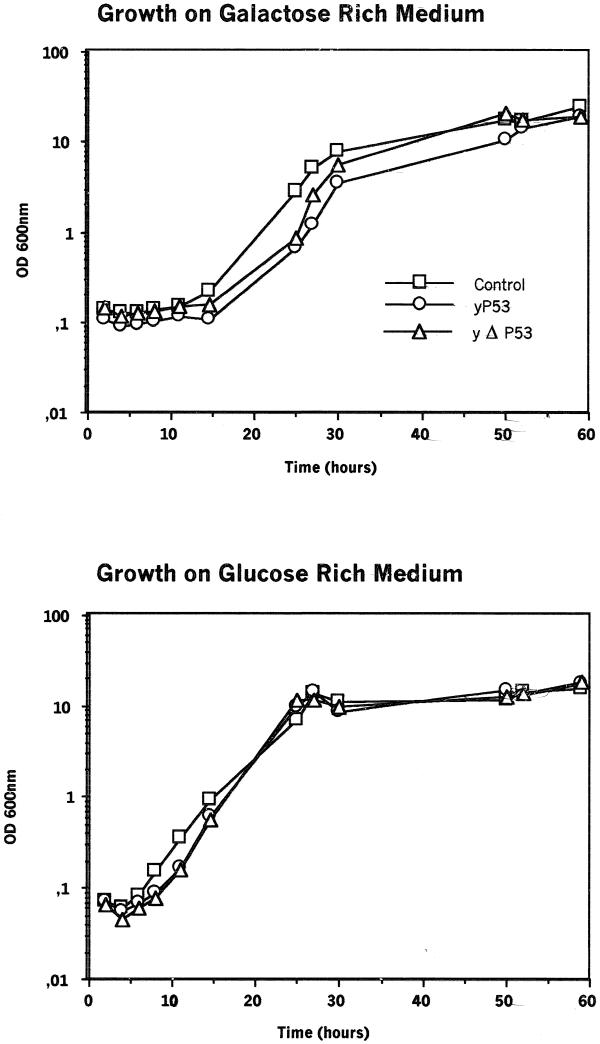
With the yeast strain used, W303-1B (a non-protease-deficient strain), a negative effect of p53 expression on yeast cell growth is clearly shown (Fig. 1). This effect is significant only on galactose where the Gal10/cyc1 promoter is induced. It is significant during the latency and the exponential phases, after which the yp53 strain follows the same growth rate as the control W303/YepDP8-1 and reaches the same plateau at stationary phase but slightly later on (Fig. 1). This effect could be seen on rich as well as on minimal media (data not shown). By northern blotting, we visualised the p53 mRNA; however, the corresponding protein was smaller than expected (only 40 kDa) and was expressed at a low level after galactose induction (see below and Fig. 2A).
Figure 1.
Effect of human p53 on yeast growth. The yeast strains yp53, yΔp53 and the control (W303/YepDP8-1) were pre-cultivated on minimal medium with 0.5% glucose for 2 days and used to inoculate rich media with either 2% galactose (inducing condition) or 2% glucose (repressive condition). We then measured optical density at 600 nm.
Figure 2.
