Abstract
The association of invasive ovarian carcinoma risk with the functional polymorphism rs2228570 (aka rs10735810; FokI polymorphism) in the vitamin D receptor (VDR) gene was examined in 1820 white non-Hispanic cases and 3479 controls in a pooled analysis of five population-based case-control studies within the Ovarian Cancer Association Consortium. Odds ratios (ORs) and 95% confidence intervals (CIs) were estimated using unconditional logistic regression. Carriers of the rare T allele were at increased risk of ovarian carcinoma compared to women with the CC genotype in all studies combined; each copy of the T allele was associated with a modest 9% increased risk (OR=1.09; 95% CI:1.01–1.19; p=0.04). No significant heterogeneity among studies was observed (p=0.37) and, after excluding the dataset from the Hawaii study, the risk association for rs2228570 among replication studies was unchanged (OR=1.09; 95% CI: 1.00–1.19; p=0.06). A stronger association of rs2228570 with risk was observed among younger women (aged < 50 years versus 50 years or older) (p=0.04). In all studies combined, the increased risk per copy of the T allele among younger women was 24% (OR=1.24; 95% CI: 1.04–1.47; p=0.02). This association remained statistically significant after excluding the Hawaii data (OR= 1.20; 95% CI: 1.01–1.43; p=0.04). No heterogeneity of the association was observed by stage (p= 0.46), tumor histology (p=0.98), or time between diagnosis and interview (p=0.94). This pooled analysis provides further evidence that the VDR rs2228570 polymorphism might influence ovarian cancer susceptibility.
Keywords: invasive ovarian carcinoma, vitamin D receptor gene (VDR), single nucleotide polymorphism, pooled analysis, case-control study
Introduction
In the past decade, there has been growing interest in a causal role of vitamin D in the incidence of chronic disease, including cancer. 1 In an early ecologic study, Lefkowitz and Garland 2 reported higher ovarian cancer mortality associated with lower regional sunlight exposure in the US. This result was confirmed in several subsequent ecologic analyses. 3 The plausibility that dietary vitamin D is involved in ovarian cancer etiology is enhanced by its inverse association with breast 4,5 and colon cancers, 6,7 malignancies with possible etiologic similarities to ovarian cancer. 8 Although dietary studies of vitamin D and disease risk are limited because of the influence of sunlight exposure, at least one case-control study reported an inverse association of dietary vitamin D with ovarian cancer risk. 9 A possible association of ovarian cancer risk with vitamin D exposure is supported by laboratory investigations demonstrating that vitamin D and its synthetic analogs inhibit growth and induce apoptosis in ovarian cells in culture and in animal models of ovarian cancer. 10–12
Most of the actions of vitamin D are mediated by the vitamin D receptor (VDR), a nuclear transcription factor. 10,13,14 VDR is present in normal ovarian epithelium, human ovarian tumors, and in human ovarian cancer cell lines. Overall, 83% of the normal ovarian surface epithelium is VDR-immunoreactive. 14 Among polymorphisms in the vitamin D receptor (VDR) gene described to date, rs2228570 (aka rs10735810) is the only single nucleotide polymorphism (SNP) that has been shown to affect the VDR protein structure (reviewed in Uitterlinden et al.15). This SNP is also known as the FokI polymorphism due to the presence or absence of a restriction enzyme site. 15 It is a coding non-synonymous SNP in the translational initiation codon that determines the formation of two protein variants: a longer version of the VDR protein that corresponds to the T allele and a form shortened by three amino acids corresponding to the C allele. Several in vitro studies showed that the shortened VDR form was more effective in transactivation of the vitamin D signal. 16–18 In addition, the rs2228570 SNP has not been found to be in linkage disequilibrium with any other polymorphisms in the VDR gene. 15
Previously, we reported that the VDR rs2228570 SNP may be an ovarian carcinoma susceptibility marker. 19 Among white non-Hispanic women, compared to CC, the CT and TT genotypes were associated with a more than two-fold increased risk [CT: odds ratio (OR) = 2.5; 95% confidence interval (CI):1.3–4.8 and TT: odds ratio (OR) = 2.1; 95%CI: 0.8–5.2]; and the per allele increased risk was 56% (95% CI:1.01–3.41; p for trend=0.04).19 In the present study, limited to non-Hispanic white women with invasive ovarian carcinoma, we present a replication analysis of our putative significant findings by including four additional studies within the Ovarian Cancer Association Consortium (OCAC), a forum for researchers to evaluate promising genetic associations with ovarian cancer with increased power. 20
Material and Methods
Study Design and Population
This pooled analysis of five population-based studies from the US (GEOCS, Stanford, CA and HAW, Honolulu, HI) and Europe (MALOVA, Denmark, and SEARCH and UKOPS, United Kingdom) included 1820 cases with primary histologically-confirmed invasive ovarian carcinoma and 3479 controls. Control subjects were randomly selected from the same geographical area as cases. Eligibility criteria for controls included age 18 years or older, no prior history of ovarian cancer, and having at least one intact ovary. All cases and controls were white non-Hispanic women. A detailed description of the studies has been previously published 21–25 and is summarized in Table 1. Epidemiological data were collected using structured questionnaires that included socio-demographic and health-related information, menstrual, reproductive and gynecological histories. OCAC members submitted their epidemiological data through a secure website to Duke University where the variables have been reviewed, cleaned, and merged. All studies were approved by the review boards and ethics committees of their parent institutions, and written informed consent was obtained from all participants. In addition, Duke University has Institutional Review Board approval as a data coordinating center.
Table 1.
