Abstract
A major function of proteasomes and macroautophagy is to eliminate misfolded potentially toxic proteins. Mammalian proteasomes, however, cannot cleave polyglutamine (polyQ) sequences and seem to release polyQ-rich peptides. Puromycin-sensitive aminopeptidase (PSA) is the only cytosolic enzyme able to digest polyQ sequences. We tested whether PSA can protect against accumulation of polyQ fragments. In cultured cells, Drosophila and mouse muscles, PSA inhibition or knockdown increased aggregate content and toxicity of polyQ-expanded huntingtin exon 1. Conversely, PSA overexpression decreased aggregate content and toxicity. PSA inhibition also increased the levels of polyQ-expanded ataxin-3 as well as mutant α-synuclein and superoxide dismutase 1. These protective effects result from an unexpected ability of PSA to enhance macroautophagy. PSA overexpression increased, and PSA knockdown or inhibition reduced microtubule-associated protein 1 light chain 3-II (LC3-II) levels and the amount of protein degradation sensitive to inhibitors of lysosomal function and autophagy. Thus, by promoting autophagic protein clearance, PSA helps protect against accumulation of aggregation-prone proteins and proteotoxicity.
INTRODUCTION
The accumulation of aggregate-prone proteins in neurons is a hallmark of many neurodegenerative disorders, including the polyglutamine tract expansion diseases such as Huntington's disease and spinocerebellar ataxia type 3, familial forms of Parkinson's disease and amyotrophic lateral sclerosis [caused by point mutations in α-synuclein and superoxide dismutase 1 (SOD1), respectively]. These abnormal proteins are thought to cause disease via toxic gain-of-function mechanisms. Thus, one rational approach to combating their toxicity is to reduce the cellular content of the mutant protein by accelerating their degradation.
The two major routes for protein degradation within mammalian cells are macroautophagy and the ubiquitin–proteasome system. Degradation by the macroautophagy–lysosomal pathway begins with the formation of double-layered autophagosomes that enclose portions of cytoplasm. These vacuoles ultimately fuse with lysosomes, and the cytosolic components are degraded by its various lysosomal acid hydrolases. Macroautophagy (which we call here autophagy) is a key mechanism for the clearance of many aggregation-prone (or aggregated) proteins associated with neurodegenerative diseases, including mutant forms of huntingtin, SOD1 and α-synuclein (1). Furthermore, activation of this autophagic process (e.g. by rapamycin) enhances the removal of the aggregate-prone proteins such as mutant huntingtin and attenuates its toxicity in cell and animal models (2). The ubiquitin–proteasome pathway also plays a critical role in the selective degradation of misfolded, mutated or damaged proteins. Such proteins are targeted for rapid hydrolysis by a series of enzymes that covalently attach a chain of ubiquitin molecules onto lysine residues on the protein. This polyubiquitin chain serves as a recognition motif for binding of the protein to the 26S proteasome. The ubiquitinated proteins are digested to small peptides within the core 20S proteasome particle. This barrel-shaped particle contains three types of peptidase sites that can cleave nearly all peptide bonds in proteins. The short (2–20) residue peptides typically released by the proteasome are then rapidly hydrolyzed to amino acids by cytosolic endo- and aminopeptidases.
The ubiquitin–proteasome pathway can efficiently digest soluble misfolded proteins, but once proteins such as huntingtin are aggregated, the autophagic/lysosomal process assumes primary importance in their clearance from the cytosol (3–5). However, in the case of proteins containing polyglutamine tracts, eukaryotic proteasomes can cleave only very poorly (if at all) within polyglutamine sequences (6). Consequently, in degrading huntingtin, the 26S proteasome appears to release polyglutamine-rich fragments for digestion by cytosolic peptidases (6,7). Because they lack extensive flanking sequences, such peptides have a very strong tendency to aggregate (probably even stronger than that of the full-length protein). Therefore, the rapid hydrolysis of these polyglutamine-rich peptides seems likely to be important in preventing or retarding the progression of polyglutamine disorders. Most larger peptides released by proteasomes are initially digested by endopeptidases (8–10), and the resulting shorter peptides are rapidly hydrolyzed to amino acids by various cytosolic aminopeptidases (11–14).
Surprisingly, only one cytosolic peptidase, puromycin-sensitive aminopeptidase (PSA, also termed cytosol alanyl aminopeptidase, human gene symbol NPEPPS), was found to be able to digest short polyglutamine peptides (15). PSA is a ubiquitous, 100 kDa, Zn2+ metallopeptidase present in high concentrations in the brain (especially in the striatum, the hippocampus and the cerebellum) (16,17). Although PSA was initially described as an enkephalin-degrading enzyme (18,19), its localization predominantly in the cytoplasm and its broad distribution in tissues argue against such a function. Instead, a role for PSA in digesting proteasome products to amino acids or antigenic peptides presented on MHC Class I molecules seems most likely based on its cytosolic location and its ability to digest diverse sequences (12–14,20). In fact, we have found that PSA is the dominant intracellular peptidase in degrading a large variety of dipeptides (R.H. and A.L.G., unpublished data).
These observations suggest that a loss of PSA function could lead to a toxic accumulation of fragments of normal gene products and increase the susceptibility to polyglutamine diseases. In fact, PSA-deficient mice display behavioural and neurological abnormalities (17,21) including movement disorders that perhaps are related to the failure to rapidly clear peptides released by the proteasomes which could have deleterious effects. Interestingly, the expression of PSA was found to be highly induced in PC12 cells upon expression of huntingtin exon 1 containing an expanded, but not a normal length, polyQ sequence (22). In addition, PSA is induced in the CNS in a mouse model of fronto-temporal dementia expressing a mutated tau (16). Furthermore, overexpression of PSA had a remarkable ability to protect against neurodegeneration induced by misexpression of the longest isoform of human tau in Drosophila (16).
The present studies were undertaken to test whether PSA may play an important role in the clearance of polyQ-rich fragments, polyQ-containing proteins or other mutant polypeptides associated with neurodegenerative diseases. We have tested whether altering the activity or content of PSA modifies aggregate accumulation and toxicity of different forms of mutant huntingtin exon 1 in cell culture and in vivo models of neurodegenerative diseases. Using a variety of approaches and experimental systems, we show that PSA is an important determinant of aggregate content and of huntingtin exon 1 toxicity. Surprisingly, this ability of PSA to reduce the content of aggregates formed by huntingtin exon 1, as well as another expanded polyQ protein, ataxin-3, correlated with an ability of PSA to promote autophagy. Indeed, PSA was found to enhance the clearance of a range of aggregation-prone proteins associated with neurodegenerative diseases, including some proteins lacking polyglutamine expansions, and to increase intracellular protein degradation by promoting autophagy. These studies have uncovered a surprising new function of this cytosolic peptidase in regulating autophagy and a remarkable ability of PSA activity to influence aggregate content and toxicity in a variety of neurodegenerative diseases.
