Abstract
Background
Disparities in cancer defined by race, age, or gender are well established. However, demographic metrics are surrogates for the complex contributions of genotypes, exposures, health care, socioeconomic and sociocultural environment, and many other factors. Macro-environmental factors represent novel surrogates for exposures, lifestyle and other factors that are difficult to measure but may influence cancer outcomes.
Methods
We applied a “multilevel molecular epidemiology” approach using a prospective cohort of 444 White prostate cancer cases who underwent prostatectomy and were followed until biochemical failure (BF) or censoring without BF. We applied Cox regression models to test for joint effects of 86 genome-wide association study-identified genotypes and macro-environmental contextual effects after geocoding all cases to their residential census tracts. All analyses were adjusted for age at diagnosis and tumor aggressiveness.
Results
Residents living in macroenvironments with a high proportion of older single heads of household, high rates of vacant housing, or high unemployment had shorter time until BF post-surgery after adjustment for patient age and tumor aggressiveness. After correction for multiple testing, genotypes alone did not predict time to BF, but interactions predicting time to BF were observed for MSMB (rs10993994) and percent of older single head of households (p=0.0004), and for HNF1B/TCF2 (rs4430796) and macroenvironment per capita income (p=0.0002).
Conclusions
Context-specific macro-environmental effects of genotype may improve the ability to identify groups that may experience poor prostate cancer outcomes.
Impact
Risk estimation and clinical translation of genotype information may require an understanding of both individual-level and macroenvironmental context.
Keywords: prostate cancer, gene-environment interaction, macroenvironments, multilevel analysis
Introduction
Interindividual variability in common disease etiology and outcomes likely represents the complex interactions of multiple factors including genetic susceptibility; individual behavior, lifestyle, and exposures; macroenvironment-level (e.g., area, community) contextual factors; and health care access including screening and treatment (1). The elucidation of interactions among these multiple factors may require novel, transdisciplinary approaches. We hypothesize that methods that account for the joint effects of both individual-level and macroenvironment-level effects using a multilevel strategy (2-4) may capture etiologic agents that are not seen in studies that consider one or a few factors at a time. We propose a “multilevel molecular epidemiology” (MME) approach that is grounded in multilevel analytical frameworks that have used both individual-level risk factors as well as macroenvironment-level contextual factors (5). Macroenvironment-level factors may serve as a surrogate for a wide range of exposures, lifestyle and other factors that are difficult to measure. We extend this concept to hypothesize that prediction of risk and outcome by inherited genotype information (and other biomarker data) may depend on both individual-level and macroenvironment-level context. Disease risk and outcomes are influenced by both “differences” (i.e., naturally occurring or achieved factors) as well as “inequities” (i.e., factors ascribed to groups as a function of their social position (6)). Using the MME framework, we can hypothesize that differences (e.g., genetics, race, and possibly other innate biological factors) may be acting in the context of inequities (e.g., discrimination, segregation, or access) and lead to observed disease risks and outcomes. Related models that define “gaps” occurring between discrete groups in the population (e.g., defined by genotype, ethnicity, gender, or other discrete characteristics), as well as effects that are defined by “gradients” across continuously distributed variables may influence risk and outcomes (7). Thus, we consider models that consider the effect of genotypes as well as macroenvironment-level factors.
To illustrate this approach, we have undertaken a multilevel, transdisciplinary analysis of susceptibility genotypes, individual-level factors, macroenvironment-level factors and prostate cancer outcomes. Prostate cancer is the second leading cause of cancer deaths in Americans (8), and is associated with extreme variability in outcomes, including extreme incidence and mortality differences by race, age, geography, and other factors (9, 10). In particular, prostate cancer outcomes represent a complex and multifactorial phenomenon likely to be caused by complex interaction of genetic susceptibility, individual risk factors, and macroenvironment-level factors (11). Genome-wide association studies (GWAS) have identified a number of loci that are likely to affect prostate cancer etiology or outcomes. The goal of this research is to better explain the complex, multifactorial nature of prostate cancer outcomes by considering the context dependency of genetic susceptibility as it relates to individual- and macroenvironment-level effects.