Description of the studies included in the analysis and VDR rs2228570 minor allele and genotype frequencies by case-control status
| Study Name | Case Ascertainment |
Selection of controls |
No. participants (participation rate) |
VDR rs2228570 genotype | MAF among controls |
P† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Invasive Cases |
Controls | No. (%) Cases | No. (%) Controls | |||||||||
| CC | CT | TT | CC | CT | TT | |||||||
| MALOVA22 (Malignant Ovarian Cancer Study, Denmark) |
Incident cases (30–80 years old) diagnosed 1994–1999 from Copenhagen, Frederiksberg, and surrounding counties |
Randomly selected from the general female population within the study area (aged 30–80 years) using the computerized Central Population Register |
424 (79%) |
1183 (67%) |
159 (38) | 208 (49) | 57 (13) | 475 (40) | 545 (46) | 163 (14) | 0.37 | 0.74 |
| SEARCH23 (Studies of Epidemiology and Risk Factors in Cancer Heredity: Ovarian Cancer Study, UK) |
Cases <70 years old from East Anglia, West Midlands, and Trent regions of England; prevalent cases diagnosed 1991–1998; incident cases diagnosed 1998–2004 |
Randomly selected from the EPIC-Norfolk cohort of 25000 individuals aged 45–74 based in the same geographical regions as the cases |
813 (67%) |
1224 (84%) |
296 (36) | 406 (50) | 111 (14) | 484 (40) | 552 (45) | 188 (15) | 0.38 | 0.14 |
| GEOCS24 (Genetic Epidemiology of Ovarian Cancer Study, CA, USA) |
Incident cases (20–64 years old) diagnosed from 1997– 2002 in Greater Bay Area Cancer Registry, San Francisco |
Identified through random-digit dial in the same geographic area as cases |
269 (75%) |
365 (75%) |
104 (39) | 116 (43) | 49 (18) | 146 (40) | 176 (48) | 43 (12) | 0.36 | 0.36 |
| UKOPS25 (United Kingdom Ovarian Cancer Population Study) |
Cases attending ten Major Gynaecological Oncology NHS centers in England, Wales and Northern Ireland from 2006 onwards |
Women aged 50 to 74 from the general population, participating in the United Kingdom Collaborative Trial of Ovarian Cancer Screening. |
258 (86%) |
567 (97%) |
101 (39) | 115 (45) | 42 (16) | 220 (39) | 281 (49) | 66 (12) | 0.36 | 0.10 |
| HAWAII26 (Hawaii Ovarian Cancer Study. Hawaii, USA) |
Incident cases (18–87 years old) diagnosed 1993–2007, ascertained through Hawaii Tumor Registry |
Randomly selected from the participants in the annual survey of representative households that is conducted under statutory provision . |
56 (66%) |
140 (69%) |
13 (23) | 33 (59) | 10 (18) | 57 (41) | 64 (46) | 19 (13) | 0.36 | 0.88 |
| P* | 0.15 | 0.49 | ||||||||||
| All | 1820 | 3479 | 673 (37) | 878 (48) | 269 (15) | 1382 (40) | 1618 (46) | 479 (14) | 0.37 | 0.88 | ||
| Excluding HAW | 1764 | 3339 | 660 (37) | 845 (48) | 259 (15) | 1325 (40) | 1554 (46) | 460 (14) | 0.37 | 0.90 | ||
Abbreviations: MAF, minor allele frequency; EPIC, The European Prospective Investigation of Cancer.
P for the log likelihood ratio test assessing heterogeneity of genotype distribution by study.
P from the chi-square test assessing deviation of genotype frequencies among controls from those expected under Hardy-Weinberg equilibrium.
Genotyping
Genotyping for European studies and one US study was performed in the UK at Cambridge University (SEARCH and GEOCS) and University College London (MALOVA, and UKOPS). Genotyping for the HAW was conducted at the Cancer Research Center of Hawaii, USA. In all laboratories, genotyping was performed using 5’ nuclease TaqMan allelic discrimination assay (TaqMan, Applied Biosystems). We used the following quality control criteria to measure the acceptability of the genotyping results: (1) >3% sample duplicates included, (2) concordance rate for duplicate samples ≥ 98%, (3) overall call rate (by study) >95% and (4) call rate >90% for each 384-well plate and (5) cases and controls intermixed on each plate. All five studies met each of the criteria. Gene and allele nomenclature was according to the National Center of Biotechnology Information.
Statistical analysis
Statistical analyses were performed using the SAS statistical package (SAS release 9.2, SAS Institute Inc., Cary, NC). The chi-square test for association was used to compare the allele frequency distributions among controls across studies, and the chi-square test for goodness-of-fit was used to test consistency with the Hardy-Weinberg equilibrium for each study and overall. The association of the rs2228570 polymorphism with ovarian carcinoma risk was assessed using multivariate logistic regression models. ORs and 95% CIs were estimated separately for heterozygous and homozygous variant T allele carriers, using women with the CC genotype as the reference group. We also performed genetic analyses testing a log-additive model in which genotype was categorized by three levels (0, 1 and 2) representing number of variant alleles. In addition, we compared risk among heterozygotes and homozygote T allele carriers combined (testing a dominant genetic model) and among women with the TT genotype compared to the CC and CT genotypes combined (testing a recessive genetic model). Based on the Akaike Information Criterion (AIC), the log-additive and dominant models provided better fit for the data than the recessive model.