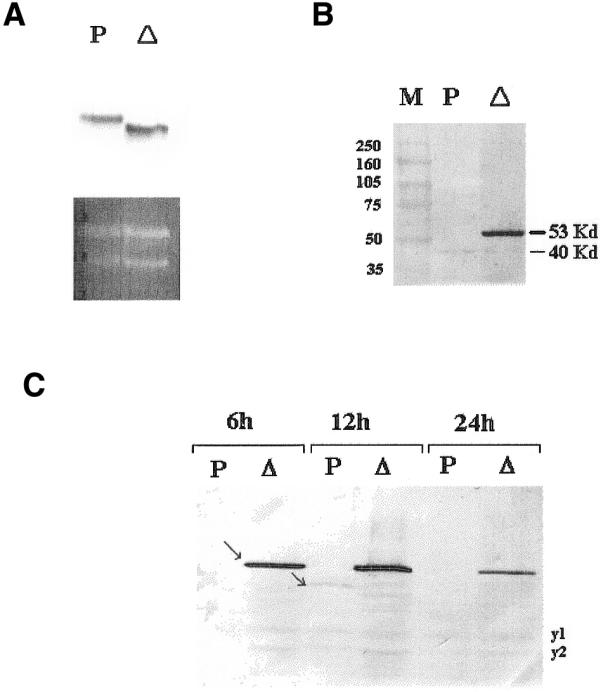
(A) Northern blot analysis of p53 transcripts in the yeast strains yp53 (P) and yΔp53 (Δ). The lower part represents the ethidium bromide-stained gel and the upper part shows the autoradiograph using p53 cDNA as a probe. (B) Western blot analysis of p53 protein expression in the two recombinant yeast strains yp53 (P) and yΔp53 (Δ) using the HR53C1 p53 monoclonal antibody. (C) Kinetics of p53 expression. The yeast strains yp53 (P) and yΔp53 (Δ) were cultivated at 28°C in rich medium with 0.5% glucose, until OD = 3, then 2% galactose was added and aliquots taken at 6, 12 and 24 h. p53 was immunoblotted for the monoclonal antibody HR53C1. Note the cross-reaction with two yeast proteins (y1 and y2), which serve as internal control of loading.
The deletion of 5′- and 3′-UTRs of human p53 cDNA increases its expression in yeast
It is well known that it is important to shorten the distance between the promoter and the ATG codon as much as possible to increase translation efficiency in a heterologous context, henceforth in yeast. For this purpose, we constructed by PCR a truncated p53 cDNA (Δp53) deleted in 134 bp of 5′-UTR and 154 bp of 3′-UTR, using two primers covering the ATG and TGA codons of the p53 ORF. The amplified ORF DNA fragment of 1190 bp was sequenced to verify that no error had occurred during Taq DNA polymerase amplification. This shortened p53 cDNA (Δp53) was inserted in YepDP8-1 and the p53 level in the two yeast strains (yp53 and yΔp53) was compared by western blot analysis. Figure 2B clearly shows that the monoclonal anti-p53 (HR53C1) recognises two proteins in the yΔp53 strain: a 53 kDa protein, highly accumulated, and a 40 kDa protein, weakly produced. Only the 40 kDa protein could be seen in the yp53 strain (Fig. 2B). The growth curves of both strains are strictly similar on glucose as well as on galactose, the negative effect of p53 expression is therefore the same in yp53 and yΔp53 (Fig. 1).
Figure 2C shows another result concerning the kinetics of recombinant p53 expression. Yp53 and yΔp53 strains were cultivated on 0.5% glucose until the end of the exponential phase, then induced by 2% galactose and aliquots were taken after 6, 12 and 24 h. The p53 is present only in yΔp53 with a maximum level reached at 12 h whereas the p40 is present in yp53 only at 12 h post-induction and is completely absent at 24 h. We cannot report in this experiment about its presence at 6 h in this strain because the amount of protein loaded on the gel was very low (notice the absence of yeast proteins y1 and y2 serving as internal controls in Fig. 2C). We verified on other gels that in fact p40 is present at 6 h post-induction in yp53 (see, for example, Fig. 4B). Interestingly enough, at 12 and 24 h, p40 could be seen in yΔp53 strain, but it was absent at 6 h post-induction.
Figure 4.