Methods
Study Participants and Individual-Level Data: The SCORE Study
Our primary goal in this analysis was to evaluate the role of genotype and macroenvironment-level context in predicting adverse clinical outcomes after a prostate cancer diagnosis. To focus these analyses and control for unmeasured confounding, we limited our sample to 444 White incident prostate cancer cases (i.e., diagnosed within 12 months of study ascertainment) who were identified through Urologic Oncology Clinics at the Hospital of the University of Pennsylvania between 1995 and 2008 who participated in the Study of Clinical Outcomes, Risk and Ethnicity (SCORE). Case status was confirmed by medical records review using a standardized abstraction form. Patients who were non-incident cases (i.e., those diagnosed more than twelve months prior to the date of study ascertainment), or had a prior diagnosis of cancer except non-melanoma skin cancer, were excluded. To further limit heterogeneity due to access and treatment, we limited the study sample to men who underwent prostatectomy as the primary treatment of their disease. No men in this sample underwent any other treatments besides prostatectomy before the occurrence of a failure event or censoring.
Individual risk factors, medical history, and prostate cancer diagnostic and treatment information were obtained by using a standardized questionnaire and review of medical records. Information collected included previous cancer diagnoses, demographic information and primary tumor characteristics including Gleason score (grade) and stage. We also created a summary tumor aggressiveness scale that reflects the highly correlated variable, tumor stage and grade to use as a single adjustment in the multivariate survival models. This composite variable was defined as non-aggressive if cases had both non-invasive cancer (stage 1-2) and low Gleason score (<7) tumors and as aggressive otherwise. All study participants provided written informed consent for participation in this research under a protocol approved by the Committee for Studies Involving Human Subjects at the University of Pennsylvania.
Biosample Collection and Genotype Analysis
Genomic DNA for the present study was self-collected by each study participant using sterile cheek swabs (Cyto-Pak Cytosoft Brush, Medical Packaging Corporation, Camarillo, CA), and processed using either a protocol modified from Richards et al. (12) as described previously (13), or using a modified protocol on the Qiagen 9604B robot with the QIAamp 96 DNA Buccal Swab Biorobot Kit (Valencia, CA). 109 variants at 30 loci identified from linkage and genome-wide association studies (GWAS) that have been independently validated as prostate cancer susceptibility genes (14) were selected for analysis. These include one of the confirmed SNPs at each of the following GWAS loci without known genes: 3p12 (15), multiple regions at 8q24 (16-18), 11q13 (15, 19), 17q24 (18); as well as those with putative candidate genes including CTBP2 (10q26 (19)), HNF1B/TCF2 (17q12 (20, 21)), JAZF1 (7p15.2 (19)), LMTK2 (7q21 (15)), MSMB (10q11 (15, 19)), and NUDT (Xp11 (15, 22)). We also considered two loci (MSR1 and RNASEL) that were identified in linkage studies (23, 24). In order to ensure adequate power for our analyses, we only considered SNPs with a minor allele frequency of 10% or greater in the failure/censoring groups. We also excluded SNPs with genotyping failure rates greater than 5% or with SNP sample size of <100 cases. No SNPs deviated significantly from Hardy Weinberg proportions. After applying these criteria, 23 SNPs were removed from consideration, leaving 86 SNPs that were included in analysis (Supplementary Table 1).
Prior to genotyping, the whole genome of the samples was amplified using the GenomePlex Complete Whole Genome Amplification kit (Sigma, St. Louis, MO). Genotyping was performed either by TaqMan™ assay using the 7900HT Fast Real-Time PCR Machine or using an Illumina GoldenGate Platform. For TaqMan™, all mixes of primers and probes were pre-designed by and purchased from Applied Biosystems. We included one negative control and three positive controls for each assay and each 384-well plate and duplicated at least 8% of samples for each assay. We designed an Illumina GoldenGate assay considering only SNPs with SNP scores of >0.59 and including at least 10% duplicated samples. Quality control for both assays was as following: excluding all individuals that failed for at least 15% of the SNPs attempted, followed by excluding any SNP with a call rate <95% and any SNPs that had >2% discordance between genotypes in duplicate samples.
Macroenvironment-level Data
In the U.S., census tracts are commonly used to define macroenvironments, despite limitations in accurately delineate macroenvironment boundaries (25), to allow replication across studies (26). Therefore, we used the 2000 census tract boundaries to group individuals by residence (27). The 2000 census was used because it represents approximately the median of our SCORE accrual period (1995-2008). Census tracts are U.S. Census Bureau defined, standardized, and relatively permanent geographical units. Census tracts are constructed specifically to include on average 4,000 people that are intended to reflect fairly homogeneous population characteristics, economic position, and living conditions. Federal, state, and local governments routinely use census tracts as administrative units (28).
We geocoded the residential addresses of our patients using ArcGIS or American Fact Finder (http://factfinder.census.gov/home/saff/main.html?_lang=en), and methods outlined by The Public Health Disparities Geocoding Project (http://www.hsph.harvard.edu/thegeocodingproject/webpage/monograph/geocoding.htm).
