Abstract
Recent genome-wide association studies have identified a novel polymorphism rs1042725 in HMGA2 gene for human adult height, a highly heritable complex trait. Replications in independent populations are needed to evaluate a positive finding and determine its generality. Thus, we performed a replication study to examine the associations between polymorphisms in HMGA2 and adult height in two US Caucasian populations (an unrelated sample of 998 subjects and a family-based sample of 8,385 subjects) and a Chinese population (1,638 unrelated Han subjects). We confirmed the association between rs1042725 in HMGA2 and adult height both in the unrelated and family-based Caucasian populations (overall P = 4.25×10−9). Another two SNPs (rs7968902 and rs7968682), which were in high linkage disequilibrium with rs1042725, also achieved the significance level in both Caucasian populations (overall P = 6.34×10−7, and 2.72×10−9, respectively). Our results provide a strong support to the initial finding. Moreover, SNP rs1042725 was firstly found to be associated with adult height (P = 0.008) in the Chinese population, and the effect is in the same direction as in the Caucasian populations, suggesting that it is a common variant across different populations. Our study further highlights the importance of the HMGA2 gene involved in normal growth.
Keywords: replication, adult height, HMGA2, association
Introduction
Human adult height is an important physical index to reflect the processes of growth and development, and is associated with several common complex diseases, including cancers,(Gunnell et al., 2001) cardiovascular disease,(Davey Smith et al., 2000) and osteoporosis.(Hemenway et al., 1995) As a classic, polygenic quantitative trait, adult height is under strong genetic influence with heritability estimated up to 90%.(Silventoinen et al., 2000, Silventoinen et al., 2003, Macgregor et al., 2006, Perola et al., 2007) Identifying the genetic determination for adult height will provide insights into the mechanism of growth and development, and the genetic architecture of other human disorders in general.
Recent advances in SNP genotyping technologies and analytic methods have provided new opportunities for genome-wide association studies (GWAS) to explore common variants for adult height. The first GWAS of height from 4,921 European individuals identified a novel variant rs1042725 in high mobility group-A2 (HMGA2) oncogene that are associated with variation in height (P = 4×10−8).(Weedon et al., 2007) Subsequently, another two GWAS provided confirmatory evidence for the association of this variant with height.(Weedon et al., 2008, Lettre et al., 2008) Replication of genetic associations in independent populations is essential to reduce the false-positive results and to further explore the role of these variants in the complex traits. However, these studies are performed by the same groups, and the samples are mainly from northern Europe. Replications in multiple additional populations by independent groups are needed to evaluate the credibility and generality of this finding.
Therefore, the aim of this study was to attempt to replicate the associations between polymorphisms in the HMGA2 gene and adult height both in unrelated and family-based US Caucasian populations. Since genetic and environmental backgrounds vary from different ethnic groups, we further examined the associations across ethnicities in a Chinese population to see whether the variants identified are common or ethnic specific.
Materials and Methods
Subjects
The study was approved by the required Institutional Review Board or Research Administration of the involved institutions. Signed informed-consent documents were obtained from all study participants before entering the study.
Unrelated Caucasian sample
A random sample of 998 unrelated healthy subjects (500 women and 498 men) was identified from our established and expanding database containing more than 10,000 subjects. All of the identified subjects were US Caucasians of Northern European origin, living in Omaha, Nebraska, and its surrounding regions in Midwestern USA. The height for each subject was measured without shoes using a standard wall mounted stadiometer in the clinic by nurses.
Family-based Caucasian sample
The family-based sample came from Framingham Heart Study (FHS), which is a longitudinal study of 14,277 phenotyped subjects to identify the risk factors for cardiovascular disease. Details and descriptions about the FHS have been reported before.(Dawber et al., 1963, Dawber et al., 1951) Subjects eligible for this investigation are drawn from the FHS SNP Health Association Resource (SHARe) project,(Cupples et al., 2007) for which genotyping is conducted in over 9,300 subjects from the three generations of subjects (including over 900 families). We have the adult height measurements on 8,385 phenotyped Caucasian subjects, 1,307 from the Original cohort (521 men and 786 women), 3,189 from the second generation cohort (1,491 men and 1,698 women), and 3,889 from the third generation cohort (1,821 men and 2,068 women). The original cohort participants had height measures at exam 1, the second generation cohort participants were measured at exam 5/6, and the third generation cohort participants were measured at exam 1.
