Abstract
Origins of DNA replication are licensed through the assembly of a chromatin-bound prereplication complex. Multiple regulatory mechanisms block new prereplication complex assembly after the G1/S transition to prevent rereplication. The strict inhibition of licensing after the G1/S transition means that all origins used in S phase must have been licensed in the preceding G1. Nevertheless mechanisms that coordinate S phase entry with the completion of origin licensing are still poorly understood. We demonstrate that depletion of either of two essential licensing factors, Cdc6 or Cdt1, in normal human fibroblasts induces a G1 arrest accompanied by inhibition of cyclin E/Cdk2 activity and hypophosphorylation of Rb. The Cdk2 inhibition is attributed to a reduction in the essential activating phosphorylation of T160 and an associated delay in Cdk2 nuclear accumulation. In contrast, licensing inhibition in the HeLa or U2OS cancer cell lines failed to regulate Cdk2 or Rb phosphorylation, and these cells died by apoptosis. Co-depletion of Cdc6 and p53 in normal cells restored Cdk2 activation and Rb phosphorylation, permitting them to enter S phase with a reduced rate of replication and also to accumulate markers of DNA damage. These results demonstrate dependence on origin licensing for multiple events required for G1 progression, and suggest a mechanism to prevent premature S phase entry that functions in normal cells but not in p53-deficient cells.
Keywords: Cdk2, CAK, p53, origin licensing, checkpoint
Introduction
DNA replication in human cells initiates at thousands of origins to ensure that each chromosome is completely replicated each cell cycle. The combined action of the Origin Recognition Complex (ORC) with the Cdc6 and Cdt1 proteins results in the recruitment and chromatin loading of a poised, but inactive DNA helicase, the Minichromosome Maintenance Complex (MCM). MCM loading at an origin completes the assembly of a prereplication complex (preRC) and marks that origin as “licensed” for DNA replication.1–5
From the onset of S phase until late mitosis, multiple overlapping mechanisms prevent new origin licensing by inducing the degradation of preRC components, blocking their interaction with one another, or inhibiting their association with chromatin.6–9 In this manner, cells are protected from the consequences of unscheduled origin licensing and the inappropriate rereplication that could then arise. Once the G1/S phase boundary has been crossed, cells do not have the opportunity to license additional origins. Therefore if too few origins were licensed in the preceding G1, cells could become trapped in S phase with partially replicated chromosomes. Ultimately, insufficient origin licensing could lead to chromosome fragmentation and cell death, or rearrangements and genome instability, making it crucial that enough origins are licensed in G1 prior to S phase entry.
Given the potential danger of entering S phase before licensing is complete, it would be advantageous for cells to delay entry into S phase until at least a minimum number of origins have been licensed. Indirect evidence for the existence of such an “origin licensing checkpoint” was first derived from the observation that licensing inhibition in normal cells induces an apparent G1 arrest, but tumor-derived cells activate a robust apoptotic response.10–12 These findings suggested that genetic alterations in cancer cells perturb a relationship between origin licensing and S phase entry that protects normal cells from attempting an S phase that is doomed to fail. However, the mechanisms that prevent S phase entry in normal cells with insufficiently licensed origins have not been fully elucidated.
In the present study, we provide evidence for a link in normal cells between origin licensing in G1 and Cdk2 activation by phosphorylation on T160. We demonstrate that a G1 arrest induced by insufficient origin licensing is accompanied by loss of Cdk2 activation and Rb hyperphosphorylation. In normal cells, the p53 tumor suppressor is required for the dependence of Rb and Cdk2 phosphorylation on origin licensing. In the absence of p53, licensing-deficient cells enter S phase and subsequently acquire markers of DNA damage. Moreover two cancer cell lines with documented deficiencies in p53 regulation fail to link Cdk activation with origin licensing and die by apoptosis with the same DNA damage markers as normal cells depleted of p53. Together these findings establish a relationship between origin licensing and Cdk2 activation that protects normal cells, but not cancer cells, from inappropriate S phase entry.
Results
Cdc6 depletion induces apoptosis in cancer cells, but not in normal human fibroblasts
To investigate the relationship between origin licensing and S phase entry, we examined the effects of suppressing origin licensing in two cancer cell lines, HeLa and U2OS, and in immortalized diploid fibroblasts NHF1-hTert (hereafter abbreviated as NHF1).15 Transfection with siRNA targeting the essential replication licensing factor, Cdc6, resulted in substantial depletion of Cdc6 protein to levels nearly undetectable by immunoblotting (Fig. 1A). In the HeLa and U2OS cells, we observed morphological changes associated with cytotoxicity and a substantial number of floating cells after transfection with cdc6 siRNA but not a control siRNA targeting GFP. In contrast, we observed no such effects of the cdc6 siRNA on the NHF1 cells (data not shown). Flow cytometric analysis of DNA content indicated that cdc6 siRNA, but not control siRNA, induced a substantial increase in the number of HeLa and U2OS cells with sub-G1 DNA content, a marker of apoptosis (Fig. 1B). Cdc6-depleted HeLa and U2OS cells also displayed high levels of cleaved caspase-3, but Cdc6-depleted NHF1 cells did not (Fig. 1C).
Figure 1.
Cdc6 depletion induces apoptosis in HeLa and U2OS cells but not normal human fibroblasts. (A) HeLa, U2OS or NHF1-hTert (NHF1) cells were transfected with a total of 75 nM of control siRNA (targeting GFP) or cdc6 siRNA and incubated for 72 h. Whole cell lysates were subjected to immunoblot analysis for endogenous Cdc6 and tubulin. (B) Portions of the samples in A were analyzed by flow cytometry after staining with propidium iodide for DNA content (x-axis). The percentage of sub-G1 cells is gated and shown on the histograms. The percentage of S phase cells for each population is listed below the corresponding histograms. (C) NHF1, HeLa and U20S cells transfected with the indicated siRNAs for 96 h were evaluated for caspase-3 cleavage by flow cytometry. The bar graph represents the average of three independent experiments. (D) NHF1 cells were transfected with control or cdc6 siRNA for 72 h, labeled for 1 h with BrdU, and analyzed by flow cytometry with anti-BrdU antibody to detect DNA synthesis (y-axis) and with propidium iodide for DNA content (x-axis). (E) Whole cell extracts and chromatin-bound fractions from the cells in (D) were evaluated for MCM, Cdc6, Orc2 and tubulin by immunoblotting.
We noted that the population of cells with an S phase DNA content was not markedly reduced in Cdc6-depleted HeLa or U2OS cells compared to control cells whereas the Cdc6-depleted NHF1 cells accumulated a G1 DNA content (Fig. 1B). To determine if indeed the Cdc6-depleted NHF1 cells had arrested in G1, we analyzed BrdU-labeled cells by flow cytometry 72 h after transfection with control siRNA (targeting GFP) or cdc6 siRNA. Compared to control cells, Cdc6 depletion resulted in a decreased number of S phase cells and a concomitant increase in the number of G1 cells (Fig. 1D). (The small population of cells incorporating BrdU after cdc6 siRNA transfection presumably includes cells with residual Cdc6 protein, and even these incorporated lower levels of BrdU than control cells.) To confirm that the depletion of Cdc6 was sufficient to inhibit replication licensing in these cells, we monitored the association of the MCM subunit, Mcm2, with chromatin by an established chromatin fractionation protocol.13,16 As expected, Cdc6 depletion resulted in a reduction of chromatin-bound Mcm2, illustrating a deficiency in origin licensing (Fig. 1E, top row, compare lanes 3 and 4).