To obtain census tract-level data about demographic, education, socioeconomic, and relationships/social isolation measures, we linked the geocoded addresses from our dataset with variables from the Geographical Comparison Tables (GCTs) of the United States Census Bureau's American Factfinder (http://factfinder.census.gov/servlet/GCTSelectedDatasetPageServlet?_lang=en&_ts=247057910048). The GCTs were accessed for 24 counties in and around Philadelphia: Kent, New Castle, and Sussex Counties in Delaware; Atlantic, Burlington, Camden, Cape May, Gloucester, Hunterdon, Mercer, Monmouth, Ocean, Salem, and Warren Counties in New Jersey; and Berks, Bucks, Chester, Delaware, Lehigh, Luzerne, Monroe, Montgomery, Northampton, and Philadelphia Counties in Pennsylvania. The 444 study participants resided in 342 census tracts. A subset of variables in the GCT were selected for analysis to represent various classes of macroenvironment factors including: aging and social isolation (% of residents age 65 years or older, % of households with a single resident age 65 years or older), education (% college graduates among those 25-34 years old), macroenvironment housing quality (% of houses vacant and not for sale, rent, or vacation), and socioeconomic status (% unemployed individuals age 16 years or older and per capita income in $1000).
Statistical Methods
In order to test the joint effects of genotype and macroenvironment context, we modeled time to biochemical failure (BF) after treatment of prostate cancer by prostatectomy with a Cox proportional hazards model. Time to BF was chosen as the primary outcome of interest because it afforded sufficient power to consider these interactions, and was defined as having experienced a post-prostatectomy PSA value of more than 0.2 mg/dl. We also allowed outcomes of interest to be diagnosis of metastases. However, no men in our sample were found to have metastasis after initial diagnosis and before BF. Censored observations had an end of follow up without BF. Failure time was defined as time from surgery until BF or censoring.
Covariates included in all models as potential confounder variables were age at diagnosis, and tumor aggressiveness. Main effects of interest were susceptibility genotypes and macroenvironment-level context variables, as described above.
For tests of genotype effects in the context of macroenvironment-level factors, we evaluated the first order interaction of these factors by using a Cox proportional hazards model with an interaction between census tract characteristics as a continuous variable and SNP per-allele effects. We also considered the effect of clustered sampling of individuals within census tracts on the standard error and 95% confidence interval (CI) estimates associated with model parameters by using the robust variance estimation approach of Lin and Wei (29). Two-sided p-values were corrected for multiple hypothesis tests using the False Discovery Rate (FDR) method of Benjamini and Hochberg (30). All analyses were performed in STATA (version 10.1, STATA Corporation, College Station, TX).
Results
Of the 444 White prostatectomy cases followed prospectively from the time of surgery, 397 (89%) did not experience BF during a mean (median) follow up period of 26.8 (21) months, while 47 (11%) men experienced BF during a mean (median) follow up period of 22.5 (19.1) months. Six men in the cohort died;death was not used as a censoring event because their psa status was unknown at the time of death. As expected, tumor stage (1+2 vs. 3+4; HR=3.12, 95%CI: 1.75-5.56), Gleason grade (<7 vs. 7+; HR=6.22, 95%CI: 2.79-13.89), and a combined metric of tumor aggressiveness (OR=8.11, 95%CI: 2.91-22.60) were significant predictors of time to BF in univariate analyses. Other individual level factors including age at diagnosis, marital status, and education were not significant predictors of time to BF (results not shown). All subsequent analyses were adjusted for age at diagnosis and tumor aggressiveness to evaluate the potential additional impact of genotype and/or macroenvironment factors on time to BF after these traditional outcome predictors were considered.
The effects of macroenvironment-level factors on time to BF are presented in Table 1. Three macroenvironment-level factors were significantly associated with BF: a demographic measure (% of residents age 65 or older; HR=1.02, 95%CI 1.00-1.04, p=0.049); a measure of macroenvironment degradation (% vacant housing; HR=1.28, 95%CI: 1.09-1.51; p=0.002); a measure of socioeconomic status (% unemployed individuals age 16 years or older; HR=1.08, 95%CI: 1.00-1.17, p=0.044). Because all variables selected from the GCTs were percentiles, except per capita income, the hazard ratio estimates represent a change in relative risk for each unit of one percent increase in the macroenvironment variable. For per capita income, the HR is change for each unit of $1000 increase in per capita income.