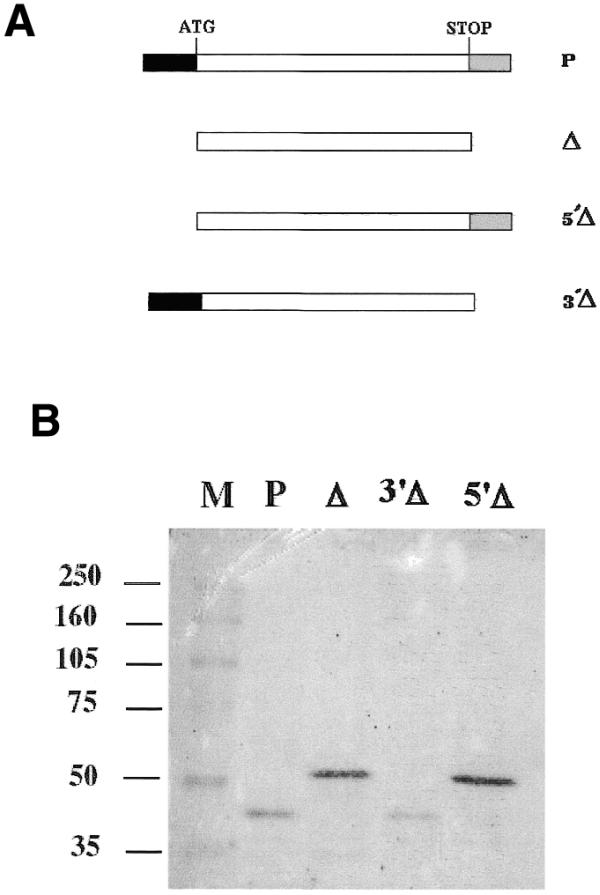
(A) Schematic representation of p53 constructs used in this study. The p53 ORF (open), 5′-UTR (black) and 3′-UTR (grey) are represented. (B) Western blot of p53 protein expression in the recombinant yeast strains yp53 (P), yΔp53 (Δ), y5′Δp53 (5′Δ) and y3′Δp53 (3′Δ). The cultures were stopped at 6 h galactose post-induction. HR53C1 was used as p53 primary monoclonal antibody.
According to these results, we conclude that the presence of both 5′- and 3′-UTRs, or only one of them, has two effects on human p53 expression in yeast: (i) the level of expression is very low and (ii) the p53 protein is truncated. Inversely, the deletion of both 5′- and 3′-UTRs leads to the accumulation of high levels of a full-length 53 kDa protein. These results prompt the following questions: (i) is the difference in the p53 protein level related to a variation in the p53 mRNA in strains yp53 and yΔp53? (ii) Is the 40 kDA protein really a truncated form of p53? (iii) Is there a link between the effect on the expression level and on protein length? (iv) Is this effect related to either the 5′- or the 3′-UTR, or to both?
Comparative analysis of p53 mRNA and protein in yp53 and yΔp53 strains
To determine whether the difference in p53 protein level in yp53 and yΔp53 reflected a difference in the abundance of p53 mRNA, we performed northern blot analyses. Thirty micrograms of total RNA extracted from both strains was hybridised with 32P-labelled p53 cDNA probe. The results show that the relative amount of p53 mRNA is quite similar in both yeast strains suggesting that the deletion of the 5′- and 3′-UTR portions has no effect on p53 transcription (Fig. 2A). In contrast, the UTRs affect the translation process since p53 is very abundant in the yΔp53 whereas in the yp53 strain, only the p40 form was observed and weakly expressed (Fig. 2B). In order to verify that the shorter protein corresponds to a truncated form of p53, we used two anti-p53 antibodies recognising either the N-terminal (HR53C1) or the C-terminal part (Ab1) of the p53 protein. Figure 3 shows that HR53C1 reacts with both p53 and p40 proteins while the Ab1 antibody recognised only the p53 in the yΔp53 strain. Therefore, we conclude that the p40 is a p53 deleted in its C-terminal region.
Figure 3.

Western blot analysis of p53 protein expression in the two recombinant yeast strains yp53 (P) and yΔp53 (Δ) using HR53C1 (left) and Ab1 (right) p53 monoclonal antibodies. HR53C1 and Ab1 recognise the N- and the C-terminal regions of p53, respectively. The cultures were stopped at 12 h post-induction.