RESULTS
Decrease in PSA activity increases the aggregation and toxicity of expanded huntingtin exon 1
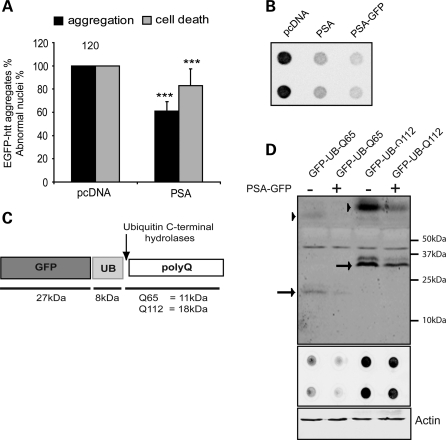
To test whether PSA activity might affect the cellular content of polyglutamine aggregates and toxicity, we studied the effects of knocking down PSA expression using RNA interference (RNAi) (Supplementary Material, Fig. S1A) and of pharmacological inhibitors of PSA activity in HeLa, HEK293A and HEK293T cells. Depletion of this enzyme in HeLa cells expressing mutant huntingtin with 74 polyglutamine repeats (Q74) exon 1 fused to GFP increased the percentage of cells that displayed macroaggregates as well as the number of apoptotic nuclei (Fig. 1A). Similarly, PSA knockdown in cells expressing a huntingtin–GFP construct with 103 glutamines enhanced the percentage of cells with aggregates (Supplementary Material, Fig. S1B). This result was corroborated when levels of insoluble macroaggregates in whole cell lysates were measured using the filter trap assay (Fig. 1B). Surprisingly, even though cells expressing the non-expanded Q23 huntingtin exon 1 GFP do not normally contain visible aggregates, decreasing PSA expression also led to the appearance of intracellular accumulations, similar to the inclusions we observed with expanded huntingtin exon 1; however, these may not necessarily have the same structure as the expanded polyQ aggregates. This was accompanied by an increase in the frequency of apoptotic nuclei (Fig. 1C). Thus, PSA appears to play a role in clearance of both expanded and normal length huntingtin exon 1.
Figure 1.
PSA knockdown and inhibition increase aggregation and toxicity in cells expressing normal and polyglutamine-expanded huntingtin exon 1. (A) HeLa cells were treated with PSA siRNA and then transfected with plasmids encoding exon 1 of huntingtin with polyglutamine tract length of 74, tagged with EGFP (htt-exon-1-GFP). The proportion of transfected (GFP-positive) cells containing GFP aggregates was measured (black bars) and cellular toxicity was assessed by measuring the proportion of transfected cells with apoptotic nuclei, visualized by DAPI staining (grey bars). Aggregation and toxicity were increased by the knockdown of PSA. Data shown are normalized to control levels of aggregation and cell death and combined from four independent experiments, each performed in triplicate. Error bars represent SEM, as for all graphs in this study. In control conditions, the proportion of cells with GFP-positive macroaggregates and apoptotic nuclei were typically 6 and 4%, respectively. ***P < 0.001, *P < 0.05 by odds ratio. (B) Aggregation was also measured by filter trap assay in HEK 293T cells using htt-exon-1-Q103-GFP. Lysates from cells treated with PSA siRNA contained more insoluble aggregates trapped on the filter membrane as shown by the increase in GFP staining. (C) Effect of PSA knockdown on aggregation of htt-exon-1-GFP with a polyglutamine repeat length of 23 was measured, exactly as in (A); data are shown normalized to control. The proportions of control cells with macroaggregates and apoptotic nuclei were 2 and 1%, respectively. ***P < 0.001 (D–F). Small molecular inhibitors of PSA also increased aggregation of polyglutamine proteins with a range of repeat lengths. 293A cells were transfected with htt-exon-1-GFP with 23Q (D), 41Q (E) or 74Q (F). PSA was inhibited with PAQ-22 (dark grey lines) or bestatin (light grey lines) to either 50 or 80% of its endogenous activity: 50% inhibition: 0.05 μm bestatin, 0.12 μm PAQ-22; 80% inhibition: 0.5 μm bestatin, 1 μm PAQ-22. The number of transfected cells containing GFP-positive aggregates was counted at the time points indicated. ***P < 0.001, *P < 0.05 by two-way ANOVA followed by post hoc Fisher's LSD test. (G) In intact WT MEF cells, 80% of Ala-amc cleavage was inhibited by minimal effective concentration of both PAQ (10 μm) and bestatin (5 μm). In PSA−/− MEF cells, PAQ and bestatin inhibit only 10 and 20% of the residual Ala-amc cleavage activity, which must be by other aminopeptidases. **P = 0.001. (H) Bestatin (0.5 μm) treatment increased aggregation of HA-tagged huntingtin exon 1 with 74Q in HeLa cells, but not following knockdown of PSA by siRNA. *P < 0.05, ns, not significant by odds ratio.
In order to determine whether this effect of PSA on formation of aggregates is due to its enzymatic activity, we employed the general aminopeptidase inhibitor, bestatin, and the specific PSA inhibitor, PAQ-22. In 293A cell extracts, both bestatin and PAQ-22 are potent inhibitors of PSA aminopeptidase activity [assayed fluorometrically with the alanine amino methylcoumarin (Ala-amc) substrate] with IC50s of 0.07 and 0.08 μm, respectively (Supplementary Material, Fig. S1C). By measuring the potency of these inhibitors of PSA in intact 293A cells (Supplementary Material, Fig. S1D) at concentrations where these drugs do not alter cellular viability for several days (Supplementary Material, Fig. S1E), we were able to assess how the level of PSA activity influences aggregate content in 293A cells expressing huntingtin exon 1 GFP with either 23 [wild-type (WT)], 41 or 74 glutamines (Fig. 1D–F and Supplementary Material, Fig. S1F). PSA inhibition increased aggregate content 2–4-fold not only with expanded polyQ, where aggregates are more abundant (Q74 and Q41), but also with the WT (23Q) huntingtin exon 1 GFP. In fact, the relative stimulation appeared larger with the 23Q construct, perhaps because the appearance of inclusions is easier to detect with this construct, since it fails to aggregate significantly under normal circumstances. It is noteworthy that large increases in aggregate content were seen with only a 50% decrease in enzyme activity using either inhibitor.
We confirmed the relative specificity of these inhibitors by comparing their activities in mouse embryonic fibroblasts (MEFs) from WT and PSA-null mice. In intact cells, the cleavage of Ala-amc was five times higher in WT MEFs than in MEFs from PSA knockout mice. In WT MEFs, 80% of the Ala-amc cleavage was sensitive to both PAQ (10 μm) and bestatin (5 μm), whereas PAQ and bestatin had negligible effects on its hydrolysis in the PSA knockout MEFs (Fig. 1G). Further evidence that bestatin increased aggregate number through PSA inhibition was the finding that, following RNAi knockdown of PSA, this inhibitor had no effect in cells expressing huntingtin exon 1 Q74 GFP (Fig. 1H). Together, these findings indicate that PSA is involved in the clearance of both normal and polyQ-expanded huntingtin exon 1 and reducing its activity by 50% increases huntingtin accumulation and toxicity.
PSA overexpression reduces aggregates and toxicity of mutant huntingtin exon 1
Consistent with the deleterious effects of PSA knockdown and inhibition, we found that PSA overexpression (Supplementary Material, Fig. S2A) decreased the frequency of expanded huntingtin exon 1 GFP aggregates and the associated cell death in neuroblastoma cells (Fig. 2A). Furthermore, in HeLa cells, PSA overexpression not only reduced the fraction of cells with inclusions, it also reduced the cells’ content of insoluble huntingtin exon 1 GFP and the sizes of the inclusions (Fig. 2B and Supplementary Material, Fig. S2B and C). Because of the inability of the proteasome to cleave efficiently within polyQ sequences, polyQ-rich peptides are likely to be generated and released during the degradation of polyQ proteins (6). In an attempt to mimic these isolated polyQ sequences in the absence of a linked protein such as huntingtin, we expressed a GFP–ubiquitin–polyQ fusion protein (23). In cells, the GFP–ubiquitin and the C-terminal polyQ sequence are efficiently cleaved by deubiquitinating enzymes, releasing pure polyQ polypeptide (Fig. 2C). When 293T cells were transfected with GFP–ubiquitin–polyQ containing 65 or 112 glutamines, with or without PSA–GFP, PSA overexpression reduced the levels of both the soluble polyQ polypeptides and insoluble polyQ aggregates (assayed by filter trap) (Fig. 2D and Supplementary Material, Fig. S2D and E), in accord with our findings on expanded huntingtin exon 1 (Fig. 2A and B and Supplementary Material, Fig. S2B and C). However, it was not possible to assess whether raising PSA levels had similar protective effects in cells expressing WT huntingtin exon 1 GFP due to their very low frequency of aggregates and cell death.