Table 1. Macroenvironment-level variables and Hazard Ratio (HR) associations with biochemical (PSA) failure after prostatectomy adjusted for age at diagnosis and tumor aggressiveness among incident White prostate cancer cases treated with prostatectomy.
| Macroenvironment Variable | HR | 95% CI |
|---|---|---|
| %Residents age 65 or older | 1.02 | 1.00-1.04 |
| % Older Single Head of Household | 1.02 | 0.98-1.07 |
| %Vacant Housing | 1.28 | 1.09-1.51 |
| %College Graduates Ages 25-34 | 0.99 | 0.98-1.01 |
| %Unemployed age 16 or older | 1.08 | 1.00-1.17 |
| Per Capita Income (per $1,000) | 1.01 | 0.98-1.03 |
While the genotypes studied here had previously been identified as being strongly associated with prostate cancer etiology in case-control studies(14), these loci have largely not been studied for a role in prostate cancer outcomes. Therefore, we evaluated the per-allele effect of the 86 variants on time to BF after adjustment for age at diagnosis, and tumor aggressiveness (Figure 1). Four loci were associated with time to BF at the p<0.05 level of significance: Chr8q24 (rs4871799, p=0.018), KLK3 (rs1506684, p=0.033), OATP1B1 (M233I; rs7311358, p=0.044), and KLK3 (rs2569739, p=0.049). However, none of these remained significant after correcting the significance level for multiple testing by FDR.
Figure 1. Main Effect of 86 GWAS SNPs on time to biochemical (PSA) failure after prostatectomy adjusted for age at diagnosis and tumor aggressiveness.
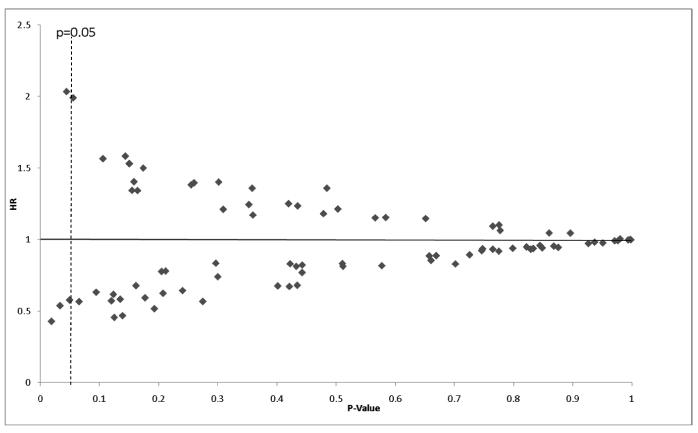
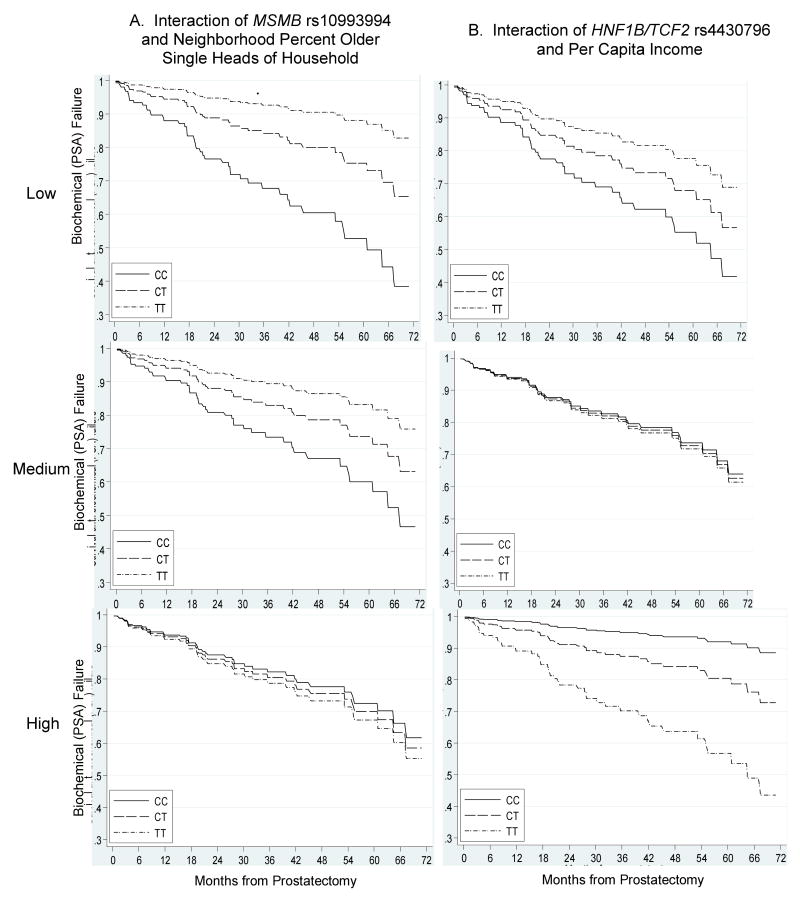
Finally, we evaluated whether genotype interacted significantly with macroenvironment-level effects to influence time to BF. Figure 2 presents the results of per-allele by census tract variable interactions. Numerous interactions reached a significance level of p<0.05, but after correction for multiple hypothesis testing, only two highly significant interactions remained. First, we identified an interaction between MSMB rs10993994 and a measure of macroenvironment social isolation (% census tract older single heads of household). For each percent increase in the macroenvironment percent of heads of household aged 65 or older living alone, there was a 10% increase in time to BF among men who carried the TT genotype at this locus, while there was no increase in time to BF among men who carried any other genotype (HR=1.10, 95%CI: 1.06-1.14, interaction p=0.0004; Figure 3).