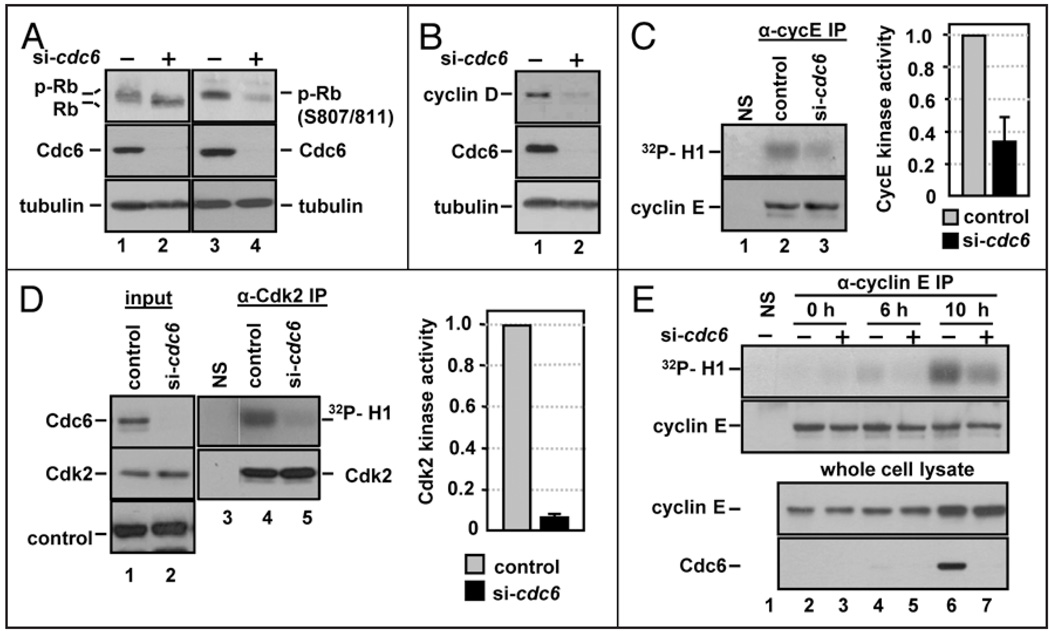
If the Cdc6-depleted normal cells were indeed delaying the G1/S transition, as suggested by the flow cytometry data, these observations would be consistent with a checkpoint-like relationship between origin licensing and S phase entry. Since Cdc6 is an essential DNA replication factor however, we considered assays of BrdU incorporation to be insufficient to accurately determine the cell cycle position of Cdc6-depleted cells. To address this question, we examined molecular markers of the G1/S transition. If Cdc6-depleted cells were in G1, we would expect the retinoblastoma protein, Rb, to be hypophosphorylated and Cdk activity, specifically cyclin D and cyclin E-stimulated Cdks, to remain low. On the other hand, if Cdc6-depleted cells entered S phase, then we would expect high Cdk activity and Rb hyperphosphorylation. We examined the phosphorylation state of Rb by SDS-PAGE mobility and with a phosphospecific antibody. Cdc6 depletion induced an increase in the gel mobility of total Rb protein indicating general hypophosphorylation (Fig. 2A, compare lanes 1 and 2) and a decrease in specific phosphorylation of Rb at serines 807 and 811 (Fig. 2A, compare lanes 3 and 4) in comparison to control cells. Phosphorylation of Rb is carried out by both cyclin D-Cdk4/6 and cyclin E/Cdk2,17–20 therefore we examined whether loss of cyclin D or cyclin E protein could account for the lack of Rb phosphorylation. NHF1 cells transfected with cdc6 siRNA displayed a decrease in cyclin D protein levels (Fig. 2B), consistent with what we and others have recently observed in both Mcm7-depleted and Cdc6-depleted cells.21 In contrast, cyclin E levels remained constant in Cdc6-depleted cells (Fig. 2C).
Figure 2.
Cdc6 depletion prevents cyclin E/Cdk2 activation during G1. (A) Whole cell extracts from NHF1 cells transfected with control or cdc6 siRNA as in Figure 1D were probed with antibodies to total Rb or with a phosphospecific antibody that recognizes Rb phosphorylated at S807 and S811 and for total Cdc6 and tubulin. (B) Whole cell extracts from siRNA-transfected NHF1 were probed with antibodies to cyclin D1, Cdc6 and tubulin. (C) Extracts of siRNA-transfected NHF1 cells were subjected to immunoprecipitation with normal rabbit serum (“NS” lane 1) or with anti-cyclin E antibody (lanes 2 and 3). The precipitates were divided and analyzed by immunecomplex kinase assay with purified histone H1 and [γ-32P]ATP, followed by SDS-PAGE (top row) or analyzed for cyclin E protein by immunoblotting (bottom row). The bar graph reports average cyclin E-associated H1-kinase activity in cells depleted of Cdc6 relative to the corresponding control cells in three independent experiments. (D) Extracts of siRNA-transfected NHF1 cells were treated as in (C), except that the immunoprecipitations and immunoblot utilized anti-Cdk2 antibody. The bar graph reports average Cdk2 H1-kinase activity in cells depleted of Cdc6 relative to the corresponding control cells in three independent experiments. (E) NHF1 cells were transfected with a total of 100 nM of control or cdc6 siRNA for 12 h, then incubated for 72 h in medium containing low serum (0.1% FBS). Cells were released from growth arrest by addition of 10% FBS, labeled for 1 h with BrdU immediately prior to each collection point at 0, 6 and 10 hours after serum addition. Immunecomplex kinases assays were performed as in (C).
Given the overall poor Rb phosphorylation, we reasoned that—in addition to Cdk4 activity—Cdk2 activity must also be low in Cdc6-depleted cells even though cyclin E levels were not. To test that notion directly, we compared the histone H1 kinase activity in both cyclin E and Cdk2 immunoprecipitates from lysates of control and Cdc6-depleted cells. Cdc6 depletion caused substantial reductions in both in cyclin E-associated kinase activity (Fig. 2C, compare lanes 2 and 3) and Cdk2 activity (Fig. 2D, compare lanes 4 and 5). To determine if the effect of Cdc6 depletion on cyclin E-associated kinase activity occurred during G1, we synchronized siRNA transfected cells by serum withdrawal shortly after siRNA transfection and before any detectable loss of Cdc6 protein had occurred (data not shown). We serum-stimulated the quiescent cells and analyzed whole cell lysates for cyclin E protein and cyclin E immunoprecipitates for histone H1 kinase activity. Cdc6 protein was undetectable in serum-starved cells (t = 0 h), but serum stimulation induced Cdc6 protein in control cells by 10 h (Fig. 2E, lane 6) at a timepoint close to the G1/S transition (data not shown, but see also Fig. 4C). Importantly, cyclin E-associated kinase activity was lower in the Cdc6-depleted cells compared to control cells during G1 (Fig. 2E, 6 h).
Figure 4.