To establish potential confounders, the distribution of the rs2228570 genotype among cases and controls was examined by the following variables associated with ovarian cancer risk: age, gravidity, parity, menopausal status, history of tubal ligation, hysterectomy, and use of contraceptive and menopausal hormones. All models in the pooled analysis and by study were adjusted for age and menopausal status, the only variables that were associated with rs2228570 genotype. Heterogeneity of effects by study was examined using two different methods. First, we included study site as a fixed effect covariate and evaluated heterogeneity of the association of the rs2228570 SNP with risk by study, using a Wald test of the genotype-study interaction term. Second, we included study site as a random effect using SAS GLIMMIX procedure. The results were the same; only the fixed effect results are presented. Ovarian cancer defined by stage (FIGO stages I-II versus III-IV), histological type (serous, mucinous, endometrioid, clear cell, and other), and time between diagnosis and interview (<6 months versus ≥ 6 months) were compared against controls in a polytomous logistic regression model. Heterogeneity of the association of the rs2228570 genotype with risk by age, menopausal status, stage, histological type, and time between diagnosis and interview was evaluated using the Wald test comparing group-specific parameters for the rs2228570 genotype in the logistic regression models. Analyses were conducted for each study separately and for all studies combined. Data were analyzed with and without the inclusion of HAW study to test the independent association of the rs2228570 genotype with risk in the replication studies. All p-values were based on two-tailed tests. We evaluated statistical significance at the 5% level. Although the initial study was small, the addition of four independent studies increased the statistical power to 90% under a log-additive model to detect an OR of 1.15 or higher. We also estimated an overall odds ratio for the TT compared to the CC genotype group combining all published studies and the studies presented in this manuscript using random effect meta-analysis techniques. 26
Results
The mean age of cases (56.7 years; SD=10.5; range: 20–91) and controls (56.6 years; SD=10.7; range 20–86) was similar. The allele distribution among controls did not significantly deviate from Hardy-Weinberg equilibrium in each study or in all studies combined (p’s ≥ 0.10) (Table 1). Minor allele frequencies among controls ranged from 0.36 to 0.38 with no statistically significant differences in genotype distribution by study (p=0.49) (Table 1).
Table 2 presents the rs2228570 association with ovarian cancer risk among cases by study and in all studies combined. In all studies combined, the T allele of the rs2228570 variant was significantly associated with invasive ovarian carcinoma risk (per allele OR=1.09; 95% CI: 1.01–1.19; p=0.04 and dominant OR=1.14; 95% CI: 1.01–1.28; p=0.03). The OR for the association of the T allele with risk remained unchanged when the Hawaii data were excluded, although the confidence interval included unity (per allele OR=1.09; 95% CI: 1.00–1.18; p=0.06). No significant heterogeneity in the association of the rs2228570 genotype with risk across studies was observed in the log additive or dominant models.
Table 2.
Association of the VDR rs2228570 SNP with invasive ovarian carcinoma risk among non-Hispanic white women by study
| Heterozygotes and rare allele homozygotes* | Log-additive model | Dominant model* | ||||||
|---|---|---|---|---|---|---|---|---|
| Study | CT OR (95% CI)† |
TT OR (95% CI)† |
P (2 d.f.) | Per allele OR (95% CI)† |
P for trend (1 d.f.) |
Any T allele OR (95% CI)† |
P (1 d.f.) | |
| HAW | 2.20 (1.05–4.63) | 2.15 (0.80–5.76) | 0.10 | 1.55 (0.98–2.47) | 0.06 | 2.19 (1.07–4.47) | 0.03 | |
| MALOVA | 1.14 (0.90–1.46) | 1.04 (0.73–1.47) | 0.54 | 1.05 (0.89–1.23) | 0.57 | 1.12 (0.89–1.41) | 0.34 | |
| SEARCH | 1.24 (1.02–1.52) | 1.02 (0.76–1.35) | 0.08 | 1.06 (0.93–1.21) | 0.41 | 1.18 (0.98–1.21) | 0.08 | |
| GEOCS | 1.01 (0.71–1.43) | 1.61 (0.99–2.62) | 0.12 | 1.21 (0.96–1.52) | 0.11 | 1.13 (0.81–1.58) | 0.46 | |
| UKOPS | 0.82 (0.59–1.14) | 1.26 (0.79–2.01) | 0.15 | 1.04 (0.83–1.30) | 0.75 | 0.90 (0.66–1.23) | 0.51 | |
| All | 1.13 (1.00–1.29) | 1.16 (0.97–1.39) | 0.10 | 1.09 (1.01–1.19) | 0.04 | 1.14 (1.01–1.28) | 0.03 | |
| All excluding HAW | 1.11 (0.97–1.26) | 1.14 (0.95–1.37) | 0.21 | 1.09 (1.00–1.19) | 0.06 | 1.11 (0.99–1.26) | 0.08 | |
| P‡ for heterogeneity | 0.14 | 0.19 | 0.39 | 0.30 | ||||
| HAW (initial published study including 15 borderline malignancy and 56 invasive cases) 19 |
2.45 (1.25–4.81) | 2.06 (0.82–5.21) | 0.03 | 1.56 (1.01–3.41) | 0.04 | 2.36 (1.23–4.53) | 0.01 | |
CC genotype was used as the reference category.
Odds ratios (OR) and 95% confidence intervals (CI) from the unconditional logistic regression models adjusted for age, menopausal status, and, in combined analyses, by study.
P for heterogeneity of the association of the rs2228570 SNP with risk by study was estimated using a Wald test of the genotype-study interaction term.
Note: statistically significant associations are presented in bold font (P<0.05).
In Table 2 (last row), we included previously published results for the non-Hispanic white women from the HAW study that included 15 women with borderline epithelial ovarian tumors. Exclusion of cases with ovarian tumors classified as borderline malignant from the HAW study weakened the association of the T allele with risk. However the association remained significant in the dominant genetic model when only invasive tumors were included (OR=2.19; 95%CI: 1.07–4.47; p=0.03). In addition to the HAW study, statistically significant increased ovarian cancer risk associated with the T allele was observed among the SEARCH study heterozygotes (OR=1.24; 95% CI:1.02–1.52; p for pair-wise comparison with the CC genotype = 0.03).