Chinese Sample
The Chinese sample consisted of 1,638 unrelated subjects including 810 males and 828 females. The subjects were recruited from northern Chinese Han adults living in Xi’an City, Shanxi province. Height measurements were made as for the US Mid-West Caucasian sample.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. For the unrelated Caucasian and Chinese sample, SNP genotyping was performed using the Affymetrix Human Mapping 500K array set (Affymetrix, Santa Clara, CA, USA), which has been finished in our previous experiments.(Yang et al., 2008, Liu et al., 2008) The experiment procedure was followed by the Affymetrix protocol strictly. For the family-based FHS sample, genotyping was performed using approximately 550,000 SNPs (Affymetrix 500K mapping array plus Affymetrix 50K supplemental array). Nineteen SNPs including rs1042725 within the HMGA2 gene were successfully genotyped in all samples. SNPs that deviated from Hardy-Weinberg equilibrium (HWE, P < 0.01) and had a minor allele frequency (MAF) < 0.01 were discarded in the each sample set. Thus, 16 SNPs for each of the sample sets were included separately for subsequent association analyses.
Statistical analyses
For the unrelated samples, a linear regression implemented in PLINK(Purcell et al., 2007) was fitted to test for association assuming an additive inheritance model. We used the genotype as an additive covariate and height as a response, including age and gender as covariates in the regression model simultaneously. For the family-based sample, significant covariates like age and gender were used to adjust for the raw height data. Height residuals was normally distributed and used to perform the association tests using the QFAM method implemented in PLINK. We performed 10,000,000 permutation procedures to generate the empirical P values. Multiple testing was adjusted by adopting the conservative Bonferroni correction. The significance threshold was set at a P value of less than 3.13×10−3 (0.05/16 SNPs that used in association analyses).
Meta-analysis statistics were generated using the weighted Z-scores (a standard normal deviate, the statistic associated with a P value) to quantify the overall evidence for association with adult height variation. The individual Z-score was weighted by the square root of the sample size of each study. We added the individual weighted Z-score derived from each sample together and divided by the square root of the sum of the sample sizes to obtain an overall Z-score and an associated combined P value.(Rosenthal, 1991)
Haploview v4.0(Barrett et al., 2005) was utilized to characterize linkage disequilibrium (LD, r2) pattern and plot the haplotype block map.
Results
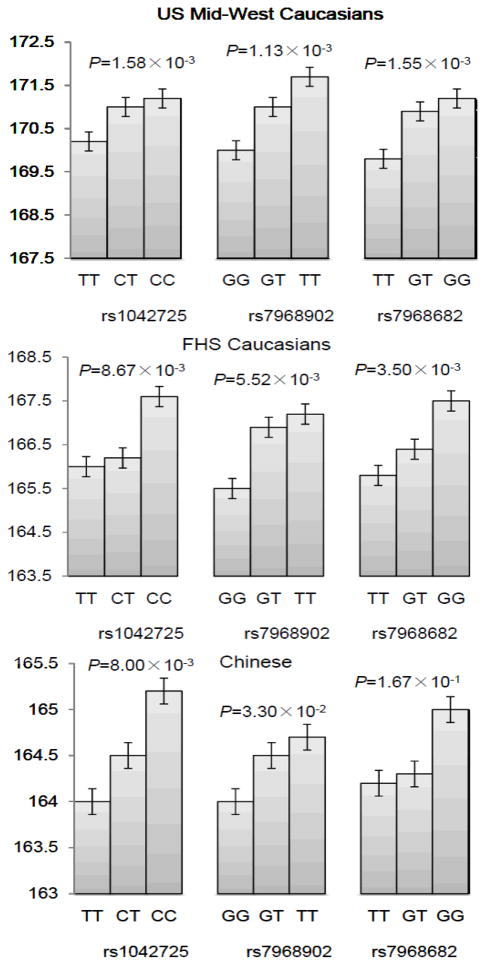
The basic characteristics of the study subjects are presented in Table 1. We summarized the association results of HMGA2 with adult height variation in Table 2. Significant association of rs1042725 with height was successfully replicated both in the unrelated (P = 1.58×10−3) and family-based Caucasian samples (P = 3.0×10−7), even after adjusting for multiple testing (significance threshold: P < 2.94×10−3). The allele effect was in the same direction as in the previous studies.(Weedon et al., 2007, Lettre et al., 2008, Weedon et al., 2008) Each copy of the C allele at rs1042725 was associated with an increase in height of ~0.9 and 0.7 cm, respectively in the unrelated and family-based Caucasian samples. Besides rs1042725, another two common variants, rs7968902 and rs7968682, achieved the significance level both in the unrelated and family-based Caucasian samples (rs7968902: P = 1.13×10−3 and 3.4×10−5, respectively; rs7968682: P = 1.55×10−3 and 2.0×10−7, respectively). These two variants were in strong LD with rs1042725 (r2 of 0.71 and 0.89 estimated in the US Mid-West Caucasian sample, Figure 1). The association of rs7968682 with height was also found in the original report.(Weedon et al., 2007) The phenotype differences among subjects with different genotypes for these three SNPs are depicted in Figure 2. When we combined all Caucasian subjects together, the statistical evidence in favor of association greatly increased for these three SNPs (rs1042725: P = 4.25×10−9; rs7968902: P = 6.34×10−7; and rs7968682: P = 2.72×10−9). The proportion of variance in height explained by these three SNPs was about 1%, 1.07%, and 1.06%, respectively, estimated in the unrelated Caucasians after adjustment for age and gender.