Deletion of either 5′- or 3′-UTR and its effect on recombinant p53 synthesis
To identify which UTR was involved in the translational control of p53, we constructed two other recombinant strains, y5′Δp53 and y3′Δp53, harbouring plasmids containing the p53 cDNA minus the 134 bp of the 5′-UTR or the 154 bp of the 3′-UTR, respectively (Fig. 4A). By northern blot analysis, we showed that no quantitative differences were observed at the p53 mRNA level (data not shown). However, the p53 full-length protein was detected only in the y5′Δp53 strain at a high level, concomitant with a low level of the p40 while only the truncated form (p40) was present in y3′Δp53 at a low level of expression (Fig. 4B). Note that in Figure 4B, the p40 was not detected in yΔp53 or in y5′Δp53; the reason is that the cells were collected after 6 h galactose induction. As stated above, at such time of induction, no p40 can be seen (Fig. 2C).
DISCUSSION
We carried out the expression of human p53 in yeast using a multicopy 2µ based vector and showed that the recombinant protein affects the cell growth rate particularly during the latency and the beginning of exponential phases. The yp53 strain reaches the same plateau at stationary phase as the control but after a certain lag. It was previously shown that p53 alters the cell growth in the fission yeast S.pombe (25). However, in the yeast S.cerevisiae, Nigro et al. (24) have previously reported that the growth inhibitory effect is observed only in a protease deficient strain and after shortening the 5′-UTR, when they expressed the human p53 in a centromeric low copy number yeast vector. This effect also became significant if the p53 was co-expressed with the human CDCH2 gene in a non-protease-deficient strain. Our strain is a non-protease-deficient strain but we used is a multicopy vector.
In this paper, we provide evidence for translational control of human p53 expression in the yeast S.cerevisiae. We have shown that 5′- and 3′-UTR deletion does not affect transcription but considerably enhances the translation efficiency of human p53 cDNA in yeast. The p53 mRNA level in both yp53 and yΔp53 strains is quite similar, hence the observed qualitative and quantitative differences on the recombinant p53 protein must result from alterations of the translational process. Indeed, qualitatively, the anti-p53 antibody (HR53C1) recognises a 40 kDa protein in both strains yp53 and yΔp53 and a 53 kDa protein only in yΔp53. Quantitatively, the level of the 53 kDa protein in yΔp53 is much higher than p40 in yp53.
Using a monoclonal antibody recognising the C-terminal part of p53, we showed that the p40 corresponds to a truncated form of p53. A similar pattern of translation products with p53 and p40 proteins was previously obtained when murine p53 mRNA was translated in vitro (20). According to Mosner et al. (20), the p40 is a premature translation termination product, but they did not explore the reasons for such a result.
By constructing two other recombinant strains y5′Δp53 and y3′Δp53 harbouring plasmids containing the p53 cDNA minus the 5′-UTR or the 154 bp of the 3′-UTR, respectively (Fig. 4A), we showed that only the 5′-UTR was involved in the translational control of p53. Indeed, no quantitative differences were observed at the p53 mRNA level, while at the protein level, the p53 full-length protein was detected only in the y5′Δp53 strain at a high level, concomitant with a low level of the p40 and only the truncated form (p40) was present in y3′Δp53 at a low level of expression (Fig. 4B). This indicated that: (i) the 3′-UTR is not involved in the generation of the shorter form of p53 or in the level of expression; (ii) the effect on the expression level and the protein length are connected.
The 5′-UTR is responsible for both effects, with the participation of a distal part of the coding sequence since even in the absence of 5′-UTR, the truncated form exists.
