Abstract
Unlike mammals, teleost fish mount a robust regenerative response to retinal injury that culminates in restoration of visual function1, 2. This regenerative response relies on Müller glia (MG) dedifferentiation into a cycling population of progenitor cells. However, the mechanism underlying this dedifferentiation is unknown. Here we report that genes encoding pluripotency factors are induced following retinal injury. Interestingly, the proneural transcription factor Ascl1a and the pluripotency factor Lin-28 are induced in MG within 6 hrs following retinal injury and are necessary for MG dedifferentiation. We demonstrate that Ascl1a is necessary for lin-28 expression and that Lin-28 suppresses let-7 miRNA expression. Furthermore we show that let-7 represses expression of regeneration-associated genes like, ascl1a, hspd1, lin-28, oct4, pax6b and c-myc. Interestingly, hspd1, oct4 and c-myca exhibit basal expression in the uninjured retina and let-7 may inhibit this expression to prevent premature MG dedifferentiation. The opposing actions of Lin-28 and let-7 miRNAs on MG differentiation/dedifferentiation are similar to that of embryonic stem cells3 and suggest novel targets for stimulating MG dedifferentiation and retina regeneration in mammals.
Despite structural and functional similarities between the teleost and mammalian retina, disease or injury of the mammalian retina leads to irreparable vision loss, while the injured teleost retina mounts a regenerative response that restores lost sight1, 2. Key to successful regeneration are MG, which dedifferentiate and assume progenitor properties following retinal injury4–7. In mammals, MG generally respond to injury by reactive gliosis that is accompanied by hypertrophy; rarely do these cells re-enter the cell cycle and regenerate new neurons8–11. These data suggest that a key difference between the regenerative responses of fish and mammals is the ability of MG to dedifferentiate in response to retinal injury. Because zebrafish mount a robust regenerative response following retinal injury, they provide a useful model system for uncovering the underlying mechanisms of MG dedifferentiation. These mechanisms are likely to suggest novel strategies for improving MG dedifferentiation and retina regeneration in mammals.
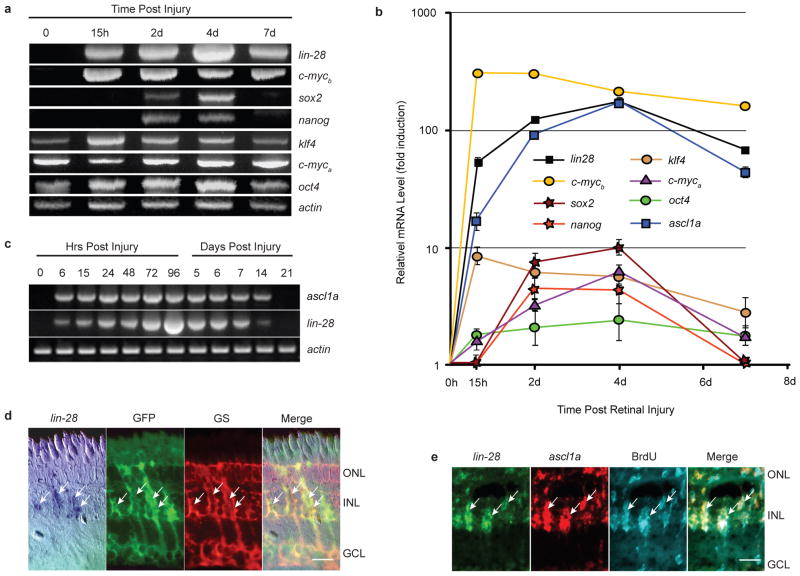
We recently reported that the proneural basic-helix-loop-helix transcription factor Ascl1a is necessary for MG dedifferentiation and retina regeneration12. However, the mechanism by which Ascl1a mediates its effects remained unknown. We reasoned that MG dedifferentiation may share some aspects with reprogramming of somatic cells by pluripotency factors13, 14. Therefore we investigated if pluripotency factors were regulated during retina regeneration. For this analysis, total RNA was isolated from uninjured and injured retinas at 15 hrs post injury (hpi) when MG dedifferentiation is beginning; 2 days post injury (dpi) when MG begin proliferating, 4 dpi when MG proliferation is maximal, and 7 dpi when MG proliferation has stopped and cellular differentiation has begun4, 12. RT-PCR (Fig. 1a) and Real-time PCR (Fig. 1b) showed pluripotency factors klf4, oct4 and c-myca were expressed in the uninjured retina and transiently increased around15 hpi. In contrast, pluripotency factors lin-28, sox2, nanog and c-mycb were undetectable in the uninjured retina and induced either within 15 hpi (lin-28, c-mycb) or by 2 dpi (sox2, nanog). We were particularly intrigued by the lack of detectable lin-28 and mycb expression in the uninjured retina and their rapid and high induction following retinal injury. Here we focused on the role lin-28 plays in retina regeneration.
Figure 1.
ascl1a and lin-28 mRNAs are rapidly induced in dedifferentiating MG following retinal injury. (a) RT-PCR shows induction of pluripotency factor mRNAs following retinal injury. (b) Real-time PCR quantification of pluripotency factor mRNA levels during retina regeneration. Data represent means ± s.d. (n=3 individual fish; compared to uninjured retina, P=0.0001 or less for lin28, c-mycb, and ascl1a expression at all time points post retinal injury; P=0.0001 for klf4 at 15 hpi, and 2 and 4 dpi; P=0.02 for klf4 at 7 dpi; P=0.0001 or less for sox2, c-myca and nanog at 2 and 4 dpi; P= 0.0425 for c-myca at 7 dpi; P=0.0178 and 0.0069 for oct4 at 2 and 4 dpi, respectively). Note Y-axis is fold induction in log scale and normalized to 0 hr time point that is assigned a value of 1. (c) RT-PCR shows ascl1a and lin-28 are coordinately induced following retinal injury. (d) In situ hybridization and immunofluorescence shows lin-28 RNA co-localizes with 1016 tuba1a:gfp transgene expression in glutamine synthetase (GS)+ MG at 4 dpi. Scale bar is 10 microns. (e) lin-28 and ascl1a double fluorescence in situ hybridization and BrdU immunofluorescence show lin-28 and ascl1a are co-expressed in proliferating MG-derived progenitors at 4 dpi. Three hrs prior to sacrifice, adult fish, whose retina was injured 4 days earlier, received an intraperitoneal injection of BrdU. Scale bar is 10 microns. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; BrdU, bromodeoxyuridine; GS, glutamine synthetase; GFP, green fluorescent protein.
A more detailed RT-PCR analysis showed lin-28 induction within 6 hrs post retinal injury, which paralleled that of ascl1a (Fig. 1c). In situ hybridization was used to visualize lin-28 RNA expression, and immunofluorescence was used to identify glutamine synthetase (GS)-expressing MG at different time points following retinal injury in wild-type and 1016 tuba1a:gfp transgenic fish. These latter fish express GFP and incorporate BrdU in MG-derived progenitors located at the injury site4 (Supplementary Information, Fig. S1). Since BrdU is only metabolically available for the first 4 hrs following intraperitoneal injection15, our observation that BrdU injection at 4 dpi identified an expanding population of cells at 5–7 dpi (Supplementary Information, Fig. S1) suggests that BrdU-labeled cells are proliferating. In situ hybridization showed lin-28 expression is restricted to MG and MG-derived GFP+ progenitors at 1and 4 dpi, respectively, that are located at the injury site (Fig. 1d; Supplementary Information, Fig. S2). Although lin-28 and ascl1a gene expression was first detected at 6 hpi, this expression peaked around 3–4 dpi. At this latter time point, ascl1a and lin-28 were co-expressed in proliferating MG-derived progenitors (Fig. 1e). Quantification showed that approximately 83% of the ascl1a+ cells also expressed lin-28, and 91% of the lin-28+ cells also expressed ascl1a. Of the BrdU+ cell population, 80% expressed lin-28, and 82% expressed ascl1a (Supplementary Information, Fig. S3).