Table 1.
Characteristics of the study subjects
| Unrelated Caucasians | Family-based Caucasians | Chinese | |
|---|---|---|---|
| Number | 998 | 8,385 | 1,638 |
| Gender (M/F) | 498/500 | 3,833/4,552 | 810/828 |
| Age (years) | 50.3 (18.3) | 45.5 (11.4) | 34.5 (13.2) |
| Weight (kg) | 80.2 (17.8) | 76.5 (17.5) | 60.2 (10.5) |
| Height (cm) | 170.8 (9.8) | 168.7 (9.5) | 164.3 (8.2) |
Note: Data are shown as mean (standard deviation, SD.).
Table 2.
Summary association results for 19 SNPs in HMGA2
| SNP | Physical Position | Genic Position | Alleles | Unrelated Caucasians | Family-based Caucasians | Chinese | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | P value | Effect size (cm) | MAF | P value | Effect size (cm) | MAF | P value | Effect size (cm) | ||||
| rs7977687 | 64502340 | 5′UTR | G/A | 0.041 | 0.171 | 0.975 | 0.045 | 0.133 | 0.485 | 0.196 | 0.120 | 0.647 |
| rs2272046 | 64510728 | intron2 | G/T | 0.028 | 0.425 | 0.699 | 0.025 | 0.041 | 0.840 | 0.088 | 0.571 | 0.192 |
| rs12310312 | 64521025 | intron3 | A/G | 0.045 | 0.084 | 1.172 | 0.062 | 0.030 | 0.619 | 0.212 | 0.029 | 0.524 |
| rs2583937 | 64521731 | intron3 | C/T | 0.069 | 0.931 | 0.051 | 0.097 | 0.804 | 0.056 | 0.114 | 0.618 | 0.154 |
| rs2272047 | 64523002 | intron3 | C/T | 0.049 | 0.084 | 1.147 | 0.058 | 0.027 | 0.641 | 0.211 | 0.030 | 0.241 |
| rs2612060 | 64529769 | intron3 | G/A | 0.068 | 0.823 | 0.132 | 0.099 | 0.923 | 0.022 | 0.115 | 0.788 | 0.082 |
| rs17101839 | 64531835 | intron3 | T/C | 0.024 | 0.115 | 1.500 | 0.049 | 0.023 | 0.754 | - | - | - |
| rs11175944 | 64542662 | intron3 | C/G | 0.064 | 0.045 | 1.149 | 0.089 | 3.65×10−3 | 0.693 | 0.211 | 0.018 | 0.566 |
| rs7959396 | 64543214 | intron3 | C/A | 0.040 | 0.245 | 0.838 | 0.044 | 0.057 | 0.627 | 0.211 | 0.018 | 0.569 |
| rs7973574 | 64558999 | intron3 | G/A | 0.028 | 0.520 | 0.555 | 0.046 | 2.49×10−3 | 1.141 | 0.036 | 0.412 | 0.434 |
| rs10878346 | 64607140 | intron3 | A/G | 0.226 | 0.223 | 0.420 | 0.246 | 1.0×10−6 | 0.974 | 0.148 | 0.267 | 0.308 |
| rs1156095 | 64613521 | intron3 | G/A | 0.167 | 0.638 | 0.185 | - | - | - | 0.032 | 0.232 | 0.661 |
| rs11175973 | 64620042 | intron3 | G/T | - | - | - | - | - | - | - | - | |
| rs1480468 | 64628384 | intron3 | A/G | - | - | - | - | - | - | - | - | |
| rs11175978 | 64629135 | intron3 | T/C | - | - | 0.052 | 5.0×10−6 | 1.993 | 0.092 | 0.025 | 0.763 | |
| rs1042725 | 64644614 | 3′ UTR | T/C | 0.499 | 1.58×10−3 | 0.906 | 0.486 | 3.0×10−7 | 0.683 | 0.803 | 0.008 | 0.614 |
| rs7968902 | 64649337 | 3′ UTR | T/G | 0.429 | 1.13×10−3 | 0.938 | 0.414 | 3.4×10−5 | 0.533 | 0.153 | 0.033 | 0.586 |
| rs1480464 | 64656838 | 3′downstream | T/C | 0.196 | 0.153 | 0.518 | 0.190 | 0.379 | 0.142 | 0.252 | 0.064 | 0.418 |
| rs7968682 | 64658147 | 3′downstream | G/T | 0.499 | 1.55×10−3 | 0.939 | 0.468 | 2.0×10−7 | 0.761 | 0.107 | 0.167 | 0.438 |
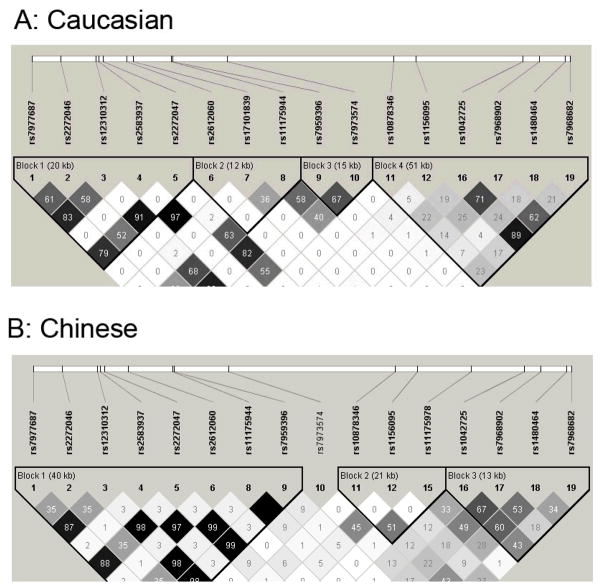
Figure 1.
Linkage disequilibrium diagram of the HMGA2 gene.
Pairwise LD, measured as r2, was calculated from unrelated Caucasians and Chinese data using Haploview program.
Figure 2.