Here, we propose an explanation based on a certain structure adopted by the mRNA in the absence and presence of the 5′-UTR (see below and the model on Fig. 5). We suggest that a particular structure could be established at the terminal part of the p53 ORF that consequently prevents the ribosome from completing the p53 mRNA translation. In fact, in the absence of 5′-UTR, this structure (probably a stem–loop) should have weak free energy that could be easily overrun by the ribosome. In the presence of the 5′-UTR, such a structure is stabilised (which could be either a defolded or a structured form) leading to a complex tertiary structure that cannot be read through by the ribosome. Such a structure could be a ‘pseudoknot’ [Fig. 5A, (1)] or a coaxial stacking of two adjacent stems [Fig. 5A, (2)]. Moreover, the engagement of the 5′-UTR in this structure would have a negative effect on the initiation of translation since it is already established that the eukaryotic ribosome has to slide along the 5′-UTR to reach the initiator ATG codon. Consequently, according to this model, in the absence of the 5′-UTR (Fig. 5B), the presence of the full-length p53 protein at a much higher level than the p40 means that more complete than incomplete translation occurred and reflects the fact that the single ‘stem–loop’ structure is read through more often than not. In the presence of the 5′-UTR, only the p40 could be seen at a low level of expression, reflecting the negative effect of the complex structure on both the initiation and progression of translation.
Figure 5.
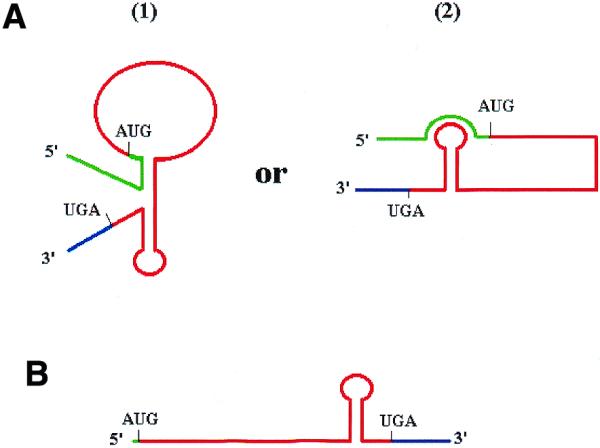
Putative model of p53 post-transcriptional regulation in a heterologous yeast context. (A) In the presence of the 5′-UTR, a complex structure is formed [two possibilities are represented: (1) a ‘pseudoknot’ structure, (2) a coaxial stacking of two stems] and strongly inhibits either the initiation or the progression of the ribosome across the stem–loop structure. Therefore, only the p40 truncated protein is produced at a low level. (B) In the absence of 5′-UTR, only a weak stem–loop is formed that could often be bypassed by the ribosome, giving rise to a large quantity of the 53 kDa protein, but sometimes the ribosome is arrested and led to a weak quantity of p40 protein. Due to the absence of structure at the 5′ end, the initiation of translation is highly improved. Complete 5′-UTR (green); p53 ORF (red); first 154 bases of 3′-UTR (blue).
Many results demonstrate the involvement of UTRs in translational control of gene expression (29). The importance of the p53 5′-UTR in mediating translational control has been shown for murine p53 where the authors gave much evidence to support a model in which the wild-type p53 interacts with its own mRNA by stabilising secondary structure in the 5′-UTR to inhibit its translation (20). It is noteworthy that previous analyses predicted a capacity of the p53 mRNA to form a stable stem–loop structure between nucleotides –216 and –108 in exon 1 of the 5′-UTR and between –216 and +284 with a calculated free energy of –56 and –170 kcal/mol, respectively (20,30,31). Furthermore, it is well known that ‘stem–loop’ structures with free energies of about –50 kcal/mol inhibit translation both in vivo (32) and in vitro (33). In addition, Nigro et al. (24) have also reported the negative effect of the 5′-UTR on the efficient expression of human p53 in yeast: they showed that an improvement in protein expression and a negative effect on growth resulted from the removal of the 5′-UTR.