Lin-28 is an RNA binding protein whose expression is associated with embryonic stem cells and cancer16. In zebrafish, RT-PCR suggests that lin-28 is a transiently expressed zygotic transcript whose expression peaks around 5 hours post fertilization (hpf) (Supplementary Information, Fig. S4). We were interested in determining if lin-28 expression in the developing retina marked proliferating retinal progenitors similar to what we observed in the adult regenerating retina. Surprisingly, at 24 hpf in situ hybridization and BrdU labeling revealed lin-28 expression in differentiated cells of the hindbrain and spinal cord with undetectable expression in the retina (Supplemental Information, Fig. S4). These results, along with our analysis of pluripotency factor gene expression (Fig. 1a), suggests that MG-derived retinal progenitors are not identical to embryonic retinal progenitors, but rather share characteristics with embryonic stem cells and raise the intriguing possibility that MG-derived progenitors may not be restricted to regeneration of only the retina if placed in the appropriate environment.
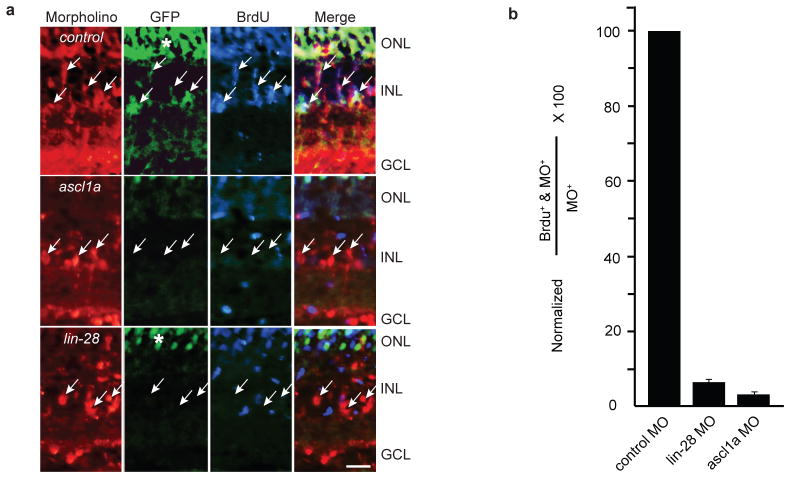
Because Lin-28 is associated with embryonic stem cells and cancer, we were interested in determining if Lin-28 expression was necessary for injury-induced MG dedifferentiation. For this analysis we knocked down the expression of Lin-28 in the adult retina using electroporated morpholino-modified antisense oligonucleotides (MOs); a well characterized approach for suppressing mRNA translation in zebrafish12, 17–19. Because antibodies to zebrafish Lin-28 are not available, we confirmed the efficacy of the lin-28-targeting MOs by co-injecting a plasmid encoding a lin-28-GFP chimeric transcript into single cell zebrafish embryos with and without the lin-28-targeting MO and assaying GFP expression 24 h later (Supplementary Information, Fig. S5). We previously demonstrated that a non-specific control MO electroporated into the injured fish retina had no appreciable effect on the regenerative response, while 2 different MOs targeting ascl1a inhibit Müller glia proliferation following retinal injury12. We confirmed these results here and showed that MO-mediated knockdown of Lin-28 expression dramatically suppressed 1016 tuba1a:gfp transgene expression and MG proliferation (Fig. 2). Quantification indicates that only about 6% of the lin-28 MO #1 and 9% of the lin-28 MO #2 positive cells were also BrdU+ (Fig. 2b; Supplementary Information, Table S1). Hence, both ascl1a and lin-28 are necessary for MG dedifferentiation and proliferation.
Figure 2.
Ascl1a and Lin-28 knockdown inhibit 1016 tuba1a:gfp transgene expression and MG-derived progenitor proliferation. (a) Control, ascl1a or lin-28 lissamine-tagged MOs were electroporated into the retina of 1016 tuba1a:gfp transgenic fish at the time of retinal injury and 3 hrs prior to sacrifice, at 4 dpi, fish received an intraperitoneal injection of BrdU. Arrows point to cells harboring lissamine-tagged MO. GFP and BrdU immunofluorescence show Ascl1a and Lin-28 knockdown suppress 1016 tuba1a:gfp expression and proliferation of MG-derived progenitors. (b) Quantification of the total number of MO+ cells that are in S phase as indicated by BrdU uptake. Data represent means ± s.d. (n=3 individual fish; compared to control MO, lin-28 MO and ascl1a MO P=0.000178 or less). (*) identifies autofluorescence in ONL. Scale bar is 10 microns and applies to all photomicrographs.
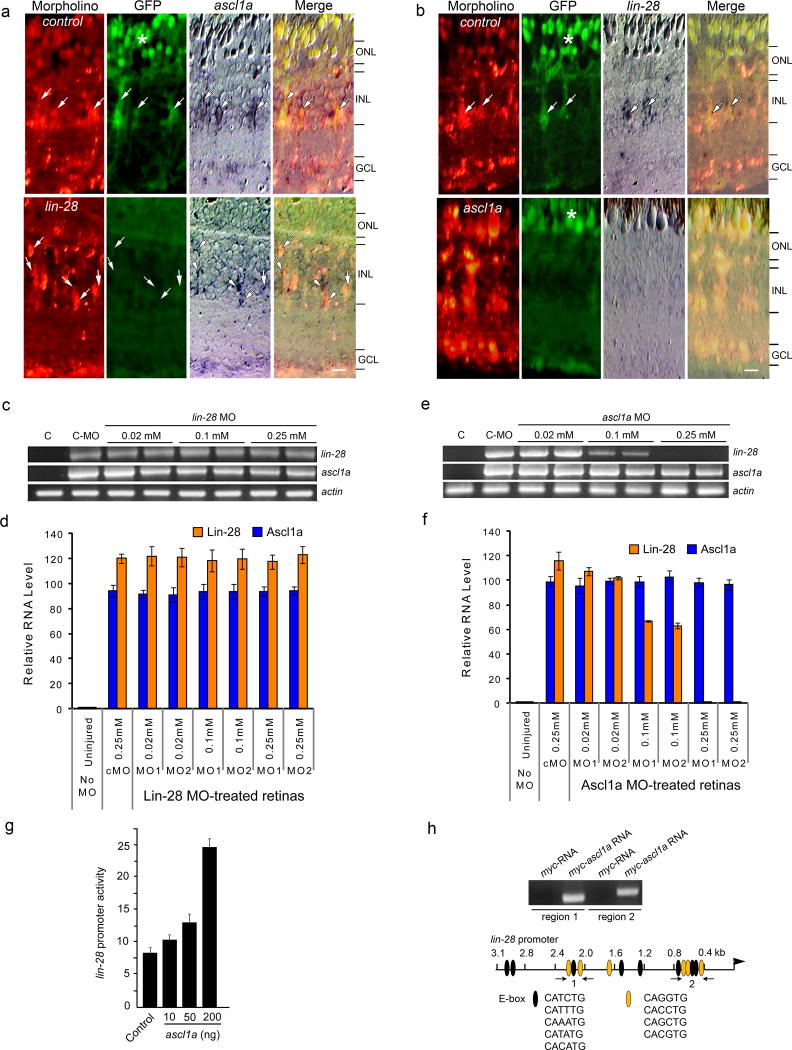
We next investigated if there is any hierarchical regulation or interdependency between Ascl1a and Lin-28. Introduction of control MO into injured retinas of 1016 tuba1a:gfp fish had little effect on the expression of GFP, ascl1a or lin-28 (Fig. 3a,b), while lin-28-targeting MOs specifically suppressed GFP expression when assayed at 4 dpi (Fig. 3a). Interestingly, Ascl1a knockdown suppressed both GFP and lin-28 mRNA expression (Fig. 3b). To further investigate ascl1a and lin-28 interdependency, we examined their expression in MO-treated retinas at 2 dpi, a time correlating with MG dedifferentiation but prior to their reentry into the cell cycle4. Similar to the in situ hybridization studies (Fig. 3a,b), whole retina RT-PCR showed that ascl1a-targeting MOs suppress lin-28 expression in a concentration-dependent manner, while lin-28-targeting MOs had little effect on ascl1a expression (Fig. 3c–f). Real-time PCR indicates that ascl1a-targeting MOs at 0.1mM and 0.25mM cause about a 40% and 99% reduction, respectively, in lin-28 mRNA levels (Fig. 3f).