Figure 2. Per-Allele Genotype by macroenvironment variable interaction P-values for time to biochemical (PSA) failure after prostatectomy. Diamonds indicate interaction p-values adjusted for age, and tumor aggressiveness. Dotted line represents interaction p-value of 0.05; p-values indicate statistically significant effects after correction for multiple testing.
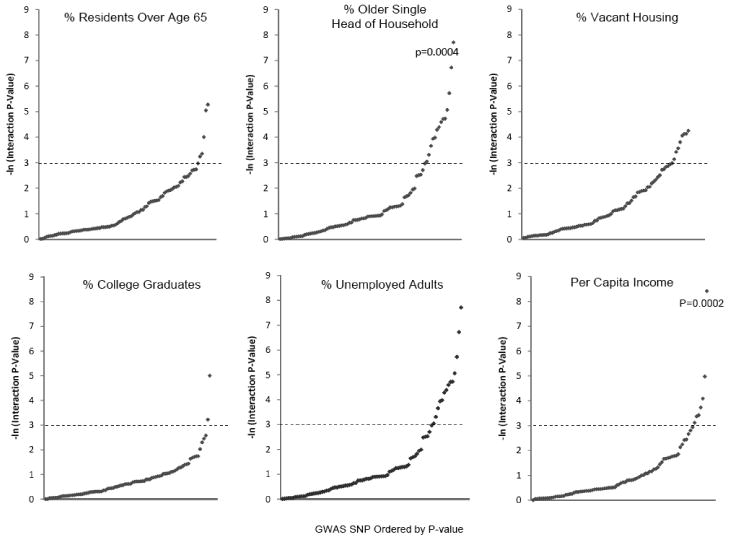
Figure 3. Time to Biochemical (PSA) Failure after Prostatectomy by Genotype and Macroenvironment Variable Exhibiting Statistically Significant Interactions after False-Discovery Rate Correction.
Second, we identified a statistically significant relationship between HNF1B/TCF2 rs4430796 and a measure of macroenvironment socioeconomic status (census tract per capita income). For each $1000 increase in macroenvironment per capita income, there was a 3% increase in time to BF among men who carried the TT genotype at this locus (HR=1.03, 95%CI: 1.02-1.04), while there was a 7% decrease in time to BF for every $1000 increase in per capita income among men who carried the CC genotype (HR=0.93, 95%CI: 0.89-0.98; interaction p=0.0002; Figure 3).
Discussion
We have identified statistically significant joint effects between genotypes known to be involved in prostate cancer etiology and macroenvironment-level effects on biochemical failure in White men. These results cannot be interpreted as having direct biological implications of macroenvironment effects. Just as race or gender are used as surrogates for differences in socio-economic status, health care access, exposures, and other factors, we interpret our statistically significant interactions as reflecting the surrogate effects causal factors in the environment that are measured by census tract-level variables. Thus, the inferences made here are not necessarily biological in nature, but may provide improved understanding of the contextual relationship of genotype effects in a given macroenvironment setting. Instead, the goal of these analyses is to identify whether information about the macroenvironment in which an individual lives provides information that is predictive of prostate cancer outcomes. Because we were able to identify significant macroenvironment effects as well as genotype by macroenvironment interactions after we considered individual factors, our data support the hypothesis that macroenvironment variables contain information that is not captured at the individual level. By providing additional surrogate metrics of factors that may be correlated with disease risk, outcomes, and disparities, the present results may provide information that moves research in these areas away from more misclassified variables (e.g., race), and toward variables that may be both less misclassified as well as point toward specific areas in which targeted interventions may be developed to reduce disparities.