Furthermore, specific sequences within the 3′-UTR have also been shown to repress translation (34,35). Indeed, a protein that binds specifically to the 3′-UTR of protamine 2 mRNA and represses its translation has been identified (36). In the case of p53, it was shown that in leukaemic bast cells, the p53 protein level does not correlate with the p53 mRNA suggesting that the p53 gene might be regulated at the translational level in AML cells (22). These authors have demonstrated by in vitro transcription-translation experiments that the p53 3′-UTR contains a negative regulatory domain (an Alu-like element). In addition, the full-length p53 3′-UTR, when present in cis of heterologous transcripts, repressed its translation (22). A negative regulatory element capable of inhibiting translation in vivo resides on a 330 nt region at the end of the human p53 3′-UTR, which interacts with RNA binding factors (23). Note that in all our constructs, such an Alu-like element is absent from the 3′-UTR part.
Acknowledgments
ACKNOWLEDGEMENTS
We are particularly grateful to M.Ozturk for the generous gift of p53 cDNA and the p53 monoclonal antibody (HR53C1), D. Pompon for providing us with the YepDP1-8 plasmid, and E.Petrochilo for the critical reading of our manuscript. We wish to thank R.Ellouz, instigator of this project. This work was supported by the International Center of Genetic Engineering and Biotechnology (ICGEB) grant CRP/TUN94-02 (c2).
References
- 1.Nigro J.M., Baker,S.J., Preisinger,A.C., Jessup,J.M., Hostetter,R., Cleary,K., Bigner,S.H., Davidson,N., Baylin,S., Devilee,P., Glover,T., Collins,F.S., Weston,A., Modali,R., Harris,C.C. and Vogelstein,B. (1989) Mutations in the p53 gene occur in diverse human tumour types. Nature, 342, 705–708. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M., Rice,K., Greenblatt,M.S., Soussi,T., Fuchs,R., Sorlie,T., Hovig,E., Smith-Sorensen,B., Montesano,R. and Harris,C.C. (1994) Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res., 22, 3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 3.Levine A.J. (1997) P53, the cellular gatekeeper for the growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 4.Lane D.P. (1992) P53, guardian of the genome. Nature, 358, 15–16. [DOI] [PubMed] [Google Scholar]
- 5.El-Diery W.S, Tokino,T., Velculescu,V.E, Levy,D.B., Parsons,R., Trent,J.M., Lind,D., Mercer,W.E., Kinzler,K.W. and Vogelstein,B. (1993) WAF-1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 6.Dulic V., Kaufmann,W.K, Wilson,S.J., Tlsty,T.D., Lees,E., Harper,J.W., Elledge,S.J. and Reed,S.I. (1994) P53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell, 76, 1013–1023. [DOI] [PubMed] [Google Scholar]
- 7.Zhan Q., Pan,S., Bae,I., Guillouf,C., Lieeberman,D.A., O’Connor,P.M. and Fornace,A.J.,Jr (1994) Induction of bax by genotoxic stress in human cells correlates with normal p53 status and apoptosis. Oncogene, 9, 3743–3751. [PubMed] [Google Scholar]
- 8.White E. (1996) Life, death, and the pursuit of apoptosis. Genes Dev., 10, 1–15. [DOI] [PubMed] [Google Scholar]
- 9.Smith M.L., Chen,I.T, Zhan,Q., Bae,I., Chen,C.Y., Gilmer,T.M., Kastan,M.B., O’Connor,P.M. and Fornace,A.J.,Jr (1994) Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science, 266, 1376–1380. [DOI] [PubMed] [Google Scholar]
- 10.Momand J., Zambetti,G.P., Olson,D.C., George,D. and Levine,A.J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 11.Ko L.J. and Prives,C. (1994) P53: puzzle and paradigm. Genes Dev., 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- 12.Levine A.J., Momand,J. and Finlay,C.A. (1991) The p53 tumour suppressor gene. Nature, 351, 453–456. [DOI] [PubMed] [Google Scholar]