There was a significant interaction between genotype and age (p=0.04 for the log-additive and p=0.03 for the dominant model) (Table 3). The association of the T allele with risk among younger women (< 50 years old) was statistically significant with the highest risk among T allele homozygotes (OR=1.47; 95% CI: 1.02–1.12; p=0.04); no statistically significant associations were observed among older women (≥ 50 years old). Figure 1 presents a forest plot for the association of the rs2228570 genotype with risk among women < 50 years old by study. Although the association of the rs2228570 genotype with risk among premenopausal women was stronger than among postmenopausal women, the interaction of genotype and menopausal status was not statistically significant (p=0.11) (Table 3). No statistically significant heterogeneity of effects was observed by histology (p= 0.98), stage of disease (p=0.46), or time between diagnosis and interview (6 months versus more than 6 months) (p=0.94) (data not shown).
Table 3.
Association of the VDR rs2228570 SNP with ovarian carcinoma risk among non-Hispanic white women by age and menopausal status subgroups
| Genotype | Subgroups by age |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Age < 50 years | Age ≥ 50 years | P for interaction† |
|||||||
| No. (%) cases |
No. (%) controls |
OR (95% CI)* | P | No. (%) cases |
No. (%) controls |
OR (95% CI)* | P | ||
|
All studies combined |
|||||||||
| CC | 137 (32) | 408 (41) | 1.00 | 1.00 | |||||
| CT | 225 (52) | 468 (46) | 1.34 (1.03–1.76) | 536 (39) | 974 (39) | 1.04 (0.90–1.21) | |||
| TT | 69 (16) | 133 (13) | 1.47 (1.02–2.12) | 0.04 | 653 (47) | 1150 (47) | 1.05 (0.85–1.29) | 0.82 | 0.08 |
| Per T allele copy | 1.24 (1.04–1.47) | 0.02 | 200 (14) | 346 (14) | 1.03 (0.93–1.13) | 0.56 | 0.04 | ||
| CT+TT | 1.37 (1.08–1.76) | 0.01 | 1.05 (0.92–1.21) | 0.53 | 0.03 | ||||
|
Excluding HAW study |
|||||||||
| CC | 134 (33) | 394 (41) | 1.00 | 526 (39) | 931 (39) | 1.00 | |||
| CT | 213 (52) | 451 (46) | 1.30 (1.00–1.70) | 632 (47) | 1103 (47) | 1.03 (0.89–1.19) | |||
| TT | 64 (15) | 130 (13) | 1.37 (0.94–2.00) | 0.10 | 195 (14) | 330 (14) | 1.04 (0.85–1.29) | 0.91 | 0.14 |
| Per T allele copy | 1.20 (1.01–1.43) | 0.04 | 1.02 (0.93–1.13) | 0.66 | 0.08 | ||||
| CT+TT | 1.32 (1.02–1.70) | 0.03 | 1.03 (0.90–1.18) | 0.69 | 0.04 | ||||
| Subgroups by menopausal status |
|||||||||
| Premenopausal | Postmenopausal | P for interaction† |
|||||||
|
All studies combined |
|||||||||
| CC | 173 (33) | 483 (39) | 1.00 | 500 (39) | 899 (40) | 1.00 | |||
| CT | 278 (52) | 578 (47) | 1.31 (1.04–1.65) | 600 (46) | 1040 (47) | 1.05 (0.90–1.22) | |||
| TT | 81 (15) | 179 (14) | 1.17 (0.84–1.62) | 0.08 | 188 (15) | 300 (13) | 1.13 (0.91–1.40) | 0.53 | 0.21 |
| Per T allele copy | 1.13 (0.97–1.31) | 0.13 | 1.06 (0.96–1.17) | 0.27 | 0.33 | ||||
| CT+TT | 1.27 (1.02–1.59) | 0.03 | 1.07 (0.93–1.23) | 0.37 | 0.11 | ||||
|
Excluding HAW study |
|||||||||
| CC | 169 (33) | 468 (39) | 1.00 | 491 (39) | 857 (40) | 1.00 | |||
| CT | 266 (52) | 554 (46) | 1.29 (1.02–1.63) | 579 (46) | 1000 (47) | 1.02 (0.88–1.19) | |||
| TT | 78 (15) | 176 (15) | 1.13 (0.81–1.58) | 0.10 | 181 (15) | 284 (13) | 1.11 (0.89–1.39) | 0.65 | 0.19 |
| Per T allele copy | 1.11 (0.95–1.30) | 0.19 | 1.05 (0.94–1.16) | 0.40 | 0.35 | ||||
| CT+TT | 1.25 (1.01–1.57) | 0.04 | 1.04 (0.90–1.21) | 0.58 | 0.11 | ||||
Adjusted for age (continuous) and study.
P for interaction of the rs2228570 genotype and age/menopausal status was estimated using a Wald test of the genotype-age group and genotype/menopausal status interaction terms.
Note: statistically significant associations are presented in bold font (P < 0.05).
Figure 1.