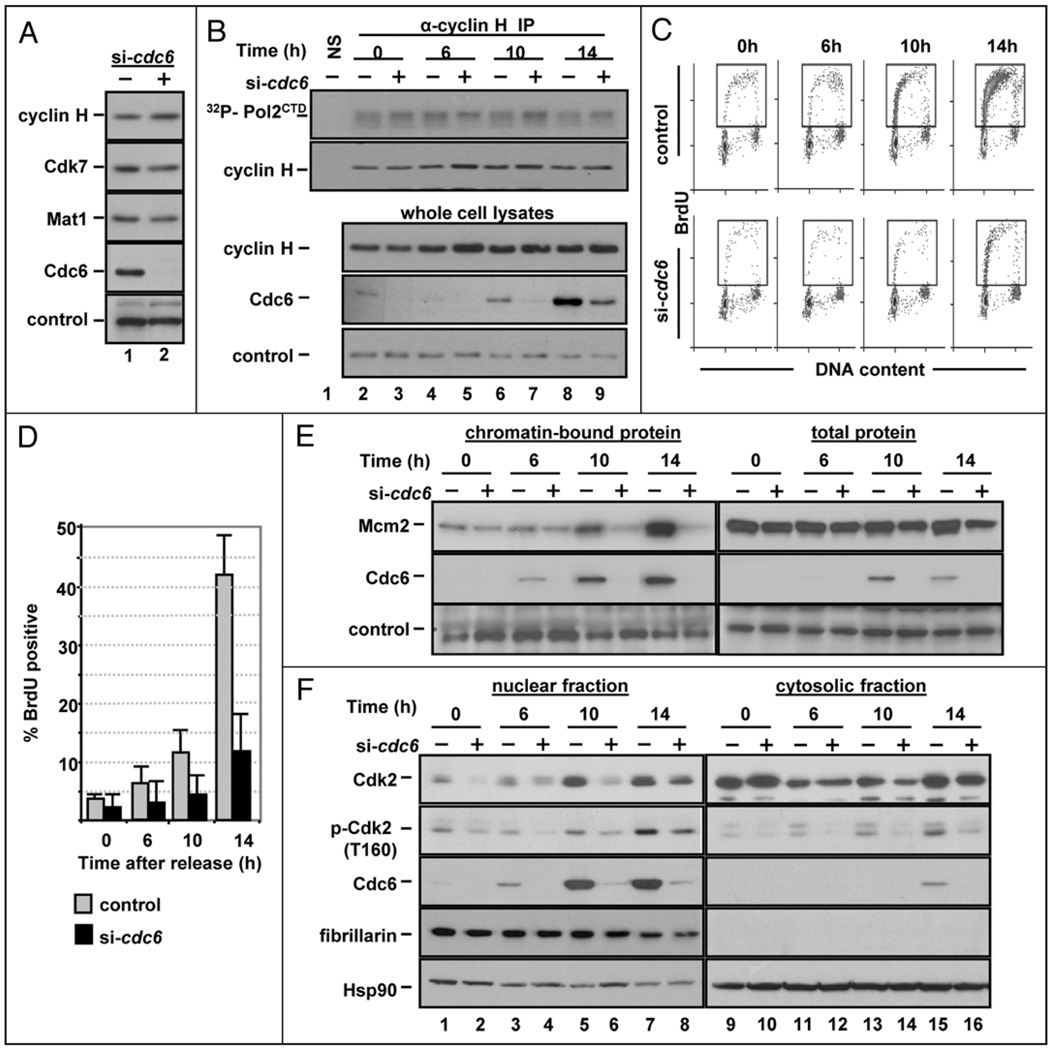
Cdc6 depletion prevents Cdk2 nuclear accumulation in G1. (A) Whole cell extracts of asynchronous NHF1 cells transfected with control or cdc6 siRNA for 72 h were probed for endogenous cyclin H, Cdk7, Mat1 and Cdc6; a non-specific band serves as a loading control. (B) NHF1 extracts were transfected as in Figure 2E and subjected to immunoprecipitation with normal rabbit serum or with anti-cyclin H antibody. The precipitates were divided and analyzed by immunecomplex kinase assay with purified histone CTD and [γ-32P] ATP, followed by SDS-PAGE (top row) or analyzed for cyclin H protein by immunoblotting (bottom row). A portion of the extracts were probed for endogenous cyclin H and Cdc6. A non-specific band served as a loading control for the whole cell extracts. (C) NHF1 cells were transfected with siRNA and synchronized by serum deprivation as in Figure 2E. BrdU incorporation was evaluated by flow cytometry; histograms from a representative experiment are shown. (D) Bar graph documenting the number of BrdU positive cells from three independent experiments as in (C). (E) Cells were treated as in (B), and Mcm2 and Cdc6 were detected in chromatin-bound fractions and in whole cell lysates; a non-specific band serves as a loading control. (F) Portions of the cells in (B) were fractionated by hypotonic lysis into cytosolic and nuclear fractions which were probed by immunoblotting for total Cdk2, phospho-Cdk2 (T160), Cdc6, fibrillarin (nuclear protein control) and HSP90 (cytosolic protein control).
We could not attribute the reduced cyclin E/Cdk2 activity to differences in the levels of cyclin E protein either in the whole cell lysates or in the immunoprecipitates (Fig. 2E, immunoblots). Cdk2 activity is blocked by association with inhibitors such as p21 and p27,22 but Cdc6 depletion had little effect on the overall abundance of either p21 or p27 in either whole cell extracts (Fig. 3A, first and second rows) or in Cdk2 immunoprecipitates (Fig. 3A, compare lanes 4 and 5). A previous report of the effects of origin licensing inhibition by depletion of the Orc2 protein suggested that licensing inhibition leads to an increase in p21 levels.10 We found however, that Orc2 depletion in NHF1 cells did not have this effect despite the fact that Orc2 was depleted enough to induce a reduction in BrdU incorporation (Fig. 3B). These results indicated that at least one mechanism other than changes in cyclin abundance or CDK inhibitor binding links origin licensing to Cdk2 activity.
Figure 3.
Licensing inhibition prevents Cdk2 phosphorylation at T160. (A) Whole cell lysates and anti-Cdk2 immunoprecipitates were probed for the CDK inhibitors, p21 and p27 by immunoblotting. (B) NHF1 cells were transfected with orc2 siRNA for a total of 120 hrs. BrdU incorporation was analyzed as in Figure 1D resulting in average (of two experiments) 29% S phase for control treated and 4.9% for orc2 siRNA treated cells. Whole cell lysates were probed for endogenous p21, Orc2 and tubulin by immunoblotting. (C) NHF1 cells were transfected with control or cdc6 siRNA for 72 h (lanes 2 and 3) or treated with 2 mM hydroxyurea for 24 h (lane 1). Whole cell extracts were immunoblotted with antibodies to detect p53 phosphorylated at S15, Chk2 phosphorylated at T68, Chk1 phosphorylated at S317, Cdk1 and Cdk2 phosphorylated at Y15 (the antibody recognizes both kinases), Cdc6 and tubulin as indicated. (D) Extracts were transfected as in (C) except, that whole cell extracts were probed for phospho-Cdk2 (T160), Cdc6 and tubulin. (E) WI-38 cells (diploid lung fibroblasts) were transfected with a total of 75 nM of control or cdc6 siRNA, incubated for 72 h and processed for flow cytometric analysis as in Figure 1D. The bar graph reports the average percentage S phase cells in three independent experiments (left). The remaining samples were subjected to immunoblot analysis, and a representative set of blots is shown (right). (F) NHF1 cells were transfected with a total of 75 nM of control or cdt1 siRNA and analyzed as in (E).
DNA damage and replication stress can trigger a well-characterized checkpoint response that blocks the G1/S transition. Important activities in this pathway include the p53 tumor suppressor, and the Chk1 and Chk2 protein kinases (reviewed in refs. 23–28). We probed cell extracts with antibodies to the phosphorylated and activated forms of p53, Chk1 and Chk2. As a positive control for detecting the phosphorylation of p53, Chk1 and Chk2, we treated NHF1 cells with hydroxyurea (HU) for 24 h. Although phosphorylation of Chk1, Chk2 and p53 were readily detectable in HU-treated cells, indicating that our antibodies were sufficiently sensitive to detect these markers (Fig. 3C, lane 1), we observed no induction of these modifications in Cdc6-depleted cells (Fig. 3C, compare lanes 2 and 3). We further evaluated cells from 24 h to 72 h after transfection with cdc6 siRNA to determine if a transient increase in DNA damage checkpoint signaling could be detected, but we found no evidence that the damage checkpoint was ever activated after cdc6 siRNA treatment (data not shown). Particularly during a DNA damage response, Cdk2 can also be regulated by Wee1-mediated inhibitory phosphorylation at amino acids T14 and Y15, and retention of phosphorylation at these sites blocks S phase entry.29–31 Using immunoblot analysis, we detected no increase in the level of this inhibitory phosphorylation in Cdc6-depleted cells compared to control cells (Fig. 3C, compare lanes 4 and 5), nor did we detect changes in abundance of the Cdc25A phosphatase responsible for removing the inhibitory phosphorylations on Cdk2 (data not shown). Therefore, the reduced cyclin E-kinase activity was not due to retention of the inhibitory tyrosine phosphorylation of Cdk2.