- 13.Hainault P., Soussi,T., Shormer,B., Hollstein,M., Greenblatt,M., Hoving,E., Harris,C.C. and Montesano,R. (1997) Database of p53 gene somatic mutations in human tumors and cell lines: update compilation and future prospects. Nucleic Acids Res., 25, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beroud C., Verdier,F. and Soussi,T. (1996) P53 gene mutation: software and database. Nucleic Acids Res., 24, 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogel A., Popliker,M., Webb,C.A. and Oren,M. (1985) P53 cellular tumor antigen: analysis of mRNAs levels in normal adult tissues, embryos and tumors. Mol. Cell. Biol., 5, 2851–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell S.A and Roth,J.A. (1994) Post translational regulation of p53 tumor suppressor protein function. Crit. Rev. Oncog., 5, 23–57. [DOI] [PubMed] [Google Scholar]
- 17.Ewen M.E. and Miller,S.J. (1996) P53 and translational control. Biochim. Biophys. Acta., 242, 181–184. [DOI] [PubMed] [Google Scholar]
- 18.Oren M. (1999) Regulation of the p53 tumor suppressor protein. J. Biol. Chem., 274, 36031–36034 [DOI] [PubMed] [Google Scholar]
- 19.Somasundaram K. (2000) Tumor suppressor p53: regulation and function. Front. Biosci., 5, 424–437. [DOI] [PubMed] [Google Scholar]
- 20.Mosner J., Mummenbrauer,T., Bauer,C., Sczakiel,G., Grosse,F. and Deppert,W. (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J., 14, 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontoura B.M.A., Atienza,C.A., Sorokina,E.A., Morimoto,T. and Caroll,R.B. (1997) Cytoplasmic p53 is associated with ribosomes. Mol. Cell. Biol., 17, 3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu L., Minden,M.D. and Benchimol,S. (1996) Translational regulation of human p53 gene expression. EMBO J., 15, 4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L. and Benchimol,S. (1997) Participation of the human p53 3′ UTR in translational repression and activation following γ-irradiation. EMBO J., 16, 4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nigro J.M., Sikorski,R., Reed,S.I and Vogelstein,B. (1992) Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischoff J.R., Casso,D. and Beach,D. (1992) Human p53 inhibits growth in Schizosaccharomyces pombe. Mol. Cell. Biol., 12, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaman J.M, Frebourg,T., Moreau,V., Charbonnier,F., Martin,C., Chappuis,P., Sappino,A.P, Limacher,J.M., Bron,L., Benhattar,J., Tada,M., Van Meir,E.G., Estreicher,A. and Iggo,R. (1995) A simple p53 functional assay for screening cell lines, blood, and tumors. Proc. Natl Acad. Sci. USA, 92, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor University Press, Cold Spring Harbor, NY.
- 29.Jackson R.J. (1993) Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell, 74, 9–14. [DOI] [PubMed] [Google Scholar]
- 30.Bienz B., Zakut-Houri,R., Givol,D. and Oren,M. (1984) Analysis of the gene coding for the murine cellular tumour antigen p53. EMBO J., 3, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bienz B., Zakut-Houri,R., Libresco,S., Givol,D. and Oren,M. (1985) The 5′ region of the p53 gene: evolutionary conservation and evidence for a negative regulatory element. EMBO J., 4, 3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNA. Mol. Cell. Biol., 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol., 115, 887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin E.B., Okkema,P.G, Evans,T.C. and Kimble,J. (1993) Translational regulation of tra-2 by its 3′ UTR controls sexual identity in C. elegans. Cell, 75, 329–339. [DOI] [PubMed] [Google Scholar]
- 35.Evans T.C., Crittenden,S.L., Kodoyianni,V. and Kimble,J. (1994) Translational control of maternal glp-1 mRNA establishes an asymmetry in the C.elegans embryos. Cell, 77, 183–194. [DOI] [PubMed] [Google Scholar]
- 36.Kwon Y.K. and Hecht,N.B. (1993) Binding of a phosphoprotein to the 3′ untranslated region of the mouse protamine 2 mRNA temporally represses its translation. Mol. Cell. Biol., 13, 6547–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]