Figure 3.
Ascl1a regulates lin-28 expression. (a, b) Lissamine-labeled control, ascl1a or lin-28-targeting MOs were electroporated into injured retinas of 1016 tuba1a:gfp transgenic zebrafish. At 4 dpi GFP, and ascl1a and lin-28 mRNA expression was detected by immunofluorescence and in situ hybridization, respectively. Control MO-treated retinas retain GFP, ascl1a and lin-28 expression (arrows), while Lin-28 knockdown suppresses GFP expression and knockdown of Ascl1a suppresses both GFP and lin-28 expression. Scale bar is 10 microns. (c,d) RT-PCR shows Lin-28 knockdown has no effect on injury-dependent induction of ascl1a mRNA at 2 dpi (c), while Ascl1a knockdown blocks injury-dependent induction of lin-28 mRNA at 2 dpi (d). Lane C is control uninjured retina. (e,f) Real-time PCR quantification of the effects of Lin-28 (e) or Ascl1a (f) knockdown on lin-28 and ascl1a mRNA levels at 2 dpi. Data are normalized to uninjured retinas and represent means ± s.d. from 3 replicas of a single experiment. (g) Ascl1a regulates lin-28 promoter activity. HEK293 cells were transfected with lin-28:luciferase and the indicated amounts of cmv:ascl1a along with SV40:Renilla luciferase for normalization. Normalized promoter activity is reported as means ± s.d. (n=3; compared to control, P=0.0123 for 50 ng Ascl1a expression vector and P=0.0001 for 200 ng of Ascl1a expression vector). (h) Ascl1a binds to regions of the lin-28 promoter that harbor multiple E-boxes. ChIP analysis of zebrafish embryos (single cell stage) injected with either myc-RNA or myc-ascl1a mRNA. Immunoprecipitated chromatin was assayed by PCR using primers (arrows) flanking putative Ascl1a binding sites 1 and 2 (ethidium bromide stained gel shown). The predicted fragments sizes of 287 bp and 323 bp were amplified. The 3.1 kb lin-28 promoter diagramed below the gel shows consensus E-box binding sites (ovals). Orange ovals are putative Ascl1 binding sites.
The above results suggest that Ascl1a is required for injury-dependent lin-28 expression. To determine if Ascl1a can directly regulate lin-28 expression, we cloned the zebrafish lin-28 promoter and generated a lin-28:luciferase expression vector. Transfection of HEK293 cells with this vector and either a control or a cmv:ascl1a expression vector showed a concentration-dependent regulation of lin-28 promoter activity by Ascl1a (Fig. 3g). Inspection of the lin-28 promoter sequence identified 14 E-boxes (CANNTG) of which 6 appeared similar (CACCTG, CAGGTG, CAGCTG, CACGTG) to previously reported Ascl1 binding sites (CAGCTG and CAGGTG)20, 21 (Fig. 3h). Two of these sites are clustered in the distal promoter region (region 1 flanked by arrows in Fig. 3h) and 3 of these sites are clustered in a more proximal promoter region (region 2 flanked by arrows in Fig. 3h). To determine if Ascl1a can bind to regions 1 and 2, we performed a ChIP experiment where we injected mRNA encoding either Myc or Myc-Ascl1a into single cell zebrafish embryos and 30 hrs later immunoprecipitated Myc:DNA and Myc-Ascl1a:DNA complexes with an anti-Myc antibody. This ChIP experiment showed that Ascl1a can bind to lin-28 promoter regions 1 and 2, consistent with the idea that Ascl1a directly regulates lin-28 promoter activity (Fig. 3h).
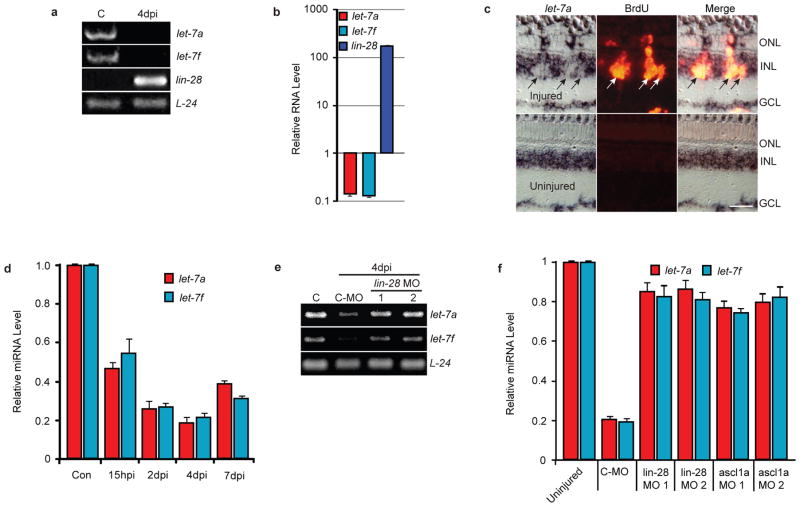
Lin-28 inhibits let-7 miRNA formation and stimulates its degradation22–24. Interestingly, Lin-28 is associated with cell proliferation, while let-7 miRNAs are associated with cellular differentiation3, 16, 22. Therefore, it seemed reasonable to hypothesize that injury-dependent lin-28 induction would lead to let-7 miRNA suppression in dedifferentiated MG. Indeed, these RNAs were regulated in an opposing fashion in MG purified from uninjured and injured retinas (Fig. 4a), with lin-28 mRNA increasing 170-fold and let-7 miRNAs decreasing by 85% (Fig. 4b). Consistent with these data, in situ hybridization at 4dpi with an LNA-modified oligonucleotide probe targeting mature let-7a miRNA showed a dramatic injury-dependent reduction of let-7a expression in MG-derived progenitors that were identified by BrdU incorporation (Fig. 4c). Quantification of let-7 miRNA-negative cells that co-labeled with BrdU showed that 81 ± 1.52% of the BrdU-positive cells were also let-7-negative (n=3). Quantification of injury-dependent let-7 miRNA expression in uninjured and injured retinas showed a ~50% reduction at 15 hpi that increased to an ~80% at 4 dpi; by 7 dpi, let-7 miRNAs begin to return to control levels (Fig. 4d). Consistent with the idea that Ascl1a regulates Lin-28 expression and that Lin-28 controls let-7 miRNA levels22–24, Lin-28 or Ascl1a knockdown almost completely abrogated injury-dependent let-7 miRNA suppression (Fig. 4e,f).