Figure 2.
PSA overexpression reduces toxicity and aggregate content in cells expressing expanded polyQ huntingtin exon 1 and expanded polyglutamine tracts. (A) Effect of PSA overexpression on aggregate content in SK-N-SH cells expressing htt-exon-1-Q74-GFP. The proportions of cells with GFP-positive macroaggregates and apoptotic nuclei were decreased by the overexpression of PSA, when assayed as in Figure 1A and C. Data shown are normalized to control for each experiment and the average of data from four independent experiments carried out in triplicate are shown. In control conditions, typically around 30% of transfected cells contained macroaggregates and 10% had apoptotic nuclei. ***P < 0.001 by odds ratio. (B) Overexpression of PSA in 293T cells also decreased levels of insoluble aggregates of htt-exon-1-Q103-GFP, as measured by filter-trap assay. Q103 aggregates were visualized using anti-GFP antibody. (C) To mimic the polyQ sequences that may be released by the proteasome, we used a GFP–ubiquitin–polyQ construct (23). Upon expression, the GFP–UB and the polyQ peptide are efficiently cleaved by deubiquitinating enzyme(s), leading to the release of a polyQ polypeptide. (D) Levels of soluble and insoluble polyglutamine peptides were also decreased by overexpression of PSA in HEK293T cells transfected with GFP–UB–polyQ containing 65 or 112 glutamines. Levels of both soluble polyQ peptides (upper panel, arrows mark monomeric polyQ peptides and arrowheads mark high molecular weight oligomeric structures) and insoluble aggregates (lower panel) detected by an anti-polyglutamine antibody decreased, when PSA-GFP was co-transfected. Actin levels are shown as a loading control.
PSA suppresses accumulation of mutant huntingtin aggregates and toxicity in vivo
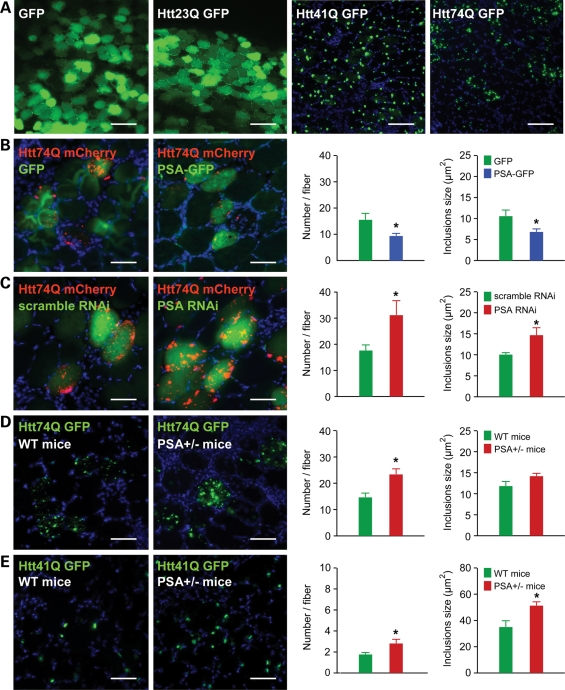
To test whether overexpression of PSA causes a similar suppression of huntingtin aggregates in mouse tissues in vivo, we electroporated GFP-tagged huntingtin exon 1 constructs bearing different polyQ lengths into the tibialis anterior muscles of adult mice. One week after electroporation, the huntingtin Q23 GFP was diffusely distributed in the electroporated fibres. However, transfection with expanded huntingtin with 41 and 74Q caused the formation of GFP-labelled inclusions in all the electroporated fibres (Fig. 3A). When a PSA-encoding plasmid was simultaneously electroporated, it decreased the number and size of inclusions formed by the expanded huntingtin exon 1 mCherry (74Q) in the tibialis anterior muscle 1 week after electroporation (Fig. 3B). On the other hand, transfecting a PSA RNAi construct enhanced the formation of mutant huntingtin exon 1 mCherry aggregates (Fig. 3C). Furthermore, when huntingtin exon 1 GFP with 74Q or 41Q was electroporated in heterozygous PSA knockout mice, more inclusions were evident than in WT mice (Fig. 3D–E). The inclusions formed by huntingtin exon 1 (41Q) were larger in muscle of heterozygous PSA knockout mice (Fig. 3E). Extracts of muscles from the heterozygous mice showed an approximately 50% reduction in PSA activity (data not shown). Thus, a decrease of only 50% in the endogenous PSA activity is sufficient to significantly enhance mutant huntingtin aggregate formation, as was also found upon treatment of 293A cells with PSA inhibitors.
Figure 3.
Effect of PSA overexpression and knockdown on the formation of polyglutamine aggregates in mouse muscle. (A) Cross-sections of mouse tibialis anterior muscle electroporated with GFP or huntingtin exon 1 with 23, 41, 74Q fused to GFP (Htt23Q, Htt41Q, Ht74Q). One week after electroporation, the formation of inclusions was observed in all fibres electroporated with Htt41Q and Htt74Q. Fibres electroporated with GFP or Htt23Q showed a diffuse fluorescence and no inclusions. Scale bar represents 100 μm. (B) PSA overexpression decreased the number and size of polyglutamine inclusions found in tibialis anterior muscle 1 week after electroporation with expanded huntingtin exon 1 (74Q) and PSA. Note that Htt74Q is seen as red (fused to mCherry) in these images. Graph shows inclusion number and size in fibres where PSA (blue bars) or GFP (green bars) is overexpressed. (C–E) Reduction in the expression of PSA by electroporation of RNAi increased the number and size of polyglutamine inclusions in mouse skeletal muscle electroporated with expanded huntingtin exon 1. (C) Tibialis anterior muscle 1 week after electroporation with Htt74Q and a plasmid expressing either a scrambled RNAi, or an RNAi targeting PSA. Reduction of PSA expression by RNAi increased the number and size of polyglutamine inclusions formed by expanded huntingtin exon 1 (74Q). (D) Electroporation of expanded huntingtin exon 1 (74Q) to tibialis anterior muscles of PSA+/− heterozygous mice led to more inclusions than in muscle from WT (PSA+/+) mice, 1 week after electroporation. These inclusions also seemed larger in PSA+/− mice, but this effect did not reach statistical significance (P = 0.09). (E) Electroporation of expanded huntingtin exon 1 (41Q) in PSA+/− heterozygous mice led the formation of more, larger inclusions than in muscle from WT (PSA+/+) mice 1 week after electroporation. Scale bar (B–E) represents 50 μm. *P < 0.05 by t-test.