Cdk2 activity requires phosphorylation at threonine 160 in the “activation loop.”32,33 Using an antibody that specifically recognizes T160 phosphorylation, we discovered that this activating modification of Cdk2 was reduced in Cdc6-depleted cells compared to control cells (Fig. 3D, first row), while total Cdk2 levels remained the same (Fig. 3D, second row). To test if these molecular phenotypes associated with Cdc6 depletion are detectable in other normal human cells, we transfected diploid human lung fibroblasts (WI-38 cells) with cdc6 siRNA, then analyzed BrdU incorporation, Cdk2 T160 and Rb phosphorylation. As in NHF1 cells, Cdc6 depletion resulted in a ~6-fold reduction in S phase cells (Fig. 3E, bar graph) and importantly, loss of both Cdk2 and Rb phosphorylation (Fig. 3E, first and second rows). We obtained similar results in another telomerase-expressing fibroblast line, NHF10 and with multiple cdc6 siRNA sequences (data not shown).
The G1 arrest and Cdk2 phosphorylation defect could be specific consequence of the loss of Cdc6 protein, or it could be a general consequence of the licensing defect caused by loss of Cdc6 function. To distinguish between these possibilities, we depleted NHF1 cells of Cdt1 which, like Cdc6 depletion, caused the predicted MCM chromatin loading defect (data not shown). Also, like Cdc6 depletion, Cdt1 depletion caused a reduction in the number of S phase cells (Fig. 3F, bar graph) with no obvious signs of cell death (data not shown). Moreover, Cdt1 depletion induced a loss of both Cdk2 and Rb phosphorylation (Fig. 3F, first and second rows, respectively) and a reduction in Cdk2 activity in vitro (data not shown). These results further confirm that the effects of the siRNAs are attributable to the origin licensing defect rather than an off-target effect of the siRNAs, since the cdc6 and cdt1 siRNAs target different components of the prereplication complex.
Cdk2 activation and nuclear accumulation during G1 are delayed in Cdc6-depleted cells
In human cells, the activating phosphorylation of all CDKs is primarily attributed to CDK activating kinase (CAK), a heterotrimeric complex of Cdk7, cyclin H and Mat1.34 A logical explanation for the reduction of Cdk2 T160 phosphorylation would be the loss of CAK activity, but the total amount of Cdk7, cyclin H and Mat1 were not reduced in Cdc6-depleted cells compared to control-treated cells (Fig. 4A). We also examined the catalytic activity of CAK in control and Cdc6-depleted cells by immunecomplex kinase assays of cyclin H immunoprecipitates using the C-terminal domain of RNA polymerase II (a standard substrate for this kinase35). Cdc6-depleted G1 cells showed no reduction in CAK activity in vitro compared to control cells using either RNA PolII (Fig. 4B) or Cdk2 as a substrate (data not shown). These observations demonstrate that neither downregulation of CAK subunit expression nor reduced CAK enzymatic activity account for reduced Cdk2 T160 phosphorylation.
We considered the possibility that a defect in Cdk2 phosphorylation by CAK could instead be explained by enzyme-substrate accessibility. Previous studies have noted that changes in Cdk2 T160 phosphorylation are often accompanied by changes in Cdk2 localization. In quiescent fibroblasts, Cdk2 resides primarily in the cytoplasm, but late in G1 it accumulates in the nucleus by a mechanism that is still incompletely understood.36–40 Since all subunits of CAK are constitutively expressed in the nucleus,41 the decrease in Cdk2 phosphorylation on T160 in quiescent cells could be explained (at least in part) to the absence of Cdk2 in the nucleus. To investigate if the defect in Cdk2 phosphorylation in Cdc6-depleted cells included altered Cdk2 nuclear accumulation, we again prepared Cdc6-depleted synchronized NHF1 cells (as in Fig. 2E) and examined not only the chromatin fractions for Cdc6 and MCM association, but also the nuclear and cytoplasmic fractions for total Cdk2 and Cdk2 phosphorylated on T160. As expected, NHF1 cells transfected with cdc6 siRNA delayed S phase entry (Fig. 4C and D). By 10 h, the rise in Cdc6 protein levels was accompanied by an increase in Mcm2 chromatin association in the control cells. As expected, cells depleted of Cdc6 were deficient in Mcm2 chromatin loading during G1 (Fig. 4E). It should be noted that despite the fact that these siRNA transfections are highly effective in suppressing Cdc6 protein expression and MCM loading, they are not likely to phenocopy a true null; the small number of cells entering S phase in the Cdc6-depleted population presumably reflects the activity of the residual Cdc6 protein in a small fraction of cells.
Consistent with previous observations,36,39,40 total Cdk2 protein was predominantly cytoplasmic in G0 cells, but the levels of nuclear Cdk2 protein increased in late G1 in control transfected cells (Fig. 4F, top row, compare lanes 1 and 5). On the other hand, Cdk2 nuclear accumulation was delayed in Cdc6-depleted cells, particularly noticeable at the time of the G1/S transition in control cells (Fig. 4F, 10 h, lane 6). These localization effects were accompanied by poor phosphorylation of Cdk2 on T160 (Fig. 4F). The observed delay in Cdk2 nuclear accumulation in Cdc6-depleted cells, coupled with a constitutively nuclear CAK, represents at least one mechanism to explain the defect in Cdk2 T160 phosphorylation in Cdc6-depleted cells.
The cell cycle arrest in Cdc6-depleted NHF1 cells requires p53
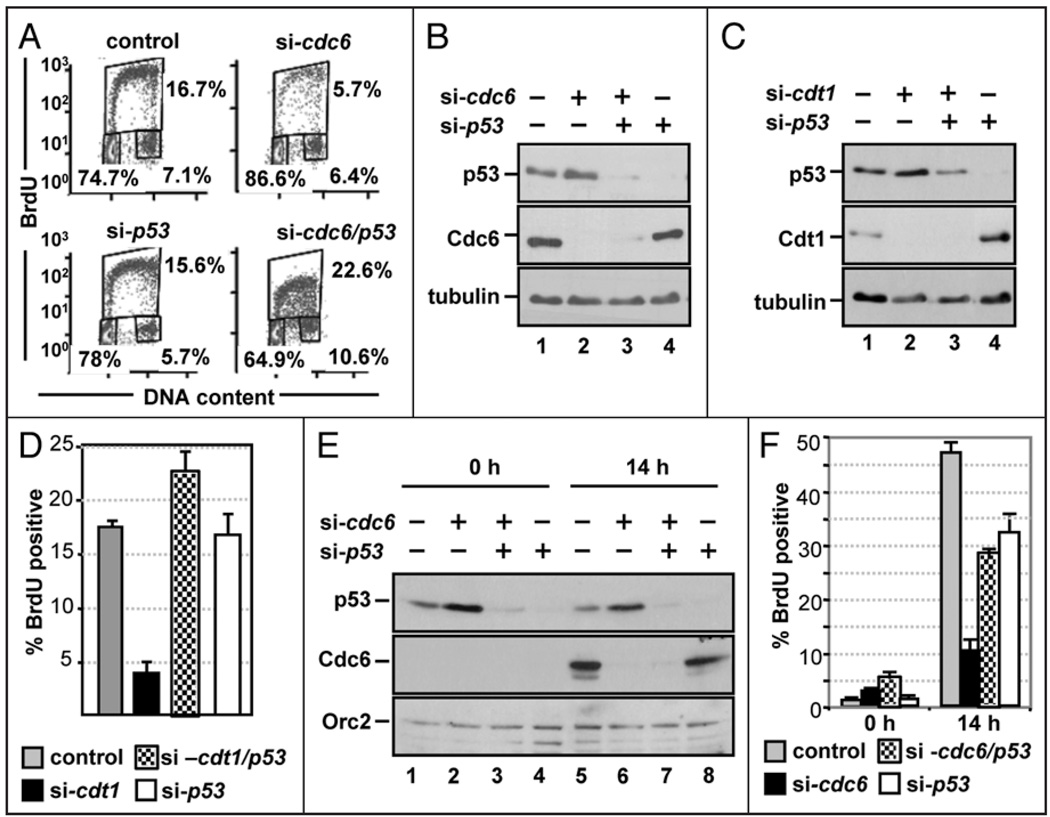
The differential response of normal and cancer cells to licensing inhibition (Fig. 1 and refs. 11 and 12) suggested that a commonly mutated pathway in cancer cells is involved in coordinating licensing with G1 progression. The p53 tumor suppressor pathway is deregulated in the vast majority of human cancers, so we considered p53 a candidate for participation in the G1 arrest induced by Cdc6 depletion. To test this idea, we performed co-depletion experiments using p53 siRNA. As before, transfection with cdc6 siRNA alone reduced the number of S phase cells from 16.7% in control cells to 5.7% in Cdc6-depleted cells (Fig. 5A, compare control and si-cdc6). Depletion of p53 alone had little effect on the cell cycle profile of normal cells under these conditions (Fig. 5A, compare control and si-p53). Co-depletion of p53 24 h after transfection with cdc6 siRNA completely prevented the cell cycle effects of Cdc6 depletion, fully restoring the number of S phase cells to control numbers (Fig. 5A, compare si-cdc6 and si-cdc6/p53). Conspicuously, the low intensity of the BrdU staining in the co-depleted cells (note the y-axis) suggested that even though the co-depleted cells entered S phase, replication was still very inefficient. Depletion of p53 did not rescue Cdc6 expression (Fig. 5B), which likely accounts for the reduced DNA synthesis rate. Virtually identical results were obtained when p53 was co-depleted with Cdt1 (Fig. 5C and D), indicating that p53 is required for the cell cycle effects of the licensing defect (i.e., loss of Cdc6 and Cdt1 functions) rather than the loss of Cdc6 or Cdt1 proteins.