Figure 4.
Lin-28 regulates let-7 miRNA levels in MG-derived progenitors. (a) RT-PCR shows let-7 miRNA expression is high and lin-28 expression is low in differentiated MG (lane C), while let-7 miRNA expression is suppressed and lin-28 expression is highly induced in MG-derived progenitors at 4 dpi. (b) Real-time PCR quantification of lin-28 mRNA and TaqMan PCR quantification of let-7a and let-7f miRNA levels in purified MG and MG-derived progenitors at 4 dpi. Data are normalized to uninjured retina and represent means ± s.d. A single sample, consisting of MG purified from 2 uninjured fish and MG-derived progenitors purified from 10 injured fish, was assayed in triplicate. (c) let-7a in situ hybridization (LNA probe) and BrdU immunofluorescence shows a dearth of let-7a miRNA in BrdU+ MG-derived progenitors of the injured retina at 4dpi. Scale bar is 20 microns. (d) TaqMan PCR quantification of let-7 miRNA levels in uninjured and injured retinas at different times after injury. Data represent means ± s.d. (n=3 fish; compared to control uninjured retina, P=0.0001 or less for let-7a and let-7f at all time points). (e) RT-PCR shows Lin-28 knockdown with 2 different MOs relieves injury-dependent let-7a and let-7f miRNA suppression. Lane C is uninjured retina. (f) Lin-28 or Ascl1a knockdown relieves injury-dependent let-7 miRNA suppression. TaqMan PCR was used to quantify let-7a and let-7f miRNA levels. Data represent means ± s.d. from 3 replicas of a single experiment.
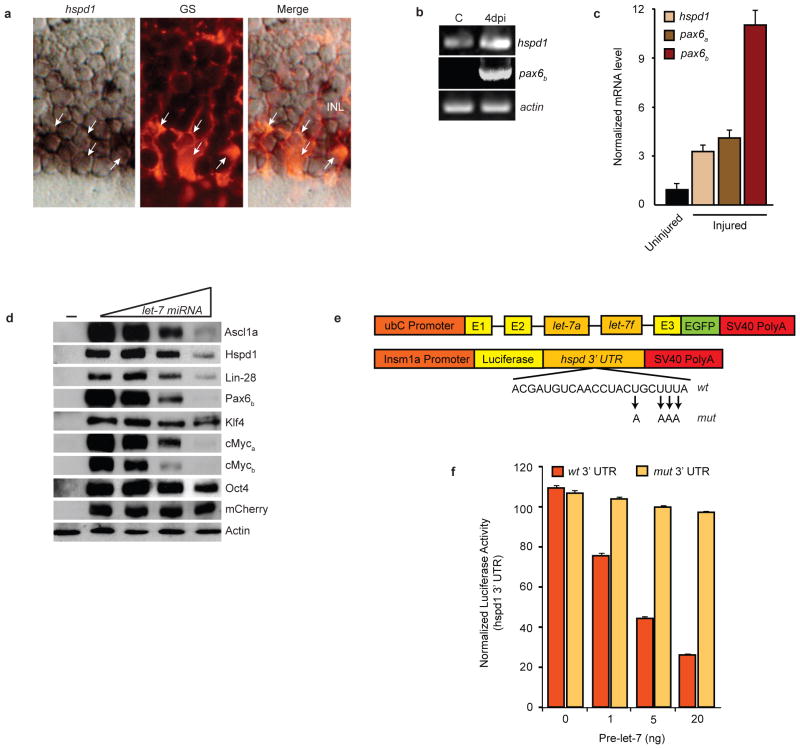
It is interesting that MG in the uninjured retina express genes associated with retinal progenitors25–28 and pluripotency (Fig. 1a,b). Let-7 is associated with cellular differentiation and may contribute to maintaining the differentiated state of MG in the uninjured retina and promoting redifferentiation of MG-derived progenitors in the injured retina by suppressing expression of regeneration and pluripotency-associated genes. Indeed, let-7 miRNAs are known to inhibit Myc and Lin-28 expression during cancer progression, embryonic stem cell development and neural commitment3, 22, 29. Interestingly, all 6 genes known to be necessary for retina regeneration (ascl1a, lin-28, pax6a, pax6b, mps1, and hspd1)12, 30, 31 harbor putative let-7 binding sites32 (Supplemental Information, Table S2). Although let-7 binding sites in the corresponding mouse genes are predicted; in general their sequences are not conserved between zebrafish and mouse. Of the genes necessary for regeneration, ascl1a, lin-28 (Fig. 1) and mps130 are undetectable in the uninjured retina, while pax6a expression4, 31, 33 is restricted to amacrine and ganglion cells. Basal hspd1 expression has been reported in the uninjured retina30 but cell-type specificity remains uncharacterized; likewise pax6b expression is not well characterized. We found that injury-dependent induction of hspd1, pax6a, and pax6b lagged behind that of ascl1a, lin-28 and c-mycb, with hspd1 increasing ~3.5-fold, pax6a increasing ~ 4.5-fold and pax6b increased ~11-fold at 4 dpi (Fig. 5c and Supplementary Information, Fig. S6). Interestingly, of all these regeneration-associated genes, only hspd1 exhibits significant basal expression in GS+ MG of the uninjured retina (Fig. 5a,b). Therefore, hspd1 may represent a let-7 target whose basal expression and injury-dependent induction are suppressed in a let-7-dependent manner.
Figure 5.
let-7 miRNAs suppress expression of regeneration and pluripotency-associated genes. (a) hspd1 in situ hybridization and glutamine synthetase (GS) immunofluorescence shows hspd1 RNA co-localizes (arrows) with GS-expressing MG in the uninjured retina. (b) RT-PCR shows hspd1 mRNA expression in uninjured retina that is increased following retinal injury, while pax6b expression is undetectable in the uninjured retina and dramatically induced following retinal injury. (c) Real-time PCR quantification of hspd1, pax6a and pax6b mRNA levels in uninjured and 4 day post injured retina (injured). Data represent means ± s.d. (n=3 fish; compared to uninjured retina, P=0.0001 or less for hspd1, pax6a and pax6b, respectively, at 4 dpi. (d) let-7-dependent suppression of zebrafish proteins that are necessary for retina regeneration or associated with pluripotency. HEK 293 cells were transfected with flag or myc-tagged regeneration or pluripotency-associated gene expression vectors (50 ng) with increasing concentrations (0, 50, 200, 500 ng) ubC:let-7a,let-7f along with pCS2:mCherry for normalization. (−) lane indicates untransfected cells. Forty-eight hrs later cells were harvested and proteins resolved by denaturing SDS-PAGE. Western blots were probed with anti-mCherry, anti-Flag or anti-Myc antibodies. let-7-dependent regulation of protein expression was quantified by densitometry (Supplementary Information, Fig. S7). Experiments were repeated 3 times with similar results. See Fig. S9 for full scans. (e) Constructs used to overexpress let-7 and investigate the function of putative let-7 miRNA binding sites in the hspd1 3’ UTR. Mutations in the let-7 binding site seed sequence are indicated. (f) The hspd1 3’ UTR let-7 binding site at position 1970 confers let-7-dependent regulation on luciferase expression. Luciferase activity from 24 hpf zebrafish embryos that were co-injected at the single cell stage with the luciferase reporter and the let-7 miRNA expression vector described in (e) along with capped Renilla luciferase mRNA for normalization. Data represent means ± s.d. (n=3 fish; compared to wt 3’UTR construct in absence of pri-let-7 miRNA or mut 3’ UTR construct with pri-let-7 miRNA, P=0.0001 or less for luciferase-wt 3’ UTR activity treated with 1, 5 and 20 ng of ubC:let-7a,let- 7f construct.