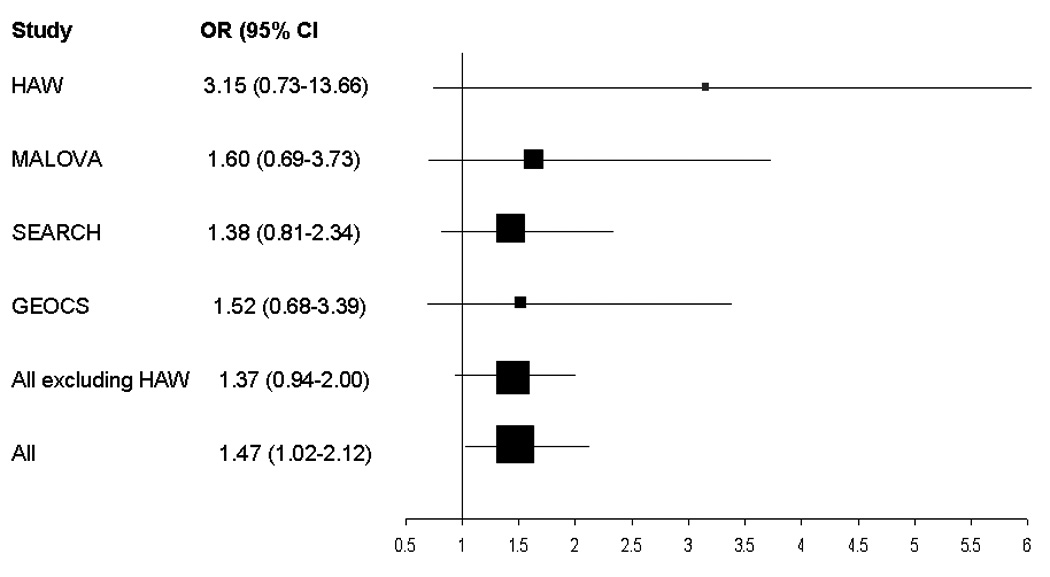
Forest plot of the ORs and 95% Cls comparing invasive ovarian carcinoma risk among women < 50 years of age associated with the VDR rs2228570 rare allele homozygotes (TT genotype) versus common allele homozygotes (CC genotype) for 4 studies included in the current pooled analysis (UKOPS study participants were excluded because all were >50 years of age). The summary OR=1.47; 95% Cl: 1.02–2.12; p=0.04.
Discussion
In this pooled analysis of five population-based case-control studies, we found a 9% increased risk of invasive ovarian carcinoma associated with each copy of the VDR rs2228570 T allele. Ovarian cancer risk associated with the T allele was higher among younger women. No heterogeneity of the genetic association was observed across tumor histological subtypes, disease stage, or subgroups by time between diagnosis and interview.
In our initial study, 19 we found a 56% increased ovarian carcinoma risk per T allele (95% CI: 1.01–3.41; p for trend=0.04). In addition, a significant association of this SNP with ovarian cancer risk was recently reported by Tworoger et al. 27 (OR for T allele homozygotes = 1.26; 95% CI: 1.01–1.57; p for trend=0.03) in a pooled analysis of 1473 cases and 2006 controls. However, the original HAW 19 study and the analysis by Tworoger et al. 27 included women with borderline malignancy tumors. Excluding women with borderline ovarian neoplasms from the analyses resulted in a widening of the confidence intervals in the HAW study (OR=1.55; 95% CI: 0.98–2.47) and in the study by Tworoger et al. 27 (OR=1.21; 95% CI: 0.96–1.53). A smaller study of 168 invasive ovarian carcinoma and 321 controls by Clendenen et al. 28 did not find a significant association. In the current pooled analysis, the risk estimate for rare allele homozygotes (OR=1.16; 95% CI: 0.97–1.39) was similar to that reported by Tworoger et al. 27 In a meta-analysis of the current studies (HAW, MAL, SEARCH, GEOCS, and UKOPS) and studies published by Tworoger et al. 27 (NHS I and II, WHS, and NECC) and Clendenen et al. 28, the summary OR comparing ovarian carcinoma risk for the VDR rs2228570 rare allele homozygotes (TT genotype) versus common allele homozygotes (CC genotype) was 1.20 (95% CI: 1.05–1.38; p=0.009).
Although the association of the rs2228570 T allele with ovarian cancer risk observed in our pooled analysis is modest, it may reflect a true underlying biological phenonemon related to disease development. The vitamin D signaling pathway is involved in a wide variety of biological processes, including cell proliferation, differentiation and apoptosis (reviewed by Deeb et al. 29), and has the potential to influence ovarian cancer development. Rs2228570 is the only VDR polymorphism that has distinct structural consequencies for the VDR protein.30 The C to T nucleotide change results in a 424-amino acid VDR protein compared with a longer 427-amino acid protein in the presence of the C allele. Functional studies showed that the shorter protein has a 1.7-fold higher transactivation of the vitamin D response element of the 24-hydrolase gene than the longer protein.15;31 In addition, Jurutka et al.17 demonstrated that the 424 amino acid VDR variant interacted more efficiently with the transcription factor TFIIB. It is important to note that rs228570 is an independent genetic marker as it is not in linkage disequilibrium with any of the other polymorphisms in the VDR gene.
A stronger association of rs2228570 with risk among younger and premenopausal women might be a result of effect modification of vitamin D expression by age 32,33 and estrogen levels.34An age-related reduction in the vitamin D receptor has been reported. 32 VDR is a known estrogen-responsive gene. 34 Estrogen has been reported to increase VDR gene expression in intestinal mucosa, uterus, and liver in studies in animal models and in T 47D human breast cancer cells. 35 Estrogen administration in women resulted in increased mRNA expression of VDR. 35 It is plausible that the role of the rs2228570 polymorphism in ovarian carcinoma risk might be more pronounced among younger, mostly premenopausal, women who have higher vitamin D levels and greater amounts of VDR.