Figure 5.
p53 is required for licensing-deficient cells to delay S phase entry. (A) NHF1 cells were transfected with a total of 75 nM of control or cdc6 siRNA, 24 h later cells were transfected with control (GFP) or p53 siRNA and incubated an additional 48 h. Cell cycle profiles were generated as in Figure 1D. (B) Immunoblots of extracts of the cells analyzed in (A) by flow cytometry with the indicated antibodies. (C) NHF1 cells were transfected with control, cdt1, p53 or cdt1 and p53 siRNA for 72 h. Immunoblots of whole cell extracts were probed to detect p53, Cdt1 and tubulin. (D) Average BrdU incorporation from three independent transfections of the experiment shown in (C). (E) NHF1 cells were transfected as in Figure 4B. Cells were released from growth arrest by addition of 10% FBS, labeled for 1 h with BrdU prior to each collection point at 0 and 14 h after serum addition. Immunoblots of whole cell lysates were probed for the indicated proteins. (F) Average BrdU incorporation from three independent transfections of the experiment shown in (B).
We further tested for dependence on p53 for the delayed S phase entry in synchronized cells following the same procedure as in Figure 4. Cdc6 expression was undetectable in quiescent cells but induced in control cells by serum stimulation, and p53 depletion did not change either of these findings (Fig. 5E). As expected, Cdc6-depleted NHF1 cells delayed S phase entry compared to control cells (Fig. 5F, gray and black bars). NHF1 cells depleted of p53 alone slightly delayed S phase entry when compared to control cells (Fig. 5F, white bars), but more importantly, cells co-depleted of Cdc6 and p53 entered S phase in numbers similar to both the control and p53-depleted cells (Fig. 5F, checkered bars). Cdc6 and Cdt1 are essential replication factors, so we attribute the DNA replication in cells co-depleted of p53 and Cdc6 or p53 and Cdt1 to the very low levels of origin licensing supported by the scant Cdc6 or Cdt1 protein in these experiments. Importantly, the rescue of the S phase population implies that in the cells transfected with just cdc6 or cdt1 siRNA there were indeed a small number of licensed origins present, but they did not fire due to p53-dependent restraint.
Cells that fail to arrest in response to origin licensing inhibition fail to downregulate Rb and Cdk2 phosphorylation
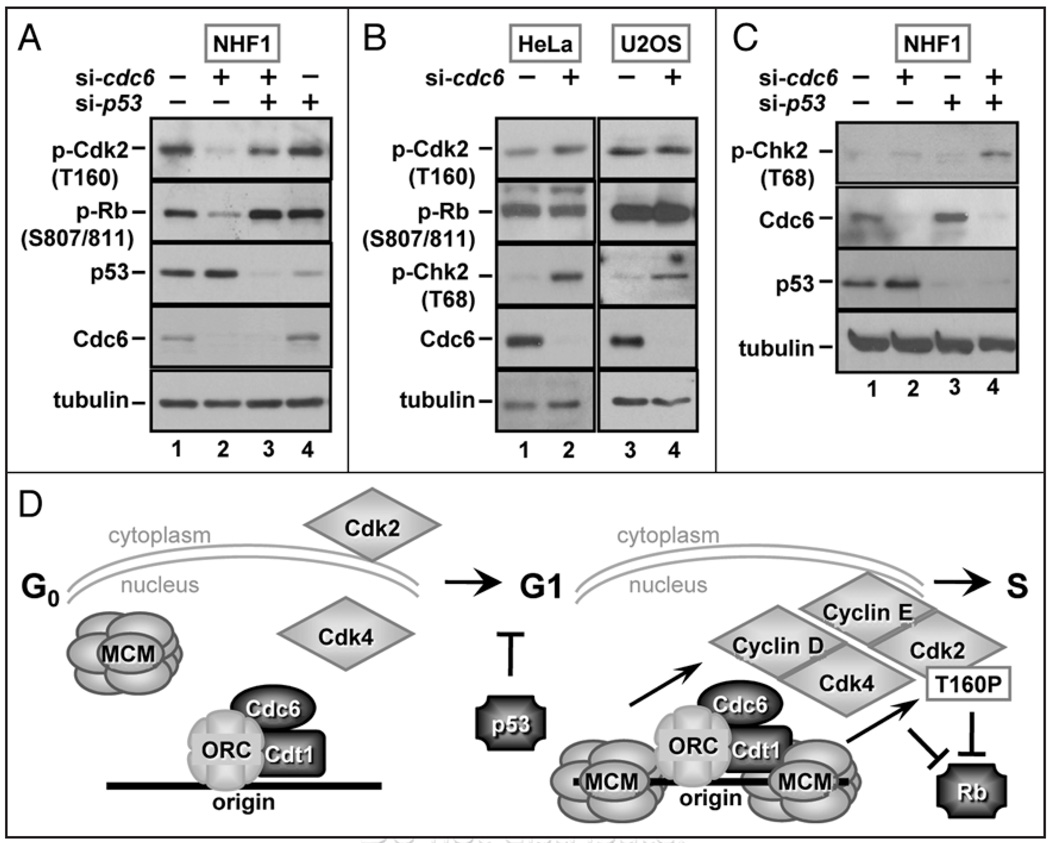
The observation that Cdc6 depletion resulted in reduced phosphorylation of both Rb and Cdk2 T160 prompted us to test if the suppression of the cell cycle defect by co-depletion of p53 also suppressed these molecular phenotypes. We probed lysates of cells transfected with siRNAs targeting Cdc6 and p53, either singly or in combination, for phosphorylation of Rb and Cdk2 on T160 as in Figures 2 and 3. As we had observed before, Cdc6-depleted cells were characterized by poor Rb and Cdk2 phosphorylation (Fig. 6A, compare lanes 1 and 2). Co-depletion of p53 with Cdc6 largely restored both Rb phosphorylation and Cdk2 phosphorylation (Fig. 6A, compare lanes 2 and 3 with lane 1), in keeping with the near-complete rescue of the S phase population (Fig. 5A, D and F). Previous reports that origin licensing inhibition induces apoptosis in a wide variety of transformed cell lines11,12 (including HeLa and U2OS, Fig. 1), suggested that these cells might be unable to induce the same G1 arrest that normal cells do. To determine if cancer cell lines were specifically defective in downregulating both Rb phosphorylation and Cdk2 T160 phosphorylation, we probed for these markers in Cdc6-depleted HeLa and U2OS cells. Despite effective Cdc6 depletion, neither HeLa nor U2OS cells showed the same loss of Rb and Cdk2 phosphorylation that we observed in the normal cells (Fig. 6B, compare lane 1 with 2 and lane 3 with 4). Thus, not only are HeLa and U2OS cells sensitive to apoptosis by Cdc6 depletion, but they are incapable of regulating the markers of a G1 arrest that the Cdc6-depleted NHF1 cells did.