Although some studies have reported associations with disease aggressiveness (19, 31), most of the loci or combinations of loci studied to date have not been associated with disease aggressiveness or outcomes (31-34). Therefore, we attempted to take a novel approach that identified contextual factors that may be associated with prostate cancer outcomes and considered information beyond genotype alone. We identified two statistically significant HNF1B/TCF2 or MSMB by macroenvironment interactions. HNF1B is the hepatocyte nuclear factor 1 homeobox B, also known as transcription factor 2 (TCF2). HNF1B/TCF2 is a member of the transcription factor superfamily that interacts with HNF1A (TCF1), HNF4A, CDH16, ONECUT1, and NR2F2. The HNF1B/TCF2 protein is involved in metabolism of glucose, cholesterol, uric acid, and is expressed not just in hepatocytes, but also in prostate and other tissues. Genotypes at HNF1B/TCF2 have been identified in prostate cancer risk (20, 22), diabetes risk (20, 35), male infertility (36), and other traits. Therefore, there is evidence that this protein is involved in a wide variety of metabolic processes that reflect potential hormonal, cardiovascular and diabetes risk factors. In our data, we found that time to BF increased with census tract per capita income in prostate cancer cases with TT genotype at rs4430796, while time to BF decreased with census tract per capita income in prostate cancer cases with CC genotype at rs4430796. Since these factors have been associated with adverse cardiovascular, diabetes, and obesity phenotypes, we also evaluated whether additional adjustment for body mass index (BMI) might in part explain our observed associations. After adjusting these analyses for obesity (i.e. BMI<30 vs. BMI≥30), there was no substantial difference in the HR effects or interaction inferences (results not shown). Therefore, if there is a relationship between HNF1B, obesity, and time to BF, it is not explained by confounding in our data.
MSMB (microseminoprotein-β) encodes PSP94 (prostate secretory protein of 94 amino acids), which is found in semen and has been proposed as a prostate cancer screening and prognosis biomarker (37, 38). A SNP in MSMB, rs10993994, has been reported in multiple GWAS to be associated with prostate cancer etiology (15, 19, 39, 40). Rs10993994 is located 57 bp upstream of the MSMB transcription start site and has been suggested to regulate PSP94 expression. We reported that men with TT genotype at rs10993994 were at increased time to BF risk if they lived in census tracts with a higher percentage of older single heads of household. This suggests that the effect of MSMB may be correlated with factors related to age or social isolation. This effect was present even after adjusting for age at diagnosis and tumor aggressiveness.
The multilevel molecular epidemiology approach is novel, yet its evaluation here is limited. First, our data included only White men who were seen at a tertiary referral hospital and underwent prostatectomy as their primary treatment. The advantage of this sample selection is that it involves a narrowly defined patient and treatment population within a single hospital which avoids some extraneous variability that may cloud the results. However, these patients are not representative of the general population, nor of patients who are not treated by prostatectomy. Therefore, these results may not reflect the same effects as might be seen in men of other ethnicities, those who receive treatment other than prostatectomy, or who are diagnosed and treated in community hospital or clinic settings. We would expect that the individual risk factor and macroenvironment-level variable distributions would be different than the distributions observed in the present study population. Similarly, genotype frequencies in this study population of White men are also likely to differ from those in non-White populations. Therefore, while we find provocative associations of time to BF with individual-level and macroenvironment-level factors and with genotype interacting with macroenvironment-level factors, these may be quite different in other populations. In particular, future studies should include African American populations that suffer from the greatest prostate cancer disparities.
Second, the present study does not fully explore the relationship of susceptibility genotypes, individual-level risk factors, and macroenvironment-level factors. Additional research may be needed to understand the relationship of these factors and the optimal approach toward their modeling. Other variables not studied here should be considered that may influence prostate cancer outcomes, including individual insurance or other metrics of health care access as well as prostate cancer screening history. Furthermore, we have used a dataset that includes a very narrowly defined sample set (i.e., White men undergoing prostatectomy within a single hospital) to limit the heterogeneity that might mask the effects seen here. However, these sample restrictions also limit the inferences because of the relatively narrow spectrum of the population being studied. Thus, broader sample definitions should be considered in the future to more fully address questions of prostate cancer disparities by race or other factors. Conceptually, the multilevel molecular epidemiology approach discussed here is conceptually tied to that of Mendelian Randomization (41), which uses observational study designs to evaluate genetic effects indirectly via exposures of interest. As such, some of the analytical approaches and pitfalls of the Mendelian Randomization framework may be applied in the future to the type of studies proposed here. In addition, we have not fully explored whether macroenvironment-level factors are a better measure of disadvantage than individual-level variables. Since the various macroenvironment-level variables are correlated with one another, and presumably with individual-level factors (many of which remain unmeasured here), it is likely that the effects of these variables do not represent independent associations. Therefore, the associations reported here may reflect similar or even identical phenomena measured through different analytical variables. Additional exploration of how correlated macroenvironment variables measure related phenomena that influence prostate cancer outcomes is required.