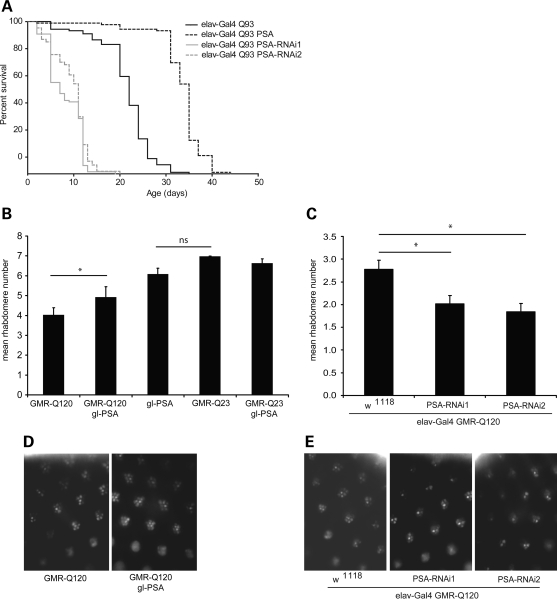
In order to test the effects of PSA on huntingtin toxicity in neurons in vivo, we used two distinct Drosophila models with different quantifiable readouts. In the first, mutant huntingtin exon 1 was expressed in the nervous system (24) and reduced lifespan. In these flies, PSA knockdown (with two different RNAi constructs) further shortened the lifespan, whereas PSA overexpression increased their lifespan (Fig. 4A). In the second model, mutant huntingtin was expressed in the photoreceptors. Fly photoreceptors that express a mutant huntingtin fragment with 120 polyglutamine repeats exhibit degeneration that is not observed in flies that express the WT fragment with 23 polyglutamine repeats (25). The compound eye of Drosophila consists of about 800 ommatidia, each composed of eight photoreceptor neurons with light-gathering parts called rhabdomeres, seven of which can be visualized by light microscopy using the pseudopupil technique (26). Neurodegeneration in these HD flies is progressive and associated with a decrease over time in the number of visible rhabdomeres in each ommatidium (25). However, PSA overexpression increased the number of visible photoreceptors (Fig. 4B and D), indicating suppression of polyglutamine toxicity. On the other hand, the expression of either of two different PSA RNAi constructs enhanced toxicity (Fig. 4C and E). Thus, as in cell culture, modulation of PSA levels alters polyQ toxicity in vivo.
Figure 4.
Effect of PSA overexpression and knockdown in Drosophila. (A) PSA expression levels alter lifespan in flies expressing htt-exon1 with 93 polyglutamine repeats in the brain. Knockdown of PSA expression levels by expression of two different PSA RNAi constructs enhanced toxicity (elav-Gal4 Q93 PSA-RNAi1 and elav-Gal4 Q93 PSA-RNAi2), compared with flies expressing mutant huntingtin alone (elav-Gal4 Q93). Overexpression of PSA (elav-Gal4 Q93 PSA) extended lifespan relative to flies expressing Q93 alone. Graph shows Kaplan–Meier survival curves, P < 0.0001 in both cases. Expression of PSA RNAi or PSA in flies expressing WT huntingtin (elav-Gal4 Q20) did not affect lifespan (Supplementary Material, Fig. S3). (B–E) Degeneration was also decreased by overexpression of PSA. Overexpression of PSA in the WT fly eye using the glass promoter (gl-PSA) caused a slight decrease in rhabdomere number compared with flies expressing Q23 alone. However, when co-expressed with GMR-Q120, PSA protected against the neurodegeneration seen in these flies (B). (C) Expression of PSA RNAi enhanced neurodegeneration in flies expressing GMR-Q120, using two RNAi lines (elav-Gal4 GMR-Q120 PSA-RNAi1 and elav-Gal4 GMR-Q120 PSA-RNAi2). Expression of PSA RNAi in flies expressing Q23 did not affect rhabdomere number (Supplementary Material, Fig. S3), *P < 0.02, 2 tailed t-test. (D) Examples of rhabdomeres in flies expressing GMR-Q120 with or without PSA (gl-PSA). (E) Examples of rhabdomeres in flies expressing mutant huntingtin GMR-Q120 when PSA is downregulated using two RNAi lines (PSA-RNAi1 and PSA-RNAi2).
PSA decreases aggregate number in cellular models of other neurodegenerative diseases
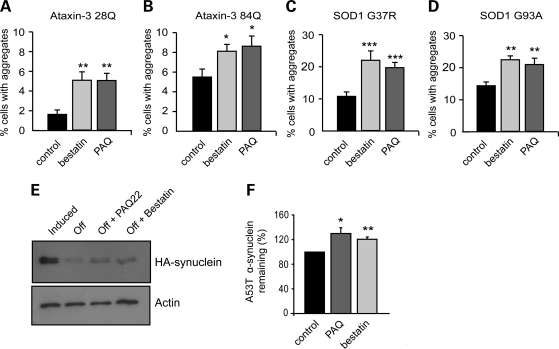
To investigate whether PSA can also promote the clearance of other aggregation-prone mutant proteins, we tested its effects on different neurodegenerative disease-associated proteins including ataxin-3, another polyglutamine-containing protein, whose expansion causes spinocerebellar ataxia type 3. In 293A cells expressing WT (28Q) and mutant (84Q) full-length ataxin-3 GFP, inhibition of PSA caused the accumulation of more aggregates than in untreated control cells (Fig. 5A and B and Supplementary Material, Fig. S4A). We also studied aggregate-prone proteins that lack polyglutamine repeat regions. In cells expressing a mutant form of SOD1, which causes an inherited form of amyotrophic lateral sclerosis, the number of SOD1 GFP inclusions was increased when PSA was inhibited (Fig. 5C, D and Supplementary Material, Fig. S4B). Furthermore, using PC12 cells, we were able to investigate the clearance of A53T mutant α-synuclein, which causes a familial form of Parkinson's disease. Its levels can be accurately measured by western blotting of the soluble fraction because this protein does not aggregate in these cells. To follow its degradation, α-synuclein expression was induced by the addition of doxycycline to the media, and then switched off by removal of the antibiotic. The degradation of A53T α-synuclein was slowed (i.e. its level was higher 16 h after synthesis was terminated) in the cells treated with the PSA inhibitors (Fig. 5E and F). Thus, this enzyme promotes the clearance and thus reduces aggregate content in a wide variety of cellular models of neurodegenerative disease.
Figure 5.
PSA inhibition enhances aggregate formation in cells expressing other aggregate-prone proteins (ataxin-3, mutant SOD1, mutant α-synuclein). (A and B) PSA inhibition increased aggregate formation in 293A cells expressing full-length ataxin-3 GFP with 28Q (A) and 84Q (B). The proportion of transfected cells with macroaggregates was also increased by inhibition of PSA in cells transfected with mutant SOD1 GFP G37R (C) or G93A (D). ***P < 0.001, **P < 0.01, *P < 0.05 by t-test. (E and F) Clearance of mutant α-synuclein (A53T) was slowed by inhibition of PSA. Stable inducible PC12 cells were treated with doxycycline to induce expression of mutant α-synuclein, and subsequently expression was turned off by removal of doxycycline from the medium. When PSA inhibitors were added after transgene expression was switched off, more α-synuclein remained present, indicating a decreased clearance. Levels of α-synuclein remaining after 16 h were assayed by western blot, a representative image is shown in (E). Densitometric quantification of this triplicate experiment is shown in (F). **P < 0.01, *P < 0.05, by ANOVA.