Figure 6.
p53 is required for downregulation of Cdk2 and Rb phosphorylation in licensing-deficient cells. (A) Whole cell extracts from control, cdc6, p53 or Cdc6 and p53 siRNA transfected NHF1 cells were probed with antibodies to detect phospho-Rb (S807/S811), phospho-Cdk2 (T160), p53, Cdc6 and tubulin. (B) HeLa and U20S cells were transfected with control or cdc6 siRNA, and whole cell extracts were subjected to immunoblot analysis with antibodies to detect phospho-Cdk2 (T160), phospho-Rb (S807/ S811), phospho-Chk2 (T68), Cdc6 and tubulin. (C) Whole cell extracts from control, cdc6, p53 or Cdc6 and p53 siRNA transfected NHF1 cells were probed for phospho-Chk2, p53, Cdc6 and tubulin. (D) Model summarizing the results described in this study.
If these cancer cell lines failed to arrest in G1, then an attempted S phase with low levels of origin licensing would be predicted to result in sparsely distributed replication forks. The partially replicated chromosomes would be more susceptible to fork collapse and double-strand breaks. To test this prediction, we probed the Cdc6-depleted cancer cells for a marker of DNA double-strand breaks, ATM-dependent phosphorylation of Chk2 on T68. In both cell lines, Chk2 phosphorylation was induced by Cdc6 depletion (Fig. 6B, third row). These observations suggest that HeLa and U2OS cells could not arrest in G1 as a consequence of an origin licensing defect, but since they could not complete S phase either, the partially replicated chromosomes accumulated DNA damage.
If this description accurately reflects what happened in the cancer cell lines, then we predicted that the Cdc6-depleted normal cells that inappropriately entered S phase due to co-depletion of p53 should also contain phosphorylated Chk2. We therefore probed lysates of the co-depleted NHF1 cells for phospho-Chk2. As before (Fig. 3C), depletion of Cdc6 alone did not induce any detectable markers of a DNA damage response (Fig. 6C, compare lanes 1 and 2), and neither did depletion of p53 (Fig. 6C, lane 3). In contrast to these single depletions, co-depletion of p53 with Cdc6 led not only to S phase entry (Fig. 5), but also to accumulation of phospho-Chk2 (Fig. 6C, lane 4). In a sense, depletion of p53 from the normal fibroblasts allowed them to phenocopy the cancer cells.
Discussion
In this study we demonstrate that the activity of cyclin E/Cdk2 in late G1 depends on the licensing of replication origins. Similar Cdk2 downregulation and Rb hypophosphorylation was observed in both Cdc6-depleted cells (that express Cdt1) and in Cdt1-depleted cells (that express Cdc6). The disruption common to both Cdc6 and Cdt1 depletion is the failure to fully load MCM complexes at origins, indicating that it is origin licensing itself that is required for Cdk2 activation and Rb phosphorylation. We conclude that the low activity of Cdk2 in Cdc6-depleted or Cdt1-depleted fibroblasts was primarily due to failure to phosphorylate Cdk2 on T160 since no other major CDK regulatory mechanism was perturbed in these cells. The physiological circumstances leading to inhibition of CDK activation by phosphorylation are not as well studied as most other forms of CDK regulation, so the finding that this event is responsive to licensing inhibition represents a novel aspect of CDK control. In further support of a link between licensing and CDK activation, Kan et al. observed a correlation between Cdc6 expression and Cdk2 phosphorylation during recovery from MMS-induced checkpoint arrest.42
We observed that expression of CAK subunits was not inhibited in licensing-deficient cells (Fig. 4A) suggesting that the downregulation of Cdk2 phosphorylation is due to other perturbations such as nuclear accumulation of Cdk2 (Fig. 4F). It should be noted however that even though there was a reduction in the nuclear pools of Cdk2 in Cdc6-depleted cells, this effect may not be the only reason Cdk2 was not phosphorylated in licensing-deficient cells. Lentz et al. noted that even a constitutively nuclear Cdk2 fusion still required one or more serum-stimulated events in late G1 for Cdk2 phosphorylation on T160.43 The results presented here suggest that origin licensing could be that event.
We also demonstrate that p53 is required for licensing-deficient cells to downregulate Cdk2 T160 and Rb phosphorylation, and p53-depleted normal cells fail to delay S phase entry when licensing is inhibited. An important conclusion from this finding is that Cdc6-depleted or Cdt1-depleted cells had a low level of origin licensing (note for instance the faint Mcm2 band in lane 4 of Fig. 1E), but these few licensed origins did not fire on schedule as long as p53 was expressed at normal levels. In other words, the normal cells treated with only cdc6 siRNA licensed enough origins for a very weak or inefficient S phase, but the presence of normal amounts of p53 prevented them from crossing the restriction point, entering S phase, and firing those few origins. During G1, MCM loading closely coincided with Cdk2 phosphorylation on T160 (Fig. 4E and F). It may be that a critical threshold of MCM loading must be reached in G1 before efficient Cdk2 activation can take place. This coordination of Cdk2 activation with licensing is suggestive of a cell cycle checkpoint relationship that serves to ensure that S phase does not begin before a critical task in G1, origin licensing, has been accomplished.
We note that a majority of cancer cells have disruptions in the regulation of both p53 and Rb; in the case of HeLa cells these disruptions include expression of the HPV E6 and E7 proteins, and in U2OS cells it is overexpression of the p53 ubiquitin ligase, MDM2,44 and loss of the p16 Cdk4 inhibitor.45 These cancer cell lines behave similarly to the p53-depleted NHF1 cells when licensing is blocked by depletion of Cdc6 in that they fail to downregulate Cdk2 and Rb phosphorylation. Many cell cycle studies have explored CDK regulation only in cancer cell lines (e.g., HeLa cells) where the dependence of Cdk2 phosphorylation on origin licensing during G1 could not have been observed.
Despite the fact that p53 is required for the cell cycle response to licensing inhibition, Cdc6-depleted cells do not activate the canonical DNA damage checkpoints since neither upstream markers (phosphorylation of Chk1 and Chk2) nor downstream targets (p53, p21 or Cdk1 and Cdk2 Y15 phosphorylation) are induced by Cdc6 depletion. One possible role for p53 in making normal cells sensitive to an origin licensing checkpoint is maintaining basal expression of a cohort of growth inhibitory genes. For example, in p53-deficient NHF1 cells, basal levels of p21 are low and Cdk2 activity is correspondingly higher (our unpublished observations).
Although our results indicate that origin licensing is required to promote Cdk2 phosphorylation, we call attention to the fact that these are not the only important events in G1 that respond to the status of origin licensing. Rb phosphorylation can be carried out not only by Cdk2, but also by cyclin D/Cdk4, and indeed it has been reported that Cdk4 and Cdk6 are principally responsible for the phosphorylation of S807 and S811 that we monitored in this study.19 If cyclin D-dependent kinase activity were unaffected by origin licensing then we would have expected Rb phosphorylation to remain high in Cdc6-depleted cells. Our demonstration that cyclin D protein levels are low (Fig. 2B) combined with our indirect measurement of Cdk4/6 activity (Rb phosphorylation in vivo) indicates that Cdk4/6 activity must also be low, and this conclusion is strongly supported by the more extensive analysis of Cdk4 activity in Liu et al.21 In that study, origin licensing inhibition by Mcm7 depletion in synchronized normal cells during G1 resulted in reduced Cdk4 activity due to low transcription of the cyclin D gene. Thus, we conclude that origin licensing is required for full activity of both early G1 CDK activity (cyclin D/Cdk4) and late G1 CDK activity (cyclin E/Cdk2).