Third, our study uses a relatively small sample size of 444 White men followed prospectively from the time of prostate cancer diagnosis. The sample size studied here included 444 men residing in 342 census tracts. While the factors studied here reflect macroenvironment-level effects, the small number of men in a single census tract limits the “multilevel” nature of the analysis. Despite the limited sample size, our study was adequately powered to detect the effects reported here. The MME approach used here involved continuously-distributed macroenvironment effects, which provides generally greater power than discrete variables. We also limit our genotype analyses to those variants with 10% or greater allele frequency to ensure reasonable statistical power, as specified a priori in our study design, and to those SNPs with a sample size of 100 or more cases. Also, we have limited our sample set to include only a narrow range of men (i.e., White men from a single hospital who have undergone prostatectomy) to minimize the potential for unmeasured confounding that may influence our results. We have taken this approach to demonstrate the MME approach. However, it is also likely that the effects of GWAS genotypes, individual-level, and macroenvironment-level effects on time to BF are small in magnitude. For example, it is possible that the sample size used here was too small to detect effects of genotypes on time to BF if they are of a similar magnitude as seen in etiology studies. In addition, while we have identified statistically significant interactions between genotypes and macroenvironment-level context, larger studies will be required to confirm these results and to extend them to other populations. Furthermore, studies with greater statistical power and longer follow up will be required to assess macroenvironment and/or genotype effects on other outcomes, including disease recurrence, or death.
Finally, macroenvironment-level variables are generally derived from administrative databases found through the US census or other community surveys, and linked back to the individual by geocoding the person to their place of residence (42). However, because macroenvironment-level effects are broad and clearly represent surrogates for both differences and inequities, the approach presented here is valid for prediction of risk or outcomes, but not necessarily as a means of identifying underlying etiology. We also considered a limited range of metrics, and only continuously distributed macroenvironment variables. Future studies should consider the optimal coding of these variables.
Using a multilevel molecular epidemiology approach, we have identified associations of candidate prostate cancer loci that are dependent on the context in which these genotype effects may be acting to predict prostate cancer outcomes. This approach could provide useful information in studies of cancer outcomes and disparities. Use of macroenvironment-level variables rather than (or in addition to) other surrogates such as age, gender, or race may provide better indices of disadvantage. Research related to cancer disparities that uses this approach may benefit from having measures other than race to compare groups that may differ in ways relevant to health disparities. These groups may represent target populations in which interventions can be designed and implemented around potentially modifiable factors. These macroenvironment-level factors may also identify novel genotype-environment interactions. However, because macroenvironment-level effects are broad and clearly represent surrogates for both differences and inequities, the approach presented here may be valid for prediction of risk or outcomes, but not necessarily as a means of identifying underlying etiology. Thus, the multilevel molecular epidemiology approach defined here may provide new avenues for research in cancer health disparities.
Supplementary Material
Acknowledgments
This study was supported by grants from the Public Health Service R29-ES08031, R01-CA85074, and P50-CA105641 to TRR and K07-CA106730 to CZJ. The authors acknowledge the support of Drs. S. Bruce Malkowicz and Alan J. Wein in the collection of data used in this study.
This study was supported by grants from the Public Health Service (R29-ES08031, R01-CA85074, and P50-CA105641 to TRR).