PSA enhances autophagy
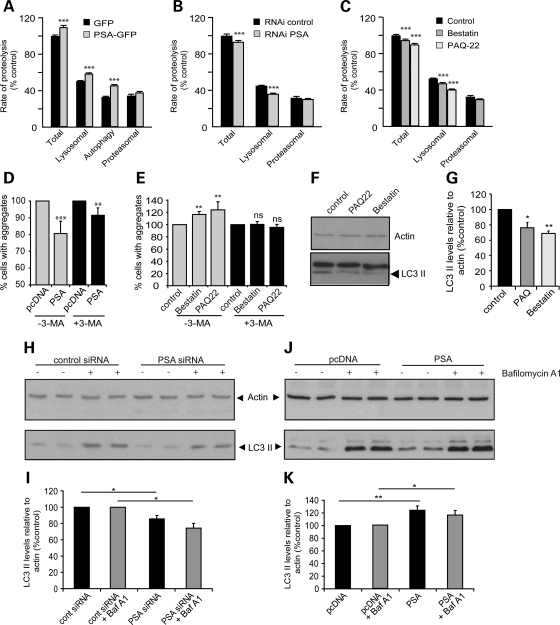
Many of the proteins found here to accumulate on PSA inhibition or to be cleared faster upon PSA overexpression are autophagy substrates, including mutant huntingtin exon 1, full-length ataxin-3, mutant SOD1 and A53T α-synuclein (27,28). In fact, a large fraction of long-lived cellular proteins, the bulk of cell constituents, are degraded by this process (29). We therefore tested whether changes in PSA activity influence the overall rate of degradation of long-lived proteins. In order to determine the rates of proteolysis by autophagy, lysosomes and proteasomes, we measured protein degradation rates in the presence or absence of specific inhibitors of the proteasome (bortezomib/velcade), lysosomal acidification (chloroquine) or autophagy [3-methyladenine (3MA)], as we previously described (29). Upon deprivation for serum and amino acids to activate autophagy, PSA overexpression increased the total rate of protein degradation by enhancing the rate of lysosomal and autophagy-dependent proteolysis but did not affect the proteasomal process (Fig. 6A). However, PSA had no measurable effect on any of these processes in complete medium (Supplementary Material, Fig. S5A–C) where autophagy makes only a minor contribution to the degradation of long-lived proteins (Supplementary Material, Fig. S5D).
Figure 6.
PSA activity enhances protein degradation by the autophagic/lysosomal pathway. (A–C) Effect of PSA modulation on different proteolytic pathways. Degradation of long-lived protein was measured in 293A cells. Proteolysis rates were measured in the presence of inhibitors of proteasomes, lysosomes or autophagy to determine flux through each of these pathways in cells starved of serum and amino acids. Proteasomes, lysosomes and autophagy inhibitors were velcade/bortezomib 1 µm, chloroquine 50 µm and 3-MA 10 mm, respectively. (A) PSA overexpression increased total, lysosomal and autophagy-dependent proteolysis. (B) PSA knockdown by RNAi decreased total and lysosomal proteolysis. (C) PSA inhibitors reduced total and lysosomal proteolysis. ***P < 0.0001. (D) Inhibiting autophagy in SK-N-SH cells expressing Q74 huntingtin exon-1 GFP using 3-MA, reduced the protective effect of PSA overexpression on aggregate content. Co-expression of PSA with htt-exon-1-GFP-74Q reduced the percentage of transfected cells with aggregates (grey bars). However, in the presence of 3-MA, added immediately after transfection, the reduction in aggregation caused by PSA expression was decreased (black bars). Control values with and without 3-MA are set to 100% in each case to allow ease of comparison, although Q74 aggregation was higher in the presence of 3-MA. Actual mean values were 24% of cells having aggregates in control and 30% in 3-MA treated cells. (E) Inhibitors of PSA caused an increase in the proportion of cells expressing htt-exon-1-GFP-Q74 with aggregates (grey bars), but if 3-MA was added at the same time as these inhibitors they did not increase the aggregate content (black bars). Actual mean values for aggregation were 19% in control cells and 32% in 3-MA treated cells. **P < 0.01, ***P < 0.001, ns, not significant, by odds ratio, relative to control. (F–K) Effect of PSA modulation on LC3-II levels. (F) Treatment of PC12 cells with PSA inhibitors also resulted in a decrease in LC3-II levels, a marker for autophagosomes. The western blot shown is a representative data set, with actin shown as a loading control. Quantification of data obtained in triplicate samples is shown in (G). (H and I) In HeLa cells treated with PSA siRNA, LC3-II levels were decreased, whereas cells overexpressing PSA showed increased levels of LC3-II (J and K). (I) and (K) show densitometric quantification of data from four experiments carried out in triplicate. LC3-II level changes were seen in the presence (grey bars) and absence (black bars) of bafilomycin A1, a lysosomal proton pump inhibitor. LC3-II levels are shown relative to actin, as a loading control in each case. Values are normalized to control, in the presence or absence of bafilomycin, to allow for comparison between different gels, although the levels of LC3-II were higher with bafilomycin than without. *P < 0.05, **P < 0.01.
On the other hand, knockdown of PSA expression using RNAi (Fig. 6B) or treatment with inhibitors of its activity (Fig. 6C) decreased the total and lysosomal proteolysis measured after nutrient deprivation, but not proteasomal degradation. Again, no significant effect of PSA was seen in complete medium (Supplementary Material, Fig. S5B and C). Thus, the activity of this cytosolic enzyme, PSA, appears to regulate the capacity of the autophagic–lysosomal pathway to degrade normal cell proteins.
In order to test whether this alteration of autophagic activity by PSA was a major contributor to the effects on protein clearance seen following PSA knockdown or overexpression, we used 3MA to inhibit autophagy (30). The decrease in aggregate content upon PSA overexpression (Fig. 6D) and the increase following by PSA inhibition (Fig. 6E) were both markedly attenuated by 3MA. These findings further suggest that most of PSA's ability to protect against expanded huntingtin exon 1 inclusions was via its ability to promote autophagy.
In order to directly assess the effects of PSA on the autophagy pathway, we measured the steady-state levels of autophagosomes using the microtubule-associated protein 1 light chain 3 (LC3). All of these experiments were performed in complete medium, to assess whether PSA could modulate basal autophagy levels. During autophagic vacuole formation, LC3 is modified post-translationally to form LC3-I, and then converted to LC3-II, which associates with autophagosome membranes. LC3-II levels, which thus reflect autophagosome number when assayed relative to the loading control, actin, were decreased by PSA inhibitors (Fig. 6F and G) and also by transfection with PSA RNAi (Fig. 6H and I). Since decreased LC3-II content could result from either impaired synthesis or enhanced degradation after lysosomal fusion, we assessed LC3-II synthesis by blocking its degradation with the lysosomal proton pump inhibitor Bafilomycin A1. As expected, this agent increased LC3-II levels in all conditions (Fig. 6H and I), but after Bafilomycin A1 treatment, the levels of LC3-II were lower in cells expressing PSA siRNA (Fig. 6H and I) than in control cells (Fig. 6J and K). Conversely, in PSA-overexpressing cells, LC3-II levels were increased over control levels. These findings imply that PSA promotes autophagy and clearance of the misfolded proteins by enhancing autophagosome formation.