To determine if let-7 can regulate the expression of proteins that are essential for regeneration we appended a sequence encoding a Myc or Flag-tag to the 5’ end of the ascl1a, hspd1, lin-28 and pax6b coding sequence and cloned them into the pCS2expression vector. Transfection of HEK 293 cells with these vectors, ± an expression vector harboring the zebrafish let-7a,let-7f pri-miRNA sequence (ubC:let-7a,let-7f ; Fig. 5e), showed a concentration- dependent suppression of Ascl1a, Hspd1, Lin-28 and Pax6b protein expression (Fig. 5d). We also investigated if let-7 could regulate the expression of pluripotency factors Klf4, cMyca, cMycb and Oct4, all of whose transcripts harbor putative let-7 binding sites (Supplementary Information, Table S2) and, except for c-mycb, exhibit basal expression in the uninjured retina (Fig. 1a). Interestingly, in addition to let-7-dependent suppression of pluripotency factor Lin-28, we also observed suppression of cMyca, cMycb and Oct4 in transfected HEK 293 cells (Fig. 5d). Densitometric quantification of Western blots suggests robust suppression of Ascl1a, Hspd1, Lin-28, Pax6b, cMyca, and cMycb, while Oct4 is modestly suppressed and Klf4 appears unaffected (Supplementary Information, Fig. S7).
We then focused on the hspd1 mRNA to determine if the predicted let-7 binding site was functional. For this analysis single cell zebrafish embryos were injected with increasing amounts of ubC:let-7a,let-7f and either insm1a:luciferase-hspd3’UTRwt or insm1a:luciferase-hspd3’UTRmut that harbor a wild type or a mutant (4 base change in putative let-7 binding site seed sequence) zebrafish hspd1 3’ UTR behind the luciferase coding sequence and under control of the zebrafish insm1a promoter (Fig. 5e). This analysis showed that the hspd1 3’ UTR conferred let-7-dependent regulation on reporter expression and that this regulation was mediated by the let-7 binding sites (Fig. 5f). These data suggest that injury-dependent let-7 regulation can have a dramatic impact on gene expression during retina regeneration.
In conclusion, our studies have uncovered a mechanism by which Ascl1a contributes to retina regeneration and suggests that Ascl1a-dependent regulation of the pluripotency gene lin-28 is crucial for stimulating MG dedifferentiation into a cycling population of progenitor cells. We propose that Ascl1a-dependent induction of Lin-28 contributes to MG dedifferentiation, in part, by decreasing let-7 miRNA levels, thus relieving repression of regeneration-associated mRNAs that are essential for MG dedifferentiation (Supplementary Information, Fig. S8). Because some pluripotency factors and regeneration-associated genes exhibit basal expression in MG of the uninjured retina and are inhibited in a let-7-dependent manner, it appears let-7 may also help maintain MG in a differentiated state in the absence of retinal injury. Because Lin-28 is an RNA binding protein that can shuttle between the nucleus and cytoplasm and may regulate other genes, including those that control the cell cycle16, 34, we suspect a much wider role for Lin-28 in retina regeneration. Likewise, Ascl1a is likely to have many additional targets in addition to lin-28 and identification of these targets should further our understanding of mechanisms underlying MG dedifferentiation and retina regeneration. A distinguishing feature of dedifferentiated MG is the expression of lin-28, which is not shared with other adult stem cell populations or with retinal progenitors generated during development and suggests MG-derived progenitors may share properties with embryonic stem cells. The opposing relationship between Lin-28 protein and let-7 miRNAs is similar to that reported in embryonic stem cells3 and cancer29, and suggests mammalian MG dedifferentiation may be enhanced by manipulating this signaling pathway.
Methods
Animals
Zebrafish were kept at 26–28 °C on a 14hr light, 10 dark cycle. Transgenic gfap:GFP zebrafish were kindly provided by Dr. David Hyde (University of Notre Dame)35 and 1016 tuba1a:gfp fish (previously 1016 α1TI:pEGFP) fish were previously described4, 12. Embryos for injections were obtained by natural mating of wild type adults.
Plasmid construction
cDNAs and genomic DNAs were amplified by RT-PCR using zebrafish adult retinal mRNA and embryonic genomic DNA, respectively. The ascl1a cDNA was amplified using ascl1a-F and ascl1a-R primers that harbor an EcoRI and XhoI site, respectively at their 5’ ends (Supplementary Information, Table S3) and cloned into pCS2+MT vector (provided by David Turner, University of Michigan) to generate cmv:myc-ascl1a. A 3.1 kb lin-28 promoter, whose 3’ end was just upstream of the translation start site, was amplified using lin-28-pro-F and lin-28-pro-R primers harboring an XhoI and ApaI restriction site, respectively, at their 5’ ends (Supplementary Information, Table S3) and cloned into a luciferase expression vector to create lin-28:luciferase. A 3 kb insm1a promoter, whose 3’ end was just upstream of the translation start site, was amplified using insm1a-pro-F and insm1a-pro-R primers harboring an XhoI and BamHI site, respectively, at their 5’ ends (Supplementary Information, Table S3) and cloned in the the pEL vector (provided by Michael Uhler, University of Michigan) that harbors a GFP-luciferase fusion to generate insm1a:luciferase. The 3’UTR of hspd1 was amplified using hspd1-UTR-F and hspd1-R primers that harbor an EcoRI and NheI site, respectively, at their 5’ ends (Supplementary Information, Table S3) and inserted at the 3’ end of the luciferase cDNA in expression vector insm1a:luciferase to generate insm1a:luciferase-hspd3’UTR. The pre-let7a and pre-let-7f miRNAs are encoded on a contiguous 1.5 kb genomic DNA fragment, which was amplified using primers Pre-let-7af-F and Pre-let-7af-R harboring an EcoRI and XbaI site, respectively, at their 5’ ends (Supplementary Information, Table S3) and cloned into the second intron of the UI4-GFP-SIBR vector36 (provided by David Turner).
Site-directed mutagenesis
We mutated 5 nucleotides in the putative let-7 binding site in the hspd1 3’ UTR using site directed mutagenesis. A typical PCR amplification contained approximately 50 ng of dsDNA (either insm1a:luciferase-hspd13’UTR), 20 pM of each of the two oligonucleotide primers (mut-hspd1-F and mut-hspd1-R; Supplementary Information, Table S3), 2 μl of 10 mM dNTP mix, 0.2 μl (1 unit) of proofreading thermo polymerase (PCR extender system, 5 Prime) and 2.5 μl of 10X reaction buffer in a reaction volume of 25 μl. Amplification conditions were: 94 °C for 5 min without enzyme, followed by 2 min on ice; Polymerase was then added and 30 cycles of denaturation at 93 °C for 30 sec, annealing at 58 °C for 1 minute and extension at 68 °C for 10 min was performed. Following PCR, the product was treated with DpnI to and mutagenized DNA purified and electroporated into XL1-Blue cells. Transformed cells were selected on LB-kanamycin plates and colonies harboring mutant sequences were identified by PCR and confirmed by DNA sequencing.
