Abstract
The simple nematode, Caenorhabditis elegans, possesses the most extensive known gene family of nicotinic acetylcholine receptor (nAChR)-like subunits. Whilst all show greatest similarity with nAChR subunits of both invertebrates and vertebrates, phylogenetic analysis suggests that just over half of these (32) may represent other members of the cys-loop ligand-gated ion channel superfamily. We have introduced a novel nomenclature system for these ‘Orphan’ subunits, designating them as lgc genes (ligand-gated ion channels of the cys-loop superfamily), which can also be applied in future to unnamed and uncharacterised members of the cys-loop ligand-gated ion channel superfamily. We present here the resulting updated version of the C. elegans nAChR gene family and related ligand-gated ion channel (lgc) genes.
Keywords: Caenorhabditis elegans, gene family, ion channel, nematode, nicotinic acetylcholine receptor
Nicotinic acetylcholine receptors (nAChRs) mediate fast chemical neurotransmission in both vertebrates and invertebrates. They are members of the ‘cys-loop’ ligand-gated ion channel (LGIC) superfamily that also includes γ-aminobutyric acid (GABA), serotonin (5-HT), glycine, glutamate and histamine receptors (Sine and Engel, 2006). Cys-loop LGICs are made up of five homologous subunits arranged around a central ion channel. The cys-loop is situated in the N-terminal extracellular ligand-binding region of each subunit and consists of two disulphide-bond forming cysteines separated by 13 amino acid residues, several of which are highly conserved to form a signature sequence (Gene Ontology ID GO:0005230). For examples of cys-loop sequences, see Fig. 1. The cys-loop plays roles in receptor assembly (Green and Wanamaker, 1997) and gating of the ion channel (Grutter et al., 2005).
Fig. 1.

Alignment of cys-loop sequences from various cys-loop LGIC subunits including: a human nAChR subunit (α7); several C. elegans nAChR subunits (ACR-16, UNC-38, ACR-8, UNC-29 and DEG-3), a C. elegans nAChR-like subunit (CUP-4), C. elegans GABA receptor (UNC-49) and glutamate receptor (GLC-1) subunits; a rat serotonin receptor subunit (5HT3a); a Drosophila melanogaster histamine receptor subunit (HisCl).
Multiple genes generate nAChR diversity. For instance, mammals and birds possess 17 genes encoding for nAChR subunits (Millar, 2003). In contrast, the genome of the simple nematode worm, Caenorhabditis elegans, contains the largest nAChR gene family described so far (Jones and Sattelle, 2004; Brown et al., 2006). Thus, C. elegans possesses 29 nAChR subunits which are divided into five ‘core’ groups based on sequence homology (Fig. 2). Generally, the groups are named after the first subunit to be characterised and the majority of the C. elegans nAChR subunit genes are denoted acr for acetylcholine receptor (Mongan et al., 1998). In addition, C. elegans possesses 32 subunits that show closest homology to vertebrate and invertebrate nAChR subunits but do not fall within the five ‘core’ groups and at present are designated ‘orphan’ subunits (Fig. 2). Unlike members of the core groups, none of these orphans are α subunits which possess two vicinal cysteine residues important for acetylcholine binding (Kao and Karlin, 1986). With the exception of CUP-4 and Y58G8A.1, which have been shown to be involved in endocytosis (Patton et al., 2005), the orphan subunits have not so far been functionally characterised and it is yet to be determined which neurotransmitters/ligands act on them.
Fig. 2.
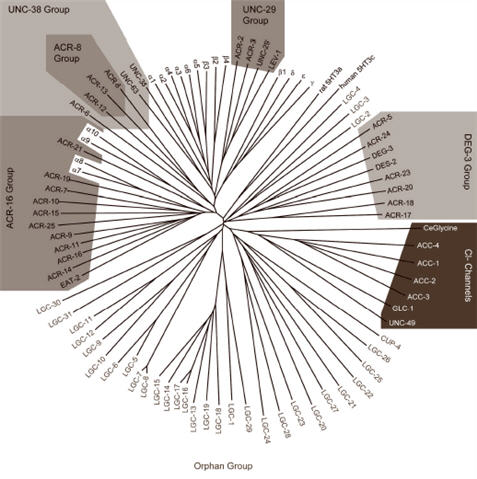
Tree showing the nAChR receptor gene family of C. elegans. Based on protein sequence homology, the worm nAChR subunits are divided into five ‘core’ groups whereas subunits showing substantial sequence homology with known nAChR subunits that do not fall within the core groups are designated ‘Orphan’ subunits. Acetylcholine-gated chloride channels, ACC-1 to ACC-4 (Putrenko et al., 2005), show greatest similarity with other anion channels such as GABA (UNC-49), glutamate (GLC-1) and glycine (T27A1.4) receptors The avian/mammalian nAChR gene family as well as mammalian serotonin-gated ion channels (5-HT3 receptors) are also included for comparison.
Here, we present an update on the genes present in the C. elegans nAChR gene family as well as introducing nomenclature for the orphan subunits. Two more subunit genes, acr-24 and acr-25, have been designated (Table 1) which are both α subunits and are members of the DEG-3 and ACR-16 Groups respectively (Fig. 2). Since the orphan subunits may represent members of the cys-loop LGIC gene superfamily other than nAChRs, they have been designated lgc, for ligand-gated ion channels of the cys-loop LGIC superfamily, following the guidelines set for genetic nomenclature for C. elegans (http://biosci.umn.edu/CGC/Nomenclature/nomenguid.htm). One subunit gene, originally denoted acr-22 (Mongan et al., 2002), has since been shown to be an orphan subunit (Jones and Sattelle, 2004) and thus has been renamed here lgc-11. As a result, there is a gap in the list of acr gene numbers. Since the last description of the C. elegans nAChR gene family (Brown et al., 2006), four new orphan subunit genes (lgc-24, lgc-26, lgc-27 and lgc-28) have been identified.