DISCUSSION
The protection conferred by PSA against polyQ and other aggregation-prone proteins was observed here in a very wide range of types of cells in culture, as well as in Drosophila and mouse models of neurodegenerative diseases. In all cases examined, cellular protection against these toxic proteins coincided with decreased content of large inclusions containing the mutant polypeptide. However, this correlation does not imply that the large aggregates are in fact the toxic species, since aggregate formation and cytotoxicity probably both depend on levels of misfolded proteins or microaggregated species whose clearance is accelerated by PSA. In addition to using various standard approaches to evaluate aggregate formation and cell protection, we have introduced a novel in vivo method—electroporation of a mutant gene into muscle fibre in adult mouse—that offers several major advantages for such studies (e.g. rapid development of inclusions, ability to use host animals of different genotypes without lengthy mating, comparisons with control cells in the same animals). Moreover, with this approach, increasing the length of the polyQ tract of huntingtin exon 1 yielded more inclusions, as observed in cell culture. In mouse muscle fibres, PSA knockdown increased while PSA overexpression reduced the content of inclusions formed by expanded huntingtin exon 1, as we found in cell culture. Surprisingly, in contrast to these marked results (obtained in three labs), an earlier study had concluded that PSA does not influence toxicity of mutant huntingtin in Drosophila (16), but that study assessed polyglutamine toxicity by inspection of external eye morphology, a rather insensitive qualitative approach, in contrast to the quantitative analysis of the number of rhabdomeres used here (Fig. 4).
The activation of autophagy by PSA
The ability of PSA to decrease aggregate content was not restricted to cells expressing expanded huntingtin exon 1, but was also observed with unattached long polyQ peptides, full-length ataxin-3, mutant SOD1 and mutant α-synuclein. This capacity of PSA to reduce the toxicity and accumulation of aggregates containing expanded huntingtin exon 1 and these other disease-associated proteins can be largely attributed to a surprising new function of PSA: its ability to promote protein degradation by autophagy. The enhancement of autophagy by PSA constitutes a novel mechanism that seems to account for the protective effects of PSA against this wide range of aggregation-prone proteins. Since LC3-II levels and the amount of protein degradation that is sensitive to 3MA or chloroquine were increased by PSA overexpression and reduced by PSA knockdown or inhibition, this enzyme somehow must promote autophagosome formation. PSA was previously shown to decrease the toxicity in Drosophila of WT and mutant form of tau, which is associated with frontotemporal dementia (16). These effects were attributed to direct degradation of tau by the aminopeptidase activity of PSA (31); however, such an exoproteolytic mechanism would be quite slow and inefficient for digesting a protein as long as tau and would generate heterogenous variants of tau of diverse lengths. Since tau has been demonstrated to be an autophagy substrate (27), it seems more likely that the enhancement of autophagy by PSA might better explain the stimulation of degradation of tau and the reduction in its toxicity upon PSA overexpression. Interestingly, the two different PSA-deficient mouse lines are both characterized by abnormal motor behaviour (17,21), which might also be a consequence of reduced autophagy, since the blockage of this process in Atg7-deficient mice leads to major behavioural deficits and widespread inclusion bodies (32). On the other hand, upregulation of autophagy via either mTOR-dependent or mTOR-independent pathways protects cells from several pro-apoptotic insults (33,34). Therefore, it seems likely that PSA protects primarily by enhancing the removal of the toxic proteins, although it possibly may also have additional antiapoptotic effects (e.g. by eliminating toxic peptides released by proteasomes as discussed below).
It is indeed unexpected that the activity of a cytosolic aminopeptidase can regulate autophagic vacuole formation. PSA inhibition was achieved using two unrelated small molecular weight compounds: bestatin, a natural, slow binding, competitive inhibitor of many aminopeptidases (35), and PAQ-22, a synthetic, non-competitive inhibitor that does not act as a substrate-mimic and binds to PSA at a distinct site (36–38). Both compounds increased the formation of aggregates and the toxicity of expanded huntingtin exon 1 at concentrations that caused only a modest (50%) inhibition of its activity. Thus, even minor variations in PSA activity, e.g. in human polymorphisms, if they exist, have the potential of influencing susceptibility to neurodegenerative disease.
PSA seems to be the major cytosolic aminopeptidase in cultured cells as well as in brain, muscle and kidney (39). It cleaves rapidly most N-terminal amino acids (especially alanine) from peptides, and PSA accounts for approximately 80% of the total soluble aminopeptidase activity in human cerebral cortex (40). Aminopeptidases such as PSA catalyse the final stages of the degradation of intracellular normal and abnormal proteins releasing amino acids from peptides generated by the catabolism of proteins by the proteasome (13,15,41). Thus, PSA potentially might link late steps in protein degradation by the ubiquitin proteasome pathway to functioning of the autophagic pathway. Certainly, regulation of any cellular process by the activity of PSA or any aminopeptidase represents an unusual mode of enzyme regulation since amino acid removal would cause irreversible changes in a protein. Alternatively, perhaps it may act to destroy a regulatory peptide that somehow inhibits autophagy. It is noteworthy that certain amino acids (especially leucine) activate mTORC1 and thus inhibit macroautophagy (42). However, the digestion by PSA of proteasome products would increase the pool of free amino acids and thus should reduce autophagy, while the exact opposite was observed. It remains possible that PSA could be involved in earlier stages in the catabolism of some proteins; in fact, there is a general slowdown in the degradation of proteins in bacteria lacking multiple aminopeptidases (41), although such effects could be indirect. For example, the in vivo half-life of a protein is determined in part by the nature of its N-terminal amino acid (termed the ‘N-end rule’). Thus, the removal of the N-terminal amino acid from certain proteins by PSA could expose destabilizing residues leading to their rapid degradation by the ubiquitin–proteasome pathway. Conceivably, PSA might in this way alter the levels (or activity) of a protein that is critical in the regulation of autophagy. However, in most globular proteins, the N-terminal amino acid is buried in the interior, so PSA would be expected to act only on proteins with ‘loose ends’ and generally aminopeptidases preferentially attack shorter peptides.
A major challenge for future work will be to establish the mechanism by which PSA promotes autophagy. The best characterized physiological inhibitor of autophagy is mTOR. However, we found no evidence for PSA inhibiting the activity of mTOR (Supplementary Material, Fig. S5E and F), and it seems quite unlikely that PSA promotes autophagy simply by causing nutrient deprivation (since its overexpression does not impede growth). In fact, PSA had clear effects on autophagic protein degradation in cells deprived of serum and amino acids, where mTOR is markedly inhibited. Autophagy is regulated by other mechanisms such as the phosphorylation of Bcl-2, which modulates autophagy via its interaction with Beclin-1 (43). However, we did not observe any effect of PSA on the levels of Bcl-2 phosphorylation (Supplementary Material, Fig. S5E and F). Thus, we believe that degradation or destabilization of an unknown inhibitory component of autophagy remains the most likely explanation of the activation of autophagy by PSA.
PSA can also act after the proteasome independently of autophagy
While these studies focused on mutant huntingtin exon 1 and other aggregation-prone proteins whose clearance seems to be due to autophagy, it is noteworthy that PSA was also found to prevent the formation of aggregates in cells expressing the WT exon 1 protein and also with the WT full-length ataxin-3. In other words, as suggested previously (15), this enzyme appears to be an important defence against aggregate formation due to non-expanded polyQ sequences as are found in various normal proteins. Our previous data also indicate that the clearance of WT huntingtin exon 1 does not depend on autophagy and is mainly via the proteasome (30,44). The proteasome would be predicted to continually produce short polyglutamine tracts from this and other polyQ proteins, since it cannot cleave between successive glutamine residues in the polyQ stretch (6). Such isolated polyglutamine tracts (even ones as short as 10Qs) are highly aggregate-prone if not cleared from the cell. Because PSA is the only non-lysosomal enzyme in tissue extracts that can degrade such polyglutamine products (15), polyglutamine stretches should accumulate and aggregate in cells when PSA is inhibited, as was found here. In cells, the isolated polyglutamine tracts not only aggregate but also sequester GFP or other tagged huntingtin constructs with flanking sequences (23,45). Because PSA is an aminopeptidase that digests soluble polypeptides one residue at a time and dissociates after each cleavage, it should degrade these aggregation-prone polyglutamine stretches only slowly. Thus, reducing its activity will have important consequences on accumulation of shorter polyglutamine stretches, whereas the more aggregate-prone mutant proteins (e.g. expanded polyQ proteins) tend to aggregate rapidly due to their inherent tendency to misfold, and their clearance by PSA appears to be more dependent on autophagy. In principle, the aggregates seen with mutant huntingtin could arise from seeds that are either the whole exon 1 fragment or the proposed post-proteasome-isolated polyQ tract. Isolated expanded polyQ tracts will seed and sequester GFP-tagged exon 1 of mutant huntingtin. Since PSA regulates overall aggregation mediated from exon 1 of huntingtin in an autophagy-dependent manner, any contribution towards aggregation of PSA acting as an aminopeptidase on isolated expanded polyQ tracts derived from larger huntingtin fragments will be very minor. Indeed, once huntingtin is engulfed in an autophagosome, it will be inaccessible to the ubiquitin–proteasome machinery and to PSA. Thus, our data show that the predominant effect of PSA in these models of HD is via autophagy.