RNA isolation, RT-PCR, Real-time PCR and TaqMan PCR
Total RNA was isolated from control and injured retinas using Trizol (Invitrogen). cDNA synthesis was performed using random hexamers and Superscript-II reverse transcriptase (Invitrogen). PCR reactions used Taq and gene specific primers (Supplementary Information, Table S3) under the following conditions: denaturation at 93 °C for 15 sec, annealing at 60 °C for 30 sec and extension at 68 °C for 60 sec. Accession numbers for mRNAs assayed by PCR are in Supplementary Information, Table S2. Real-time PCR reactions were carried out in triplicate with ABsolute SYBR Green Fluorescein Master Mix (ThermoScientific) on an iCycler real-time PCR detection system (BioRad). The Δ ΔCt method was used to determine relative expression of mRNAs in control and injured retinas and normalized to L-24 or β-actin mRNA expression levels.
miRNA quantification was performed using TaqMan microRNA probes (Applied Biosystems) according to the manufacturer’s instructions. Briefly, total RNA was reverse transcribed using stem-loop RT primers (Applied Biosystems) and Real-time PCR performed with a TaqMan PCR kit on an Applied Biosystems 7300 sequence detection system.
Cell culture, transfection and Western blots
HEK293 cells were seeded in a 24 well plate at ~40% confluence and grown in DMEM, 10% fetal bovine serum, antibiotics and antimycotics at 37 °C incubator with 5% CO2. Cells were transfected 24 hrs post plating. For assaying lin-28 promoter activity, cells were transfected with lin-28:luciferase and varying amounts of cmv:ascl1a along with an SV40:Renilla luciferase normalization vector. Forty-eight hrs later cells were harvested for luciferase assays. For examining let-7-dependent regulation of gene expression, cells were transfected with 50 ng of pCS2 vector harboring flag-tagged (hspd1, klf4, c-myca, c-mycb) or myc-tagged (ascl1a, lin-28, oct4, pax6b) cDNA, along with 0, 50, 200 or 500 ng of the ubC:let-7a/let-7f and 50 ng of the β-actin2:mCherry normalization vector. Forty-eight hrs post transfection cells were harvested and protein expression assayed on Western blots. Western blots were quantified by densitometry using NIH Image J software (Version 1.43). Normalization to mCherry expression or endogenous β-actin expression gave similar results.
mRNA synthesis and microinjections
Capped mRNAs encoding Myc and Myc-Ascl1a were synthesized in vitro from linearized cmv:myc-ascl1a using the mMESSAGE mMACHINE kit (Ambion) according to manufacturer’s directions. Approximately 50 pg of mRNA (1 nl) was injected into single cell embryos. Embryos were harvested for ChIP experiments at 30 hpf. ChIP was performed as previously described37. For examining if the hspd1 3’ UTR confers let-7-dependent regulation in zebrafish, we injected single cell zebrafish embryos with approximately 200 pl of a stock solution containing 0.1 ng/μl of Renilla luciferase mRNA (for normalization), 20 ng/μl of insm1a:luciferase-hspd13’UTRwt or insm1a:luciferase-hspd13’UTRmut and 0, 1, 5 or 20 ng/μl of ubC:let-7a/let-7f. Embryos were harvested at 24 hpf for dual luciferase reporter assays (Promega). Graph shows average of experiment performed in triplicate. Error bars are standard error of the mean.
Retinal injury, BrdU labeling, FACS and MO-mediated gene knockdown
Eye lesions have been previously described4, 12, 38. Briefly, fish were anesthetized and under microscopic visualization, the right eye was pulled from its socket and stabbed 4–8 times through the sclera with a 30g needle inserted to the length of the bevel. Lissamine-tagged MOs (Gene Tools, LLC) (~0.5 μl of a 0.1–0.5 mM) were delivered at the time of injury using a Hamilton syringe attached to the needle. MO delivery to cells was facilitated by electroporation as previously described12. The control and 2 different ascl1a-targeting MOs were previously described12. lin-28-targeting MOs were: MO #1 TGAGATGCGGATTTGCCGGGGGCAT and MO #2 ACTAGGCCATACAATTAACTGCTTT. Because antibodies that recognize zebrafish Lin-28 are not available, we used an indirect method to confirm the efficacy of the lin-28-targeting MO. For this analysis we appended the MO target sequence to the amino terminus of EGFP in the cmv:egfp expression vector and injected zebrafish embryos with this expression vector along with either lissamine-tagged control or lin-28-targeting MOs. EGFP expression, 24 h later, was used to assay MO efficacy (Supplementary Information, Fig. S6). For BrdU labeling, adult fish received a 20 μl injection of BrdU (20 mM) 3 hrs prior to sacrifice; embryos were immersed in 20 mM BrdU, 4% DMSO for 3 hrs prior to harvesting. FACS sorting of GFP+ MG was performed by dissecting uninjured retinas from 2 gfap:gfp transgenic fish or 4 days post injured retinas (10 needle pokes/retina) from 10 1016 tuba1a:gfp transgenic fish. Retinas were collected in 0.5 ml Leibovitz’s L15 media, treated for 15 min with 1mg/ml hyaluronidase at room temp and then dissociated in 0.01% trypsin for an additional 10 min with frequent trituration. A single cell suspension was confirmed by microscopy and cells were washed in L15 medium before sorting on a BC Biosciences FACSViDa 3 laser high-speed cell sorter. A total of approximately 90,000 GFP+ cells were obtained from 12 injured retinas and 235,000 GFP+ cells were obtained from 2 uninjured retinas.
Tissue preparation, immunohistochemistry and in situ hybridization
Adult fish were overdosed with tricaine methane sulfonate and eyes were enucleated, followed by removal of the lens and then immersion in fresh 4% paraformaldehyde in 0.1M phosphate buffer, pH 7.4 for ~16 hrs. After fixation, samples were cryoprotected in phosphate-buffered 20% sucrose before embedding with Tissue-Tek O.C.T. compound (Sakura, Finetek). Embedded samples were kept frozen at −80 °C until sectioned to 8 microns on a CM3050S cryostat (Leica). Sections were collected on Superfrost/Plus slides (Fisher Scientific), dried and stored at −80 °C. Immunohistochemistry was performed as previously described4, 38 using the following primary antibodies: rat anti-BrdU (dividing cell marker, 1:400; Abcam); rabbit anti-GFP (1:1000; Invitrogen); mouse anti-glutamine synthetase (GS) (Müller glia marker, 1:500; Chemicon/Millipore). Secondary antibodies were conjugated to Alexa Fluor 488 and used at the following dilutions: 1:500 for anti-mouse, 1:250 for anti-rat and 1:1000 for anti-rabbit. For BrdU staining, sections were pretreated with 2N HCl for 20 min at 37 °C and then soaked in 100 mM sodium borate for 10 min. Following immunhistochemical staining, slides were rinsed with water and allowed to dry in the dark prior to cover-slipping with 2.5% PVA (PVA-polyvinyl alcohol)/DABCO (1,4 diazabicyclo [2.2.2]octane). Slides were examined in a Zeiss Axiophot fluorescence microscope equipped with a digital camera or an Olympus FluoView FV1000 confocal imaging system.