The strengths of this investigation are the population-based nature of the studies included, histological confirmation of all cases, and stringent genotyping quality control procedures established by the OCAC. 24 Population stratification might have influenced the results of our investigation, and a false-positive association is possible. To minimize the population stratification effects, this study included only white non-Hispanic women from developed countries with comparable ovarian cancer incidence rates, and women were only compared within geographical areas. Another strength is that the sample size was large and the allele frequency was relatively high. It is also important that the rs2228570 SNP is not in LD with any other polymorphism. Although one study (SEARCH) included prevalent cases, there was no heterogeneity of effects among incident and prevalent cases (women diagnosed <6 months prior to participation versus ≥ 6 months after diagnosis) indicating survival bias was minimal. However, the power was limited to examine gene-environment interactions. Larger studies are required to explore the potential effect modification of the rs2228570 SNP-ovarian cancer risk association by BMI, exogenous hormone use, gravidity/parity, anti-inflammatory drug use, and other factors. It would also be of interest to identify differences in genetic associations between invasive and borderline malignancy neoplasms in future analysis. It may be possible in the future to explore the hypothesis that the association of the rs2228570 genotype with risk might be modified by vitamin D status, 30 but it is likely that this will need to be analysed in studies with prospectively collected serum samples.
In conclusion, VDR rs2228570 is a functional polymorphism that may play a role in ovarian carcinogenesis through regulation of cell proliferation, apoptosis, and angiogenesis. Our pooled analysis is the first report of effect modification of the rs2228570 association with risk by age, with stronger associations among younger women.
Figure 2.
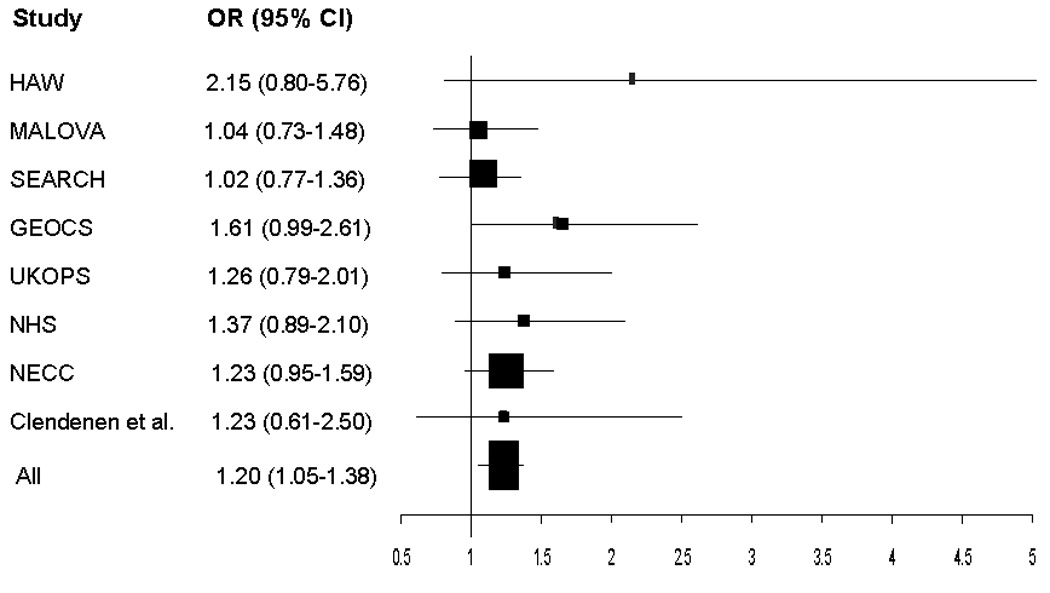
Forest plot of the ORs and 95% Cls comparing ovarian carcinoma risk for the VDR rs2228570 rare allele homozygotes (TT genotype) versus common allele homozygotes (CC genotype) for 5 studies included in the current pooled analysis (HAW, MALOVA, SEARCH, GEOCS, UKOPS) and published reports by Tworoger et al.27 (NHS I and II, WHS, and NECC studies) and Clendenen et al.28 The summary OR=1.20; 95% Cl:1.05–1.38; p=0.009.
Acknowledgements
The Ovarian Cancer Association Consortium is funded by Ovarian Cancer Research Fund provided by the family and friends of Kathryn Sladek Smith. The Hawaii study was supported in part by Public Health Service grant R01-CA-58598 and by contracts N01-CN-67001 and NO1-PC-035137 from the Department of Health and Human Services, National Institutes of Health, USA. SEARCH study is funded by Cancer Research UK. The UKOPS study is funded by the OAK Foundation. Dr. Ramus is funded by the ‘Eve Appeal’. A portion of this work was done at UCLH/UCL within the “Women’s Health Theme” of the NIHR UCLH/UCL Comprehensive Biomedical Research Centre supported by the Department of Health, UK. We thank all study participants and all members of the research teams of all the participating studies, including research nurses, research scientists, data entry personnel and consultant gynecological oncologists.
Abbreviations used
- SNP
single nucleotide polymorphism
- OR
odds ratio
- CI
95%confidence interval
- VDR
vitamin D receptor gene
- OCAC
Ovarian Cancer Association Consortium
- AIC
Akaike Information Criterion
- MALOVA
Malignant Ovarian Cancer Study, Denmark
- SEARCH
Studies of Epidemiology and Risk Factors in Cancer Heredity: Ovarian Cancer Study, United Kingdom
- GEOCS
Genetic Epidemiology of Ovarian Cancer Study, California, United States
- HAW
Hawaii Ovarian Cancer Study, Hawaii, United States
- UKOPS
United Kingdom Ovarian Cancer Population Study
Footnotes
Novelty
In this pooled analysis of five population-based case-control studies from different geographical areas (United Kingdom, Denmark, Northern California and Hawaii), we observed a significant association of the VDR rs2228570 polymorphism with invasive ovarian carcinoma, particularly among younger women (age < 50 years old). The vitamin D signaling pathway is involved in a wide variety of biological processes, including cell proliferation, differentiation and apoptosis that may influence ovarian cancer risk. This marker was selected based on its functional significance; the rs2228570 polymorphism is the only VDR polymorphism that has distinct structural consequencies for the VDR protein. Although the association of the rs2228570 T allele with ovarian cancer risk found in this pooled analysis is modest, it may reflect a true biological relation.