References
- 1.NIH Centers for Population Health and Health Disparities. From Cells to Society: Overcoming Health Disparities. Bethesda, MD: National Cancer Institute; 2007. [Google Scholar]
- 2.Longford NT. Multilevel statistical models. New York: Oxford University Press; 1995. [Google Scholar]
- 3.Raudenbush SW, Bryk AS. Hierarchical Linear Modelling. 2nd. Newbury Park, CA: Sage Publications; 2001. [Google Scholar]
- 4.Snijders T, Bosker R. Multilevel analysis: An introduction to basic and advanced multilevel modelling . London: Sage; 1998. [Google Scholar]
- 5.Holmes JH, Lehman A, Hade E, et al. Challenges for multilevel health disparities research in a transdisciplinary environment. Am J Prev Med. 2008;35:S182–92. doi: 10.1016/j.amepre.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham H. Social Determinants and Their Unequal Distribution: Clarifying Policy Understandings. Milbank Quarterly. 2004;82:101–24. doi: 10.1111/j.0887-378X.2004.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallgarda S. Social inequality in health: dichotomy or gradient? A comparative study of problematizations in national public health programmes. Health Policy. 2008;85:71–82. doi: 10.1016/j.healthpol.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Hsing AW, Devesa SS. Trends and patterns of prostate cancer: what do they suggest? Epidemiol Rev. 2001;23:3–13. doi: 10.1093/oxfordjournals.epirev.a000792. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman RM, Gilliland FD, Eley JW, et al. Racial and ethnic differences in advanced-stage prostate cancer: the Prostate Cancer Outcomes Study. J Natl Cancer Inst. 2001;93:388–95. doi: 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 10.Casey G, Neville PJ, Plummer SJ, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32:581–3. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 11.Nwosu V, Carpten J, Trent JM, Sheridan R. Heterogeneity of genetic alterations in prostate cancer: evidence of the complex nature of the disease. Hum Mol Genet. 2001;10:2313–8. doi: 10.1093/hmg/10.20.2313. [DOI] [PubMed] [Google Scholar]
- 12.Richards B, Skoletsky J, Shuber AP, et al. Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Mol Genet. 1993;2:159–63. doi: 10.1093/hmg/2.2.159. [DOI] [PubMed] [Google Scholar]
- 13.Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environmental Health Perspectives. 1999;107:517–20. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 15.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 16.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 17.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 18.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 19.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 20.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 21.Sun J, Purcell L, Gao Z, et al. Association between sequence variants at 17q12 and 17q24.3 and prostate cancer risk in European and African Americans. Prostate. 2008;68:691–7. doi: 10.1002/pros.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Zheng SL, Komiya A, et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32:321–5. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- 24.Casey G, Neville PJ, Plummer SJ, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32:581–3. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian SV, Lochner KA, Kawachi I. Neighborhood differences in social capital: a compositional artifact or a contextual construct? Health Place. 2003;9:33–44. doi: 10.1016/s1353-8292(02)00028-x. [DOI] [PubMed] [Google Scholar]
- 26.Raudenbush S. The quantitative assessment of neighborhood social environments. In: Kawachi I, B LF, editors. Neighborhoods and Health. Princeton, NJ: Princeton University Press; 2003. [Google Scholar]
- 27.Krieger N, Williams DR, Moss NE. Measuring social class in US public health research: concepts, methodologies, and guidelines. Annu Rev Public Health. 1997;18:341–78. doi: 10.1146/annurev.publhealth.18.1.341. [DOI] [PubMed] [Google Scholar]
- 28.Krieger N, Ziehler S, Hogan JW, et al. Geocoding and measurement of neighborhood socioeconomic position: A US perspective. In: Kawachi I, Berkman LF, editors. Neighborhoods and Health. Princeton, NJ: Princeton University Press; 2003. [Google Scholar]
- 29.Lin DY, Wei LJ. The robust inference for the Cox proportional hazards model. JASA. 1989;84:1074–8. [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 31.Kader AK, Sun J, Isaacs SD, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salinas CA, Koopmeiners JS, Kwon EM, et al. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–72. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 34.Waters KM, Le Marchand L, Kolonel LN, et al. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev. 2009;18:1285–9. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–93. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 36.Anahi Franchi N, Avendano C, Molina RI, et al. beta-Microseminoprotein in human spermatozoa and its potential role in male fertility. Reproduction. 2008;136:157–66. doi: 10.1530/REP-08-0032. [DOI] [PubMed] [Google Scholar]
- 37.Nam RK, Reeves JR, Toi A, et al. A novel serum marker, total prostate secretory protein of 94 amino acids, improves prostate cancer detection and helps identify high grade cancers at diagnosis. J Urol. 2006;175:1291–7. doi: 10.1016/S0022-5347(05)00695-6. [DOI] [PubMed] [Google Scholar]
- 38.Reeves JR, Dulude H, Panchal C, Daigneault L, Ramnani DM. Prognostic value of prostate secretory protein of 94 amino acids and its binding protein after radical prostatectomy. Clin Cancer Res. 2006;12:6018–22. doi: 10.1158/1078-0432.CCR-06-0625. [DOI] [PubMed] [Google Scholar]
- 39.Kote-Jarai Z, Easton DF, Stanford JL, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–61. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, Zheng SL, Wiklund F, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–5. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 42.Gatrell AC. On the spatial representation and accuracy of address-based data in the UK. International Journal of Geographical Information Systems. 1989;3 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.