Combined in situ hybridization and antibody staining were performed on retinal sections as described previously39. Double in situ hybridizations were done according to manufacturer’s instructions (Perkin Elmer). Sense control probes were generated and showed no signal above background (data not shown). In situ hybridization for let-7a miRNA expression was performed using a zebrafish let-7a LNA probe (Exiqon). The probe was diluted to 1 μM in prehybridization buffer and hybridization and wash conditions were as previously described40.
Statistical Analyses
Data were analyzed by analysis of variance followed by a post-hoc Fisher’s PLSD t-test.
Supplementary Material
1016 tuba1a:gfp transgenic fish specifically express GFP in MG-derived progenitors that have reentered the cell cycle. (a) Shown are fluorescent photomicrographs of retinal sections at 4 dpi after incubation with anti-GFP and anti-BrdU antibodies demonstrating transgene GFP (green) expression in BrdU-labeled (blue) MG (arrows point to clusters of double-labeled cells). Note the GFP+/BrdU+ cells have processes that span from the GCL to the ONL which is characteristic of and unique to MG. Also note that GFP+/BrdU+ cells are localized to the site of injury (*). (b) Quantification of BrdU+ cells at various times post injury. Fish retinas fish received an IP injection of BrdU at 4dpi and then harvested 3.5 hrs later (4 dpi), 24 hrs later (5 dpi), 48 hrs later (6 dpi) and 72 hrs later (7 dpi). Retinal sections were immunostained with anti-BrdU antibody and BrdU+ cells quantified. Graphed values are means ± s.d. (n=3; compared to day 4 post injury, P=0.0001 for days 5 and 6 post injury; P=0.0071 for day 7 post injury). Scale bar is 10 microns.
lin-28 expression is restricted to the retinal injury site and co-localizes with MG as early as 1dpi. (a) Low magnification photomicrograph of in situ hybridization shows lin-28 expression (arrows point to blue/purple in situ hybridization signal) at the injury site (*) at 1dpi. (b) High magnification of in situ hybridization for lin-28 expression (arrows) and immunofluorescence for MG-specific marker, glutamine synthetase (GS) (red), shows co-labeling in the injured retina at 1 dpi. (c) Low magnification of in situ hybridization at 4 dpi shows lin-28 expression is restricted to the injury site (*)(arrows point to in situ hybridization signal in INL). Scale bar in (a) and (c) is 50 microns and in (c) is 10 microns.
ascl1a and lin-28 are co-expressed in proliferating cells of the injured retina. Tables show the number of ascl1a+, lin-28+, ascl1a+ & lin-28+, ascl1+ & BrdU+, and lin-28+ & BrdU+ cells in the retina at 4 dpi. The graph shows the percentages of cells that co-label with the markers as indicated. Graphed data represent means ± s.d. (n=3 fish).
Transient expression of lin-28 during development. (a) RT-PCR shows lin-28 is a zygotic mRNA whose expression peaks around 5 hpf. (b) Section through a 24 hpf embryo showing in situ hybridization with a lin-28 probe (black arrows point to dark blue in situ hybridization signal in left- and right-hand panels) and BrdU labeling of proliferating cells (red/orange signal in middle panel and right-hand panels; white arrowheads point to BrdU+ cells in the retina). Embryos at 24 hpf are lying curved over the yolk and this section shows the head at the top of the section and spinal cord at the bottom; the middle portion that is not intact is a result of sectioning through the yolk. Scale bar is 100 microns.
lin-28-targeting MO effectiveness shown by knockdown of lin-28-EGFP chimeric mRNA reporter expression in MO-injected zebrafish embryos. Single cell zebrafish embryos were injected with lissamine-tagged MO (red) and a cmv:lin-28-EGFP reporter plasmid where the amino terminus of EGFP is appended with the MO target sequence. (a, d, g) Bright-field images of 24 hpf embryos injected with either a control MO (a) or one of 2 different lin-28-targeting MOs (d and g). (b, e, h) Red fluorescence shows embryos received lissamine-tagged MO. (c, f, i) GFP expression in MO-treated fish. The table below the images shows quantification of the number of MO injected embryos (identified by lissamine fluorescence) with GFP expression at 24 hpf. Approximately 50% of the embryos injected with MOs targeting lin-28 survive and these were used for quantification. Almost all embryos injected with the control MO survived. Scale bar is 100 microns.
Injury-dependent induction of hspd1, pax6a and pax6b. Real-time PCR was used to quantify hspd1, pax6a and pax6b mRNA levels in uninjured and injured zebrafish retinas at the indicated time points. Expression levels were normalized to that in the uninjured retina which was set at 1. Data represent means ± s.d. (n=3 fish; compared to uninjured retina, P=0.0001 0.0001 and 0.0009 for hspd1 at 2, 4 and 7 dpi, respectively; P= 0.0001 for pax6a at 4 dpi; P=0.0006, 0.0001 and 0.0001 for pax6b at days 2, 4 and 7 post injury, respectively).
Quantification of let-7-dependent suppression of zebrafish regeneration-associated and pluripotency factor protein expression shown in Fig. 5f. HEK 293 cells were transfected with 50 ng of Flag or Myc-tagged expression vectors harboring the indicated cDNAs (x-axis) and various amounts (0–500 ng) of ubC:let-7a,let-7f along with a β-actin2:mCherry normalization vector. Forty-eight hrs later cells were harvested for analysis of protein expression on Western blots. Western blots were quantified by densitometry using NIH image J software. Normalization to either transfected mCherry or endogenous Actin gave similar results. Western blot scans are shown in Fig. S9.
A model depicting Ascl1a-dependent regulation of lin-28 gene expression and the antagonism of let-7 miRNAs and Lin-28 in regulating MG differentiation/dedifferentiation. Proteins are printed in regular script and begin with a capital letter; mRNAs are in italic. Activation or repression pathways active in a given cell state are indicated in bold, while inactive pathways are indicated by faint line and lettering.
Western blots full scans for data presented in Fig. 5d.
Acknowledgments
We thank David Hyde for sharing gfap:gfp transgenic fish; Robert Thompson, David Turner and Michael Uhler for sharing expression vectors and reagents; James Beals for help with confocal microscopy; The UM Flow Cytometry Core for help purifying GFP-labeled MG; Vaibhav Kapuria for help with Western blots; Bernard R. Wilfred and David Turner for advice on miRNAs; Peter Macpherson for help with statistics; Tori Melendez for expert care of fish; David Turner, Jack Parent and members of the Goldman lab for their support and comments on this work. This work was supported by funds from the NIH NEI RO1 EY018132 (DG) and NIH NICHD T32HD007507 (RR).
Footnotes
Author contributions. DG and RR designed and analyzed the research and wrote the paper. DG generated the 1016 tuba1a:gfp transgenic fish. RR performed all experiments except for the following: BF assayed let-7a and let-7f miRNA levels by RT-PCR reported in Fig. 4a and e.
Competing Financial Interests. The authors declare no competing financial interests.