Impact
Ovarian cancer is a fatal gynecologic malignancy largely because of the absence of screening methods for its early detection. The discovery of common genetic variants, such as the VDR FokI polymorphism, to assist in the identification of women at increased risk of ovarian cancer holds great promise. Considering the possible value of synthetic vitamin D analog use in cancer treatment, our findings also could be useful for the future investigations of individualized therapeutic strategies.
References
- 1.Holick MF. Vitamin D: its role in cancer prevention and treatment. Prog Biophys Mol Biol. 2006;92:49–59. doi: 10.1016/j.pbiomolbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz ES, Garland CF. Sunlight, vitamin D, and ovarian cancer mortality rates in US women. Int J Epidemiol. 1994;23:1133–1136. doi: 10.1093/ije/23.6.1133. [DOI] [PubMed] [Google Scholar]
- 3.Garland CF, Mohr SB, Gorham ED, Grant WB, Garland FC. Role of ultraviolet B irradiance and vitamin D in prevention of ovarian cancer. Am J Prev Med. 2006;31:512–514. doi: 10.1016/j.amepre.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Lipkin M, Newmark HL. Vitamin D, calcium and prevention of breast cancer: a review. J Am Coll Nutr. 1999;18 Suppl:392S–397S. doi: 10.1080/07315724.1999.10718903. [DOI] [PubMed] [Google Scholar]
- 5.Cui Y, Rohan TE. Vitamin D, calcium, and breast cancer risk: a review. Cancer Epidemiol Biomarkers Prev. 2006;15:1427–1437. doi: 10.1158/1055-9965.EPI-06-0075. [DOI] [PubMed] [Google Scholar]
- 6.Flood A, Peters U, Chatterjee N, Lacey JV, Jr, Schairer C, Schatzkin A. Calcium from diet and supplements is associated with reduced risk of colorectal cancer in a prospective cohort of women. Cancer Epidemiol Biomarkers Prev. 2005;14:126–132. [PubMed] [Google Scholar]
- 7.Park SY, Murphy SP, Wilkens LR, Nomura AM, Henderson BE, Kolonel LN. Calcium and vitamin D intake and risk of colorectal cancer: the Multiethnic Cohort Study. Am J Epidemiol. 2007;165:784–793. doi: 10.1093/aje/kwk069. [DOI] [PubMed] [Google Scholar]
- 8.Tung KH, Goodman MT, Wu AH, McDuffie K, Wilkens LR, Nomura AM, Kolonel LN. Aggregation of ovarian cancer with breast, ovarian, colorectal, and prostate cancer in first-degree relatives. Am J Epidemiol. 2004;159:750–758. doi: 10.1093/aje/kwh103. [DOI] [PubMed] [Google Scholar]
- 9.Salazar-Martinez E, Lazcano-Ponce EC, Gonzalez Lira-Lira G, Escudero-De Los Rios P, Hernandez-Avila M. Nutritional determinants of epithelial ovarian cancer risk: a case-control study in Mexico. Oncology. 2002;63:151–157. doi: 10.1159/000063814. [DOI] [PubMed] [Google Scholar]
- 10.Saunders DE, Christensen C, Wappler NL, Schultz JF, Lawrence WD, Malviya VK, Malone JM, Deppe G. Inhibition of c-myc in breast and ovarian carcinoma cells by 1,25-dihydroxyvitamin D3, retinoic acid and dexamethasone. Anticancer Drugs. 1993;4:201–208. doi: 10.1097/00001813-199304000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Jiang F, Bao J, Li P, Nicosia SV, Bai W. Induction of ovarian cancer cell apoptosis by 1,25-dihydroxyvitamin D3 through the down-regulation of telomerase. J Biol Chem. 2004;279:53213–53221. doi: 10.1074/jbc.M410395200. [DOI] [PubMed] [Google Scholar]
- 12.Saunders DE, Christensen C, Lawrence WD, Malviya VK, Malone JM, Williams JR, Deppe G. Receptors for 1,25-dihydroxyvitamin D3 in gynecologic neoplasms. Gynecol Oncol. 1992;44:131–136. doi: 10.1016/0090-8258(92)90028-h. [DOI] [PubMed] [Google Scholar]
- 13.Ahonen MH, Zhuang YH, Aine R, Ylikomi T, Tuohimaa P. Androgen receptor and vitamin D receptor in human ovarian cancer: growth stimulation and inhibition by ligands. Int J Cancer. 2000;86:40–46. doi: 10.1002/(sici)1097-0215(20000401)86:1<40::aid-ijc6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 14.Villena-Heinsen C, Meyberg R, Axt-Fliedner R, Reitnauer K, Reichrath J, Friedrich M. Immunohistochemical analysis of 1,25-dihydroxyvitamin-D3-receptors, estrogen and progesterone receptors and Ki-67 in ovarian carcinoma. Anticancer Res. 2002;22:2261–2267. [PubMed] [Google Scholar]
- 15.Uitterlinden AG, Fang Y, van Meurs JB, Pols HA, van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Gross C, Krishnan AV, Malloy PJ, Eccleshall TR, Zhao XY, Feldman D. The vitamin D receptor gene start codon polymorphism: a functional analysis of FokI variants. J Bone Miner Res. 1998;13:1691–1699. doi: 10.1359/jbmr.1998.13.11.1691. [DOI] [PubMed] [Google Scholar]
- 17.Jurutka PW, Remus LS, Whitfield GK, Thompson PD, Hsieh JC, Zitzer H, Tavakkoli P, Galligan MA, Dang HT, Haussler CA, Haussler MR. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol. 2000;14:401–420. doi: 10.1210/mend.14.3.0435. [DOI] [PubMed] [Google Scholar]