The consequences of poor origin licensing may be even more global than just regulation of CDK activity however. The molecular markers we have examined in this study and in the study by Liu et al. are indicative of a cellular state that is more similar to quiescence than to G1, despite the fact that growth factor stimulated signal transduction pathways appear unaffected (our unpublished observations and ref. 21). A “pseudo-quiescent” state induced by failure to license origins may explain why bypass of the G1 arrest has thus far only been achieved by manipulations that cause pleiotropic effects such as expression of HPV E621 or depletion of p53.
Given that mammalian MCM proteins are constitutively nuclear,46–49 the events in G1 that are reduced in licensing-deficient cells must be sensitive to a difference between chromatin-bound MCM complex and the absence of chromatin-bound MCM (model in Fig. 6D). An emphasis on chromatin localization further supported by a recent proteomics analysis by Khoudoli et al.50 These investigators examined the entire collection of proteins that associate with X. laevis chromatin under normal licensing conditions in vitro and when licensing is blocked by excess geminin which inhibits Cdt1 function.14,51,52 Intriguingly, proteins affected in the geminin-treated samples included not only the predicted MCM subunits, but also factors involved in a surprising variety of nuclear processes including nuclear pore assembly, nuclear structure, chromatin remodeling, and DNA repair. If these results are applicable to human cells, then origin licensing could be a step that is critical not only for DNA replication, but also for many aspects of cell cycle progression.
Materials and Methods
Cell culture and siRNA transfection
Normal human fibroblasts immortalized with human telomerase (NHF1-hTert) and cancer cell lines (HeLa and U2OS) were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Invitrogen) containing 10% fetal bovine serum (Sigma) and 2 mM L-glutamine (Sigma). WI-38 lung fibroblasts were cultured in Minimum Eagles medium (Gibco) containing 10% fetal bovine serum (Sigma), 1X nonessential amino acids (Gibco) and 2 mM L-glutamine (Sigma). All cultures were maintained in a humidified incubator at 37°C and 5% CO2.
Log phase normal fibroblasts were plated at a density of 1.35 × 105 per 100-mm dish 24 h prior to transfection. SiRNA oligonucleotides were synthesized by Invitrogen (GFP control, 5'-GGC UAC GUC CAG GAG CGC ACC TT-3'; cdc6 siRNA- 2144, 5'-UCU AGC CAA UGU GCU UGC AAG UGU A-3'; cdc6 siRNA-2534, 5'-CAC CAU GCU CAG CCA UUA AGG UAU U-3'; p53 siRNA, 5'-AAG GAA GAC UCC AGU GGU AAU-3'; cdt1 siRNA, 5'-CCU ACG UCA AGC UGG ACA ATT-3'). Transfections were performed with a total of 75 nM siRNA duplex using Dharmafect 1 reagent (Dharmacon) according to the manufacturer’s guidelines. Equal amounts of both cdc6 siRNA molecules were used (37.5 nM each). The following day, the total volumes were brought to 10 ml with fresh medium, and cultures were typically incubated for an additional 48 h. In the case of co-depletion of p53 and Cdc6 or Cdt1, cells were transfected with p53 siRNA 24 h after cdc6 or cdt1 siRNA, and examined 72 h after the initial transfection. For Orc2 depletions, log phase normal fibroblasts were seeded at 80% confluence in 6-cm dishes and transfected with 100 nM Orc2 or control siRNA the following day. Cells were incubated for 24 h, then split into 10-cm dishes, and transfected again 24 h later. Cells were collected 72 h after the second siRNA transfection.
Cell synchronization
Asynchronously growing fibroblasts were plated at a density of 2.5 × 105 cells per 100-mm dish and transfected 24 h later, as described above, except that 100 nM siRNA was used. Twelve hours later, the transfection solution was removed and replaced with medium containing 0.1% FBS. After 72 h in low serum, cells were stimulated by the addition of 10% FBS and collected at the indicated times after stimulation.
Immunoblot analysis
Total cell lysates were prepared by resuspending pellets in lysis buffer containing 0.1% Triton X-100, 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mM orthovanadate, 1 mM β-glycerolphosphate, 5 µg/ml phosvitin, 1 µg/ml leupeptin, 1 µg/ml aprotinin and 1 µg/ml pepstatin A (all from Sigma) in phosphate-buffered saline (PBS); protein concentrations were determined by the Bradford assay (Bio-Rad). Chromatin fractions were prepared as previously described.13 Samples containing equal amounts of protein were combined with Laemmli sample buffer [10% glycerol, 0.05 M Tris (pH 6.8), 0.1% bromophenol blue, 1% SDS] containing 10% beta-mercaptoethanol and boiled. Samples were separated by SDS-PAGE and transferred to PVDF membrane (Millipore) and probed with appropriate antibodies to detect the following proteins: Cdc6 (sc-9964), p53 (sc-126), cyclin H (sc-609), cyclin D1 (sc-295), Mat1 (sc-13142) and cdk2 (M2, sc-163) from Santa Cruz Biotechnology; HSP90 (4874), phospho-Chk1 (S345), phospho-Chk2 (Th68), phospho-Rb (S807/811), phospho-Cdc2/Cdk2 (Y15), phospho-Cdk2 (T160) and phospho-p53 (S15) from Cell Signaling Technologies; Mcm2 (BM-28), Orc2, Cdc25A and Rb from BD Pharmingen; fibrillarin from Abcam; tubulin (DM1A) from Sigma; and p21 (Ab-10), Cdk7 (MO-1.1), cdk2 (2B6), p27 (DCS-72.F6) and cyclin E (HE12) from Neomarkers. Antibodies detecting p27 and cyclin E were gifts from Dr. Yue Xiong (University of North Carolina at Chapel Hill). Anti-Cdt1 antiserum was described previously.14 We confirmed the specificity of the phospho-Cdk2 (T160) antibody using myc epitope-tagged normal and T160A mutant forms of Cdk2 (gift of J. Baldassare) expressed in transfected 293 cells (data not shown).
For nuclear fractionation, cells were incubated on ice for 30 min in hypotonic lysis buffer containing 10 mM HEPES-KOH pH 7.5, 10 mM KCl, 3 mM MgCl2, 1 mM EDTA, 0.05% NP-40, 1 mM DTT, 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 0.5 mM orthovanadate, 1 mM β-glycerol phosphate, 10 µg/ml phosvitin, 1 µg/ml leupeptin, 1 µg/ml aprotinin and 1 µg/ml pepstatin A. Extracts were centrifuged at 600 ×g for 5 min and cytosolic fractions (supernatant) were transferred to new tubes. Nuclei were washed with hypotonic buffer and resuspended in Laemmli sample buffer. Cytosolic fractions were quantified by Bradford assay, and equivalent amounts of protein (typically 30 µg) were separated by SDS-PAGE. Nuclear extracts corresponding to twice the cell equivalents of the paired cytosolic extracts were loaded. Fibrillarin (nuclear) and HSP90 (cytoplasmic) were used as fractionation controls.
Cell cycle and cleaved caspase-3 analysis
Cells to be analyzed by flow cytometry were labeled with 10 µM BrdU for 1 h prior to trypsinization and ethanol fixation. Nuclei were stained with fluorescein isothiocyanate (FITC)-labeled anti-BrdU antibody (BD Biosciences) and counterstained with propidium iodide. Nuclei were analyzed using the CyAn (DakoCytomation), and cell cycle distributions were determined using Summit v4.3 software (DakoCytomation). The fraction of apoptotic cells was determined using an active caspase-3 antibody kit (BD Pharmingen), according to the manufacturer’s guidelines.