Presumably, the physiological levels of PSA have evolved so as to ensure the rapid elimination of peptides from the nucleus and cytosol, but under the present experimental conditions when an aggregation-prone protein is overexpressed, PSA is not present in large excess over what is necessary to clear the misfolded, toxic proteins or even the products of WT huntingtin exon 1. Indeed, only a 50% reduction in PSA activity using inhibitors in cell culture or the 50% reduction in expression in PSA in tissues of heterozygous PSA+/− mice led to greater content of aggregates formed by expanded huntingtin exon 1. These observations and the protection by overexpression argue that activation of autophagy by PSA is a tightly regulated process, and that only a modest increase in its activity might have beneficial effects. Increasing PSA activity thus represents a novel potential approach for therapy of neurodegenerative diseases. It is noteworthy that treatment of cells with PSA inhibitors for several days was remarkably non-toxic. In contrast, proteasome or lysosomal inhibitors are highly toxic when applied for more than 6–12h. While a low toxicity would thus be expected for agents that would moderately increase the activity or expression of PSA, a high level of PSA overexpression by itself was toxic in Drosophila (data not shown). It is also noteworthy that PSA expression is elevated in PC12 cells expressing polyQ expanded exon 1 (22) and also in some neurons expressing mutant tau (16). Thus, induction of PSA may well represent an important cellular adaptation that reduces the accumulation of toxic proteins and functions in host defences against neurodegenerative disease (15).
MATERIALS AND METHODS
Details of plasmids used as well as cell culture methodology and protocols for aggregate analysis, western blotting and filter trap assays can be found in Supplementary information.
Inhibitors
PAQ-22 was from Wako, puromycin aminonucleoside was from Biomol, PS-341 (Bortezomid or Velcade) was a gift from Millenium. Bestatin, puromycin dihydrochloride, chloroquine, 3MA, NH4Cl and staurosporin were from Sigma. Fresh stock solutions of chloroquine (50 mm) were prepared just before experiments (final concentration 50 µm). Fresh 3MA solutions (100 mm) were prepared in boiled PBS just before experiments (final concentration 10 mm).
Muscle electroporation
Adult CD1 male mice (25–35 g) were anaesthetized by intraperitoneal injection of Avertin (0.2 ml/10 g as 1.2% solution). Fur overlying the hindlimb was removed, and the surgical site was cleansed with betadine swab followed by isopropyl alcohol wipe. Using sterile technique, a 0.5–1.0 cm longitudinal incision was made in the skin overlying the tibialis anterior muscle, and the underlying muscle was exposed by blunt dissection of the surrounding skin. A paddle electrode was then introduced between the muscle and the underlying tibia. Plasmid DNA in sterile saline (40 µl, 25 µg) was injected into the muscle. A second paddle electrode was applied gently over the muscle and electroporation performed (12 V, 20 ms duration, 5 pulses, 200 ms intervals) using an ECM830 Electro Square Porator (BTX, Harvard Apparatus). The electrodes were removed and the skin was closed by suture. Post-operative analgesia was administered every 12 h for the first 24 h with buprenorphine 0.05 mg/kg SQ. Additional analgesia was administered thereafter if there was any evidence of continued animal discomfort. The animals were followed for 1 week, after which time they are euthanized by Halothane inhalation followed by cervical dislocation, and the muscles harvested, quickly frozen in liquid-nitrogen-cooled isopentane and stored at −80°C. Ten micrometer muscle cryosections were analyzed for GFP/mCherry fluorescence after fixation with 4% paraformaldehyde and Hoechst staining (10 µg/ml in PBS for 10 min) using a Nikon 80i upright microscope in the Nikon Imaging Center at Harvard Medical School. No gross evidence of necrosis or inflammation as a result of the transfection procedure was noted. Inclusion size and number were analysed with Metamorph using fixed size and intensity thresholds.
Drosophila
Fly crosses and experiments were performed at 25°C. All crosses for individual experiments were performed at the same time and under the same conditions.
For survival assays, virgins carrying either P{UAS-dPsa}8.10 [PSA (47)] or UAS-RNAi constructs that knockdown Psa [PSA-IR-R2 (referred to as PSA-RNAi1) and PSA-IR-R3 (referred to as PSA-RNAi2), National Institute of Genetics Fly Stock Center, Japan] were crossed with males expressing w; P{UAS-Q93httexon1}4F1 [Q93 (24)] and w; P{UAS-Q20httexon1} [Q20 (24)] in the nervous system [using P{GawB}elavC155, elav-Gal4 (46)]. Control flies were progeny of flies expressing Q93 or Q20 under control of elav-GAL4, crossed to w1118 males that segregated from some stocks of other UAS-RNAi lines and were therefore representative of the genetic background of these lines. The analysis was based on 100 female flies per genotype divided in groups of 10 flies per vial. Flies were transferred to new vials and counted every 2 days. Survival curves were plotted using the Kaplan–Meier estimator. The statistical significance was calculated using the log-rank test (SPSS 11.0).
Pseudopupil analysis was performed at 4 days post-eclosion as previously described (26). To evaluate the effect of PSA overexpression, virgins of genotype y w; P{GMR-HD.Q120}2.4 (GMR-Q120) or y w; P{GMR-HD.Q23} [GMR-Q23 (25)] were crossed with males y w; P{gl-dPSA}A [gl-PSA (16)]. As controls, the above flies (GMR-Q120, GMR-Q23 and gl-PSA) were crossed with w1118 isogenic males (48). To evaluate the effect of PSA downregulation on neurodegeneration, virgins of genotype elav-GAL4C155; {GMR-HD.Q120}4.62/TM3 (elav-Gal4 GMR-Q120) or elav-GAL4C155; {GMR-HD.Q23} (elav-Gal4 GMR-Q23) were crossed with the same UAS-RNAi or w1118 control males as described for the survival assay, and the progeny were scored using the pseudopupil assay. Pictures were acquired using an ×100 objective (Zeiss Axioscope2 microscope). Comparisons were performed using paired t-tests using data from five to seven independent experiments, each based on approximately 10 individuals of each genotype, in which 15 ommatidia each were scored.