- 18.Whitfield GK, Remus LS, Jurutka PW, Zitzer H, Oza AK, Dang HT, Haussler CA, Galligan MA, Thatcher ML, Encinas DC, Haussler MR. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol. 2001;177:145–159. doi: 10.1016/s0303-7207(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 19.Lurie G, Wilkens LR, Thompson PJ, McDuffie KE, Carney ME, Terada KY, Goodman MT. Vitamin D receptor gene polymorphisms and epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:2566–2571. doi: 10.1158/1055-9965.EPI-07-0753. [DOI] [PubMed] [Google Scholar]
- 20.Berchuck A, Schildkraut JM, Pearce CL, Chenevix-Trench G, Pharoah PD. Role of genetic polymorphisms in ovarian cancer susceptibility: development of an international ovarian cancer association consortium. Adv Exp Med Biol. 2008;622:53–67. doi: 10.1007/978-0-387-68969-2_5. [DOI] [PubMed] [Google Scholar]
- 21.Soegaard M, Jensen A, Hogdall E, Christensen L, Hogdall C, Blaakaer J, Kjaer SK. Different risk factor profiles for mucinous and nonmucinous ovarian cancer: results from the Danish MALOVA study. Cancer Epidemiol Biomarkers Prev. 2007;16:1160–1166. doi: 10.1158/1055-9965.EPI-07-0089. [DOI] [PubMed] [Google Scholar]
- 22.Song H, Ramus SJ, Quaye L, Dicioccio RA, Tyrer J, Lomas E, Shadforth D, Hogdall E, Hogdall C, McGuire V, Whittemore AS, Easton DF, et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006;27:2235–2242. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 23.McGuire V, Felberg A, Mills M, Ostrow KL, DiCioccio R, John EM, West DW, Whittemore AS. Relation of contraceptive and reproductive history to ovarian cancer risk in carriers and noncarriers of BRCA1 gene mutations. Am J Epidemiol. 2004;160:613–618. doi: 10.1093/aje/kwh284. [DOI] [PubMed] [Google Scholar]
- 24.Ramus SJ, Vierkant RA, Johnatty SE, Pike MC, Van Den Berg DJ, Wu AH, Pearce CL, Menon U, Gentry-Maharaj A, Gayther SA, Dicioccio RA, McGuire V, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Int J Cancer. 2008;123:380–388. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushley AW, Ferrell R, McDuffie K, Terada KY, Carney ME, Thompson PJ, Wilkens LR, Tung KH, Ness RB, Goodman MT. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol. 2004;95:672–679. doi: 10.1016/j.ygyno.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Sutton AJ, Abrams KR, Jones D, Sheldon DR, Song F. Methods for Meta-analysis in Medical Research. New York: Wiley; 2002. [Google Scholar]
- 27.Tworoger SS, Gate MA, Lee IM, Buring JE, Titus-Ernstoff L, Cramer D, Hankinson SE. Polymorphisms in the vitamin D receptor and risk of ovarian cancer in four studies. Cancer Res. 2009;69:1885–1891. doi: 10.1158/0008-5472.CAN-08-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clendenen TV, Arslan AA, Koenig KL, Enquist K, Wirgin I, Agren A, Lukanova A, Sjodin H, Zeleniuch-Jacquotte A, Shore RE, Hallmans G, Toniolo P, et al. Vitamin D receptor polymorphisms and risk of epithelial ovarian cancer. Cancer Lett. 2008;260:209–215. doi: 10.1016/j.canlet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 30.Colin EM, Weel AE, Uitterlinden AG, Buurman CJ, Birkenhager JC, Pols HA, van Leeuwen JP. Consequences of vitamin D receptor gene polymorphisms for growth inhibition of cultured human peripheral blood mononuclear cells by 1, 25-dihydroxyvitamin D3. Clin Endocrinol (Oxf) 2000;52:211–216. doi: 10.1046/j.1365-2265.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- 31.Arai H, Miyamoto K, Taketani Y, Yamamoto H, Iemori Y, Morita K, Tonai T, Nishisho T, Mori S, Takeda E. A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res. 1997;12:915–921. doi: 10.1359/jbmr.1997.12.6.915. [DOI] [PubMed] [Google Scholar]
- 32.Lanske B, Razzaque MS. Vitamin D and aging: old concepts and new insights. J Nutr Biochem. 2007;18:771–777. doi: 10.1016/j.jnutbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilad LA, Schwartz B. Association of estrogen receptor beta with plasma-membrane caveola components: implication in control of vitamin D receptor. J Mol Endocrinol. 2007 Jun;38(6):603–618. doi: 10.1677/JME-06-0040. [DOI] [PubMed] [Google Scholar]
- 34.Tang S, Han H, Bajic V. ERGDB: Estrogen Responsive Genes Database. Nucleic Acids Research. 2004;32:D533–D536. doi: 10.1093/nar/gkh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Protiva P, cross HS, Hopkins ME, Kallay E, Bises G, dreihaupt E, Augenlicht L, Lipkin M, Lesser M, Livote E, Holt PK. Chemoprevention of colorectal neoplasia by estrogen: potential role of vitamin D activity. Cancer Prevention Res. 2009 Jan;2(1):43–51. doi: 10.1158/1940-6207.CAPR-08-0103. [DOI] [PubMed] [Google Scholar]