References
- 1.Sherpa T, et al. Ganglion cell regeneration following whole-retina destruction in zebrafish. Dev Neurobiol. 2008;68:166–181. doi: 10.1002/dneu.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mensinger AF, Powers MK. Visual function in regenerating teleost retina following cytotoxic lesioning. Vis Neurosci. 1999;16:241–251. doi: 10.1017/s0952523899162059. [DOI] [PubMed] [Google Scholar]
- 3.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooto S. Potential for neural regeneration in the adult mammalian retina. Nippon Ganka Gakkai Zasshi. 2006;110:864–871. [PubMed] [Google Scholar]
- 9.Wan J, et al. Preferential regeneration of photoreceptor from Muller glia after retinal degeneration in adult rat. Vision Res. 2008;48:223–234. doi: 10.1016/j.visres.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Karl MO, et al. Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A. 2008;105:19508–19513. doi: 10.1073/pnas.0807453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda M, et al. alpha-Aminoadipate induces progenitor cell properties of Muller glia in adult mice. Invest Ophthalmol Vis Sci. 2008;49:1142–1150. doi: 10.1167/iovs.07-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 14.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zupanc GK, Horschke I. Proliferation zones in the brain of adult gymnotiform fish: a quantitative mapping study. J Comp Neurol. 1995;353:213–233. doi: 10.1002/cne.903530205. [DOI] [PubMed] [Google Scholar]
- 16.Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 18.Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 19.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, et al. Identification and analysis of the mouse basic/Helix-Loop-Helix transcription factor family. Biochem Biophys Res Commun. 2006;350:648–656. doi: 10.1016/j.bbrc.2006.09.114. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 22.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jadhav AP, Roesch K, Cepko CL. Development and neurogenic potential of Muller glial cells in the vertebrate retina. Prog Retin Eye Res. 2009;28:249–262. doi: 10.1016/j.preteyeres.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roesch K, et al. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 30.Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thummel R, et al. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Thummel R, et al. Characterization of Muller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res. 2008;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassen SC, et al. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- 36.Chung KH, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindeman LC, Vogt-Kielland LT, Alestrom P, Collas P. Fish'n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol Biol. 2009;567:75–86. doi: 10.1007/978-1-60327-414-2_5. [DOI] [PubMed] [Google Scholar]
- 38.Senut MC, Gulati-Leekha A, Goldman D. An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J Neurosci. 2004;24:7663–7673. doi: 10.1523/JNEUROSCI.2281-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jowett T. Double in situ hybridization techniques in zebrafish. Methods. 2001;23:345–358. doi: 10.1006/meth.2000.1147. [DOI] [PubMed] [Google Scholar]
- 40.Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1016 tuba1a:gfp transgenic fish specifically express GFP in MG-derived progenitors that have reentered the cell cycle. (a) Shown are fluorescent photomicrographs of retinal sections at 4 dpi after incubation with anti-GFP and anti-BrdU antibodies demonstrating transgene GFP (green) expression in BrdU-labeled (blue) MG (arrows point to clusters of double-labeled cells). Note the GFP+/BrdU+ cells have processes that span from the GCL to the ONL which is characteristic of and unique to MG. Also note that GFP+/BrdU+ cells are localized to the site of injury (*). (b) Quantification of BrdU+ cells at various times post injury. Fish retinas fish received an IP injection of BrdU at 4dpi and then harvested 3.5 hrs later (4 dpi), 24 hrs later (5 dpi), 48 hrs later (6 dpi) and 72 hrs later (7 dpi). Retinal sections were immunostained with anti-BrdU antibody and BrdU+ cells quantified. Graphed values are means ± s.d. (n=3; compared to day 4 post injury, P=0.0001 for days 5 and 6 post injury; P=0.0071 for day 7 post injury). Scale bar is 10 microns.
lin-28 expression is restricted to the retinal injury site and co-localizes with MG as early as 1dpi. (a) Low magnification photomicrograph of in situ hybridization shows lin-28 expression (arrows point to blue/purple in situ hybridization signal) at the injury site (*) at 1dpi. (b) High magnification of in situ hybridization for lin-28 expression (arrows) and immunofluorescence for MG-specific marker, glutamine synthetase (GS) (red), shows co-labeling in the injured retina at 1 dpi. (c) Low magnification of in situ hybridization at 4 dpi shows lin-28 expression is restricted to the injury site (*)(arrows point to in situ hybridization signal in INL). Scale bar in (a) and (c) is 50 microns and in (c) is 10 microns.
ascl1a and lin-28 are co-expressed in proliferating cells of the injured retina. Tables show the number of ascl1a+, lin-28+, ascl1a+ & lin-28+, ascl1+ & BrdU+, and lin-28+ & BrdU+ cells in the retina at 4 dpi. The graph shows the percentages of cells that co-label with the markers as indicated. Graphed data represent means ± s.d. (n=3 fish).
Transient expression of lin-28 during development. (a) RT-PCR shows lin-28 is a zygotic mRNA whose expression peaks around 5 hpf. (b) Section through a 24 hpf embryo showing in situ hybridization with a lin-28 probe (black arrows point to dark blue in situ hybridization signal in left- and right-hand panels) and BrdU labeling of proliferating cells (red/orange signal in middle panel and right-hand panels; white arrowheads point to BrdU+ cells in the retina). Embryos at 24 hpf are lying curved over the yolk and this section shows the head at the top of the section and spinal cord at the bottom; the middle portion that is not intact is a result of sectioning through the yolk. Scale bar is 100 microns.
lin-28-targeting MO effectiveness shown by knockdown of lin-28-EGFP chimeric mRNA reporter expression in MO-injected zebrafish embryos. Single cell zebrafish embryos were injected with lissamine-tagged MO (red) and a cmv:lin-28-EGFP reporter plasmid where the amino terminus of EGFP is appended with the MO target sequence. (a, d, g) Bright-field images of 24 hpf embryos injected with either a control MO (a) or one of 2 different lin-28-targeting MOs (d and g). (b, e, h) Red fluorescence shows embryos received lissamine-tagged MO. (c, f, i) GFP expression in MO-treated fish. The table below the images shows quantification of the number of MO injected embryos (identified by lissamine fluorescence) with GFP expression at 24 hpf. Approximately 50% of the embryos injected with MOs targeting lin-28 survive and these were used for quantification. Almost all embryos injected with the control MO survived. Scale bar is 100 microns.
Injury-dependent induction of hspd1, pax6a and pax6b. Real-time PCR was used to quantify hspd1, pax6a and pax6b mRNA levels in uninjured and injured zebrafish retinas at the indicated time points. Expression levels were normalized to that in the uninjured retina which was set at 1. Data represent means ± s.d. (n=3 fish; compared to uninjured retina, P=0.0001 0.0001 and 0.0009 for hspd1 at 2, 4 and 7 dpi, respectively; P= 0.0001 for pax6a at 4 dpi; P=0.0006, 0.0001 and 0.0001 for pax6b at days 2, 4 and 7 post injury, respectively).
Quantification of let-7-dependent suppression of zebrafish regeneration-associated and pluripotency factor protein expression shown in Fig. 5f. HEK 293 cells were transfected with 50 ng of Flag or Myc-tagged expression vectors harboring the indicated cDNAs (x-axis) and various amounts (0–500 ng) of ubC:let-7a,let-7f along with a β-actin2:mCherry normalization vector. Forty-eight hrs later cells were harvested for analysis of protein expression on Western blots. Western blots were quantified by densitometry using NIH image J software. Normalization to either transfected mCherry or endogenous Actin gave similar results. Western blot scans are shown in Fig. S9.
A model depicting Ascl1a-dependent regulation of lin-28 gene expression and the antagonism of let-7 miRNAs and Lin-28 in regulating MG differentiation/dedifferentiation. Proteins are printed in regular script and begin with a capital letter; mRNAs are in italic. Activation or repression pathways active in a given cell state are indicated in bold, while inactive pathways are indicated by faint line and lettering.
Western blots full scans for data presented in Fig. 5d.