Kinase assays
Cells were lysed for 30 min at 4°C in RIPA (50 mM Tris-HCL, pH 8.0, 200 mM NaCl, 0.5% NP-40, 1 mM dithiothreitol, 50 µg/ml of AEBSF, 10 µg/ml aprotinin, 20 mM NaF, 0.1 mM sodium orthovanadate). Lysates were clarified and subjected to immunoprecipitation with either anti-cyclin E antiserum, anti-Cdk2 (M2) or anti-cyclin H and protein A-agarose for a total of 3 h. Beads were washed twice with buffer A (20 mM Tris-HCL, pH 8.0, 250 mM NaCl, 1 mM EDTA, 0.5% NP-40), twice in buffer B (buffer A but containing 100 mM NaCl), and then once in kinase buffer (50 mM Tris-HCL, pH 7.5, 10 mM MgCl2 and 1 mM DTT). Kinase reactions were carried out in 25 µl kinase buffer containing 5 µg of histone H1 (Sigma) or the C-terminal domain of RNA Pol II (purified as a GST fusion from E. coli), 1 µM ATP, and 5 µCi [γ-32P]ATP (Perkin Elmer) and incubated at 30°C for 30 min. Reactions were stopped with 1X SDS sample buffer, boiled and separated on 10% SDS-PAGE. Gels were washed 3 times by soaking them for 15 min each in buffer C (20 mM Tris base, 200 mM glycine, 0.1% SDS, 10% glycerol, 1% sodium pyrophosphate), dried and autoradiographed. Relative phosphorylation of histone H1 or CTD was determined using the ImageJ program (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2006).
Acknowledgements
The authors thank W. Kaufmann, R. Duronio, C. Vaziri, X. Grana and members of the Cook laboratory for critical comments on the manuscript, and we thank Y. Xiong (University of North Carolina) for the gift of antibodies, J. Baldassare (University of St. Louis) for Cdk2 plasmids, Arno Greenleaf (Duke University) for the GST-RNA PolII plasmid, and particularly A. Dutta (University of Virginia) for the gift of phospho-T160 antibody and the suggestion to test Cdk2 T160 phosphorylation. We are grateful to K. Reidy, G. Cameron, K.S. Luce, K. McKernan, T. Tan and A. Williamson for technical support. This work was supported by an Environmental Pathology Training Grant T32-ES07017 (K.R.N.), National Institutes of Health Grant K01-CA094907 (J.G.C.), the American Cancer Society Research Scholars Grant GMC-111880 (J.G.C.) and in part by center grants from the National Cancer Institute (P30 CA16086) and the National Institute for Environmental Health Sciences (P30 ES10126).
References
- 1.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007 doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasanth SG, Mendez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond B Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 5.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machida YJ, Hamlin JL, Dutta A. Right place, right time and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div. 2006;1:22. doi: 10.1186/1747-1028-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7:523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 10.Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem. 2005;280:27624–27630. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- 11.Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- 12.Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook JG, Park CH, Burke TW, Leone G, DeGregori J, Engel A, et al. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci USA. 2002;99:1347–1352. doi: 10.1073/pnas.032677499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 15.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 18.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- 20.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Slater DM, Lenburg M, Nevis K, Cook JG, Vaziri C. Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle. 2009;8:125–136. doi: 10.4161/cc.8.1.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 23.Kastan MB, Bartek J. Cell cycle checkpoints and cancer. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 24.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 26.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 27.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 28.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 29.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 30.Costanzo V, Robertson K, Ying CY, Kim E, Avvedimento E, Gottesman M, et al. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol Cell. 2000;6:649–659. doi: 10.1016/s1097-2765(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 31.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, et al. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 34.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25:839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe Y, Fujimoto H, Watanabe T, Maekawa T, Masutani C, Hanaoka F, et al. Modulation of TFIIH-associated kinase activity by complex formation and its relationship with CTD phosphorylation of RNA polymerase II. Genes Cells. 2000;5:407–423. doi: 10.1046/j.1365-2443.2000.00336.x. [DOI] [PubMed] [Google Scholar]
- 36.Bresnahan W, Boldogh I, Ma T, Albrecht T, Thompson E. Cyclin E/Cdk2 activity is controlled by different mechanisms in the G0 and G1 phases of the cell cycle. Cell Growth Differ. 1996;7:1283–1290. [PubMed] [Google Scholar]
- 37.Bresnahan WA, Thompson EA, Albrecht T. Human cytomegalovirus infection results in altered Cdk2 subcellular localization. J Gen Virol. 1997;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- 38.Brown KA, Roberts RL, Arteaga CL, Law BK. Transforming growth factor-beta induces Cdk2 relocalization to the cytoplasm coincident with dephosphorylation of retinoblastoma tumor suppressor protein. Breast Cancer Res. 2004;6:130–139. doi: 10.1186/bcr762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich C, Wallenfang K, Oesch F, Wieser R. Translocation of cdk2 to the nucleus during G1-phase in PDGF-stimulated human fibroblasts. Exp Cell Res. 1997;232:72–78. doi: 10.1006/excr.1997.3507. [DOI] [PubMed] [Google Scholar]
- 40.Keenan SM, Bellone C, Baldassare JJ. Cyclin-dependent Kinase 2 nucleocytoplasmic translocation is regulated by extracellular regulated kinase. J Biol Chem. 2001;276:22404–22409. doi: 10.1074/jbc.M100409200. [DOI] [PubMed] [Google Scholar]
- 41.Tassan J, Schultz S, Bartek J, Nigg E. Cell cycle analysis of the activity, subcellular localization and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kan Q, Jinno S, Kobayashi K, Yamamoto H, Okayama H. Cdc6 determines utilization of p21WAF1/CIP1-dependent damage checkpoint in S phase cells. J Biol Chem. 2008;283:17864–17872. doi: 10.1074/jbc.M802055200. [DOI] [PubMed] [Google Scholar]
- 43.Lents NH, Keenan SM, Bellone C, Baldassare JJ. Stimulation of the Raf/MEK/ERK cascade is necessary and sufficient for activation and Thr-160 phosphorylation of a nuclear-targeted CDK2. J Biol Chem. 2002;277:47469–47475. doi: 10.1074/jbc.M207425200. [DOI] [PubMed] [Google Scholar]
- 44.Landers JE, Cassel SL, George DL. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- 45.Craig C, Kim M, Ohri E, Wersto R, Katayose D, Li Z, et al. Effects of adenovirus-mediated p16INK4A expression on cell cycle arrest are determined by endogenous p16 and Rb status in human cancer cells. Oncogene. 1998;16:265–272. doi: 10.1038/sj.onc.1201493. [DOI] [PubMed] [Google Scholar]
- 46.Kimura H, Nozaki N, Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J. 1994;13:4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte D, Burkhart R, Musahl C, Hu B, Schlatterer C, Hameister H, et al. Expression, phosphorylation and nuclear localization of the human P1 protein, a homologue of the yeast Mcm 3 replication protein. J Cell Sci. 1995;108:1381–1389. doi: 10.1242/jcs.108.4.1381. [DOI] [PubMed] [Google Scholar]
- 48.Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita M, Kiyono T, Hayashi Y, Ishibashi M. hCDC47, a human member of the MCM family. Dissociation of the nucleus-bound form during S phase. J Biol Chem. 1996;271:4349–4354. doi: 10.1074/jbc.271.8.4349. [DOI] [PubMed] [Google Scholar]
- 50.Khoudoli GA, Gillespie PJ, Stewart G, Andersen JS, Swedlow JR, Blow JJ. Temporal profiling of the chromatin proteome reveals system-wide responses to replication inhibition. Curr Biol. 2008;18:838–843. doi: 10.1016/j.cub.2008.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 52.Yanagi KI, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem. 2002;277:40871–40880. doi: 10.1074/jbc.M206202200. [DOI] [PubMed] [Google Scholar]