Comparison of height values among subjects with different genotypes for the three significant SNPs in Caucasians and Chinese.
For the family-based Caucasian sample, we selected 1,518 unrelated subjects including the parents from each family to plot this figure. Error bars denote standard error. P values were calculated by the linear model.
For the Chinese sample, SNP rs1042725 in HMGA2 was detected as nominally significant to adult height (P = 0.008). Although the allelic frequency (allele C: 0.197) was quite different from the frequency in Caucasian samples (allele C: 0.501), the effect was in the same direction as in the Caucasian samples. Each copy of the C allele was estimated to be associated with an increase in height of ~0.6 cm. SNP rs7968902 showed a marginally significant association (P = 0.033). However, no significant association was found for rs7968682. As shown in Figure 1, rs1042725 and rs7968902 were in strong LD with each other (r2 of 0.67), whereas the LD was relatively weak between rs1042725 and rs7968682 (r2 of 0.43).
Discussion
Recent GWAS reports have revealed a novel variant rs1042725 in HMGA2 for human adult height in the European populations.(Lettre et al., 2008, Weedon et al., 2008, Weedon et al., 2007) Replication of GWAS findings can be quite difficult, especially for the quantitative trait. In this study, we successfully replicated this finding both in the unrelated and family-based US Caucasian populations, which demonstrated the validity of the initial finding. We also identified another two SNPs (rs7968902 and rs7968682) in HMGA2, which were in high LD with rs1042725, significantly associated with adult height. HMGA2 was considered as a strong biologic candidate for height as a rare severe mutation in this gene alters body size in mice(Zhou et al., 1995) and in human.(Ligon et al., 2005) All of these three SNPs are located within the 3′-UTR region of HMGA2, which may directly or indirectly influence the mRNA stability of HMGA2.
It would be worth to evaluate the association in populations of different ancestry from that of the initial report, since genomic variations is greater when compared across populations, and should increase the credibility of the findings. In this study, we successfully replicated the association between rs1042725 in HMGA2 and adult height in Chinese population and the effect direction was consistent despite of the different allele frequencies. Our findings suggest that this SNP might be a common variant for adult height across different populations.
The statistical power of our study is estimated by using the program Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html). Assuming that a marker has a minor allele frequency of 0.25 and is in strong LD (D′ = 0.9) with a functional mutation that accounts for ~2% variation of height, under the conservative significance level of P = 0.0029 (Bonferroni correction threshold), our Caucasian and Chinese samples can achieve > 85% statistical power, which is large enough to detect a genetic variant under the additive model.