Table 1.
C. elegans nAChR-like subunits that have been given names by the Caenorhabditis Genetics Center (CGC) in this report
| CGC name | Sequence name | Other names | nAChR Group |
|---|---|---|---|
| ACR-24 | Y73F8A.30 | – | DEG-3 |
| ACR-25 | Y73B6BL.42 | – | ACR-16 |
| LGC-1 | T05B4.1 | – | Orphan |
| LGC-2 | Y44A6E.1 | – | Orphan |
| LGC-3 | F11C7.1 | – | Orphan |
| LGC-4 | F18G5.4 | – | Orphan |
| LGC-5 | F17E9.7 | – | Orphan |
| LGC-6 | F17E9.8 | – | Orphan |
| LGC-7 | Y57G11C.2 | – | Orphan |
| LGC-8 | Y57G11C.49 | – | Orphan |
| LGC-9 | C04C3.2 | – | Orphan |
| LGC-10 | Y73B6BL.26 | – | Orphan |
| LGC-11 | F48E3.7 | ACR-22 | Orphan |
| LGC-12 | R13A5.4 | – | Orphan |
| LGC-13 | T01H10.1 | – | Orphan |
| LGC-14 | T01H10.2 | – | Orphan |
| LGC-15 | T01H10.3 | – | Orphan |
| LGC-16 | T01H10.5 | – | Orphan |
| LGC-17 | T01H10.6 | – | Orphan |
| LGC-18 | T01H10.7 | – | Orphan |
| LGC-19 | Y105C5B.16 | – | Orphan |
| LGC-20 | B0491.4 | – | Orphan |
| LGC-21 | C33G3.3 | – | Orphan |
| LGC-22 | F15E6.2 | – | Orphan |
| LGC-23 | C15A7.1 | – | Orphan |
| LGC-24 | C15A7.4 | – | Orphan |
| LGC-25 | R03E1.3 | – | Orphan |
| LGC-26 | Y58G8A.1 | – | Orphan |
| LGC-27 | Y74E4A.1 | – | Orphan |
| LGC-28 | D2062.12 | – | Orphan |
| LGC-29 | Y45G5AL.2 | – | Orphan |
| LGC-30 | F58H7.3 | – | Orphan |
| LGC-31 | F21A3.7 | – | Orphan |
We have set up the lgc gene nomenclature in the anticipation that more cys-loop LGIC subunits with unclear identities will be revealed in the C. elegans genome. According to WormBase (http://www.wormbase.org/) release WS172, there are 206 genes that have the ‘extracellular ligand-gated ion channel activity’ motif (Gene Ontology ID GO:0005230) which is characteristic of cys-loop LGICs. Since several of these are splice variants, there are in fact less than 200 cys-loop LGIC subunits. Nevertheless, there are still many putative cys-loop LGIC subunits, the majority of which show greatest similarity to chloride-gated channels (Dent, 2006), that have yet to be named.
References
- Brown LA, Jones AK, Buckingham SD, Mee CJ, Sattelle DB. Contributions from Caenorhabditis elegans functional genetics to antiparasitic drug target identification and validation: nicotinic acetylcholine receptors, a case study. Int J Parasitol. 2006;36:617–624. doi: 10.1016/j.ijpara.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Dent JA. Evidence for a diverse cys-loop ligand-gated ion channel superfamily in early bilateria. J. Mol. Evol. 2006;62:523–535. doi: 10.1007/s00239-005-0018-2. [DOI] [PubMed] [Google Scholar]
- Green WN, Wanamaker CP. The role of the cystine loop in acetylcholine receptor assembly. J. Biol. Chem. 1997;272:20945–20953. doi: 10.1074/jbc.272.33.20945. [DOI] [PubMed] [Google Scholar]
- Grutter T, de Carvalho LP, Dufresne V, Taly A, Edelstein SJ, Changeux JP. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc Natl Acad Sci U S A. 2005;102:18207–18212. doi: 10.1073/pnas.0509024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Sattelle DB. Functional genomics of the nicotinic acetylcholine receptor gene family of the nematode, Caenorhabditis elegans. Bioessays. 2004;26:39–49. doi: 10.1002/bies.10377. [DOI] [PubMed] [Google Scholar]
- Kao PN, Karlin A. Acetylcholine receptor binding site contains a disulfide cross-link between adjacent half-cystinyl residues. J. Biol. Chem. 1986;261:8085–8088. [PubMed] [Google Scholar]
- Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Trans. 2003;31:869–874. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- Mongan NP, Baylis HA, Adcock C, Smith GR, Sansom MS, Sattelle DB. An extensive and diverse gene family of nicotinic acetylcholine receptor alpha subunits in Caenorhabditis elegans. Receptors Channels. 1998;6:213–228. [PubMed] [Google Scholar]
- Mongan NP, Jones AK, Smith GR, Sansom MS, Sattelle DB. Novel alpha7-like nicotinic acetylcholine receptor subunits in the nematode Caenorhabditis elegans. Protein Sci. 2002;11:1162–1171. doi: 10.1110/ps.3040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton A, Knuth S, Schaheen B, Dang H, Greenwald I, Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Putrenko I, Zakikhani M, Dent JA. A family of acetylcholine-gated chloride channel subunits in Caenorhabditis elegans. J Biol Chem. 2005;280:6392–6398. doi: 10.1074/jbc.M412644200. [DOI] [PubMed] [Google Scholar]
- Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]