In summary, using data from ~11,000 subjects, we have confirmed the recently found association between HMGA2 and human adult height even across ethnic boundaries, in both Caucasian and Chinese populations, which highlights the importance of this gene involved in normal growth. Follow-up molecular and functional studies are warranted to elucidate its detailed roles and identify the true functional variant.
Acknowledgments
This work was partially supported by the National Institutes of Health [R01 AR050496, R21 AG027110, R01 AG026564, P50 AR055081 and R21 AA015973]. The study was also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, HuNan Province, Xi’an Jiaotong University, and the Ministry of Education of China. The Framingham Heart Study and the Framingham SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University. The Framingham SHARe data used for the analyses described in this manuscript were obtained through dbGaP (phs000007.v3.p2, phs000008.v3.p2). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or the NHLBI.
Footnotes
Competing interest statement
The authors declare that they have no competing financial interests.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cupples LA, Arruda HT, Benjamin EJ, D’agostino RB, Sr, Demissie S, Destefano AL, Dupuis J, Falls KM, Fox CS, Gottlieb DJ, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Kathiresan S, Kiel DP, Laramie JM, Larson MG, Levy D, Liu CY, Lunetta KL, Mailman MD, Manning AK, Meigs JB, Murabito JM, Newton-Cheh C, O’connor GT, O’donnell CJ, Pandey M, Seshadri S, Vasan RS, Wang ZY, Wilk JB, Wolf PA, Yang Q, Atwood LD. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Hart C, Upton M, Hole D, Gillis C, Watt G, Hawthorne V. Height and risk of death among men and women: aetiological implications of associations with cardiorespiratory disease and cancer mortality. J Epidemiol Community Health. 2000;54:97–103. doi: 10.1136/jech.54.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci. 1963;107:539–56. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–81. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnell D, Okasha M, Smith GD, Oliver SE, Sandhu J, Holly JM. Height, leg length, and cancer risk: a systematic review. Epidemiol Rev. 2001;23:313–42. doi: 10.1093/oxfordjournals.epirev.a000809. [DOI] [PubMed] [Google Scholar]
- Hemenway D, Feskanich D, Colditz GA. Body height and hip fracture: a cohort study of 90,000 women. Int J Epidemiol. 1995;24:783–6. doi: 10.1093/ije/24.4.783. [DOI] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon AH, Moore SD, Parisi MA, Mealiffe ME, Harris DJ, Ferguson HL, Quade BJ, Morton CC. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–8. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Meyre D, Delplanque J, Pei YF, Zhang L, Recker RR, Froguel P, Deng HW. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17:1803–13. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Cornes BK, Martin NG, Visscher PM. Bias, precision and heritability of self-reported and clinically measured height in Australian twins. Hum Genet. 2006;120:571–80. doi: 10.1007/s00439-006-0240-z. [DOI] [PubMed] [Google Scholar]
- Perola M, Sammalisto S, Hiekkalinna T, Martin NG, Visscher PM, Montgomery GW, Benyamin B, Harris JR, Boomsma D, Willemsen G, Hottenga JJ, Christensen K, Kyvik KO, Sorensen TI, Pedersen NL, Magnusson PK, Spector TD, Widen E, Silventoinen K, Kaprio J, Palotie A, Peltonen L. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal . Meta-analytic procedures for social research. Beverly Hills, Calif: Sage; 1991. [Google Scholar]
- Silventoinen K, Kaprio J, Lahelma E, Koskenvuo M. Relative effect of genetic and environmental factors on body height: differences across birth cohorts among Finnish men and women. Am J Public Health. 2000;90:627–30. doi: 10.2105/ajph.90.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silventoinen K, Sammalisto S, Perola M, Boomsma DI, Cornes BK, Davis C, Dunkel L, De Lange M, Harris JR, Hjelmborg JV, Luciano M, Martin NG, Mortensen J, Nistico L, Pedersen NL, Skytthe A, Spector TD, Stazi MA, Willemsen G, Kaprio J. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 2003;6:399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, Mccarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Davey Smith G, Groop LC, Hattersley AT, Mccarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–50. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, Pan F, Chen Y, Zhang ZX, Dong SS, Xu XH, Yan H, Liu X, Qiu C, Zhu XZ, Chen T, Li M, Zhang H, Zhang L, Drees BM, Hamilton JJ, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide Copy-Number-Variation Study Identified a Susceptibility Gene, UGT2B17, for Osteoporosis. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–4. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]