Protein degradation measurement
293A cells were seeded in six-well plates and transfected 24 h later (confluence 50%). Forty-eight hours after transfection, fresh culture medium containing 5 µCi/ml l-[3,5-3H]-tyrosine (Perkin Elmer) was applied for 24 h to label long-lived proteins. Cells were then washed twice for an hour with a chase medium containing 2 mm tyrosine (in order to limit reincorporation of the 3H-tyrosine and to allow degradation of short-lived proteins, total chase duration 2 h). Cells were then washed with either culture medium (basal condition) or HBSS (serum and amino acid starvation) containing 2 mm tyrosine and the PSA, proteasome, lysosomes or autophagy inhibitors for 1 h (pre-treatment before protein degradation measurement). Cells were then washed with the same medium and 200 µl of the medium was collected after 0, 1, 2, 3 and 4h for quantitation of 3H-tyrosine release (protein degradation measurement). Proteins were precipitated with TCA (10% final concentration) and pelleted. Radioactivity in the TCA-soluble supernatant was measured using a 1900TR liquid scintillation analyser (Packard). At the end of the measurement period, cells were solubilized in 0.2 N NaOH and an aliquot was taken to measure the residual radioactivity in the cells. Total radioactivity is the sum of the residual radioactivity in the cells and the TCA-soluble radioactivities at different time points. Protein breakdown rates were expressed as 3H-tyrosine released over time as a percentage of total 3H-tyrosine incorporated. Proteasomal, lysosomal and autophagy-dependent proteolysis rates were determined precisely as done before by treating cells with 1 μm bortezomib/velcade, 50 µm chloroquine or 10 mm 3MA, respectively.
PSA activity measurement
To prepare cytosolic extracts, 293A or MEF cells were washed with ice-cold PBS twice and scraped in ice-cold lysis buffer containing 50 mm phosphate buffer (pH 7.4), 0.5 mm CaCl2, 5 mm MgCl2, 1 mm DTT and 5% glycerol, collected in an eppendorf and homogenized manually on ice. This homogenate was spun at 10 000g for 15 min to remove the nuclear fraction and then at a higher speed of 100000g for an hour to remove membranous organelles. Protein concentration was obtained using Coomassie Plus Protein Assay (ThermoFisher). PSA activity was determined by monitoring for 5–20 min the increase in fluorescence (excitation 380 nm, emission 460 nm, SpectraMax M5, Molecular Devices) caused by hydrolysis of the substrate Ala-amc (Bachem, 100 µm final) in 100 µl of lysis buffer containing 2–5 µg of protein from the cell extract. Background from the fluorescence of Ala-amc in the absence of enzyme was subtracted.
To measure Ala-amc hydrolysis by intact cells, 293A cells were plated in 96-well plate adapted for fluorescence measurement, grown in culture medium without phenol red containing various concentrations of inhibitors for 24h. Cells were washed with fresh culture medium containing Ala-amc 100 µm and inhibitors and the increase in fluorescence was monitored for 1h. Background from the fluorescence of medium containing Ala-amc in the absence of cells was subtracted.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the Medical Research Council (Programme grant to D.C.R.), and the Wellcome Trust (Senior Fellowship to D.C.R.); A.L.G.'s laboratory is supported by grants from the National Institutes of Health/National Institute for Aging (AR055255 and GM51923), Harvard Neurodiscovery Center and the Ellison Foundation; R.H. is supported by Horlait-Dapsens Medical Foundation, Belgian Neurological Society, Hereditary Disease Foundation and Fonds National de la Recherche Scientifique; E.R.'s laboratory is supported by a Vidi grant from De Nederlandse Organisatie voor Wetenschappelijk Onderzoek—ZonMW and a grant from the Dutch Cancer Foundation. We would like to thank George Jackson for sending flies. Funding to pay the Open Access Charge was provided by The Wellcome Trust.
Supplementary Material
REFERENCES
- 1.Williams A., Jahreiss L., Sarkar S., Saiki S., Menzies F.M., Ravikumar B., Rubinsztein D.C. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top Dev. Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. doi:10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 2.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 3.Verhoef L.G., Lindsten K., Masucci M.G., Dantuma N.P. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. doi:10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 4.Qin Z.H., Wang Y., Kegel K.B., Kazantsev A., Apostol B.L., Thompson L.M., Yoder J., Aronin N., DiFiglia M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Genet. 2003;12:3231–3244. doi: 10.1093/hmg/ddg346. doi:10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 5.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. doi:10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 6.Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A.L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. doi:10.1016/S1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 7.Holmberg C.I., Staniszewski K.E., Mensah K.N., Matouschek A., Morimoto R.I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. doi:10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reits E., Neijssen J., Herberts C., Benckhuijsen W., Janssen L., Drijfhout J.W., Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. doi:10.1016/S1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 9.Saric T., Beninga J., Graef C.I., Akopian T.N., Rock K.L., Goldberg A.L. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J. Biol. Chem. 2001;276:36474–36481. doi: 10.1074/jbc.M105517200. doi:10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- 10.York I.A., Mo A.X., Lemerise K., Zeng W., Shen Y., Abraham C.R., Saric T., Goldberg A.L., Rock K.L. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. doi:10.1016/S1074-7613(03)00058-X. [DOI] [PubMed] [Google Scholar]
- 11.Beninga J., Rock K.L., Goldberg A.L. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. doi:10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 12.Constam D.B., Tobler A.R., Rensing-Ehl A., Kemler I., Hersh L.B., Fontana A. Puromycin-sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J. Biol. Chem. 1995;270:26931–26939. doi: 10.1074/jbc.270.45.26931. doi:10.1074/jbc.270.45.26931. [DOI] [PubMed] [Google Scholar]
- 13.Saric T., Graef C.I., Goldberg A.L. Pathway for degradation of peptides generated by proteasomes: a key role for thimet oligopeptidase and other metallopeptidases. J. Biol. Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. doi:10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 14.Stoltze L., Schirle M., Schwarz G., Schroter C., Thompson M.W., Hersh L.B., Kalbacher H., Stevanovic S., Rammensee H.G., Schild H. Two new proteases in the MHC class I processing pathway. Nat. Immunol. 2000;1:413–418. doi: 10.1038/80852. doi:10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 15.Bhutani N., Venkatraman P., Goldberg A.L. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007;26:1385–1396. doi: 10.1038/sj.emboj.7601592. doi:10.1038/sj.emboj.7601592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karsten S.L., Sang T.K., Gehman L.T., Chatterjee S., Liu J., Lawless G.M., Sengupta S., Berry R.W., Pomakian J., Oh H.S., et al. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. doi:10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Osada T., Ikegami S., Takiguchi-Hayashi K., Yamazaki Y., Katoh-Fukui Y., Higashinakagawa T., Sakaki Y., Takeuchi T. Increased anxiety and impaired pain response in puromycin-sensitive aminopeptidase gene-deficient mice obtained by a mouse gene-trap method. J. Neurosci. 1999;19:6068–6078. doi: 10.1523/JNEUROSCI.19-14-06068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hersh L.B., McKelvy J.F. An aminopeptidase from bovine brain which catalyzes the hydrolysis of enkephalin. J. Neurochem. 1981;36:171–178. doi: 10.1111/j.1471-4159.1981.tb02392.x. doi:10.1111/j.1471-4159.1981.tb02392.x. [DOI] [PubMed] [Google Scholar]
- 19.Hersh L.B., Smith T.E., McKelvy J.F. Cleavage of endorphins to des-Tyr endorphins by homogeneous bovine brain aminopeptidase. Nature. 1980;286:160–162. doi: 10.1038/286160a0. doi:10.1038/286160a0. [DOI] [PubMed] [Google Scholar]
- 20.Botbol V., Scornik O.A. Peptide intermediates in the degradation of cellular proteins. Bestatin permits their accumulation in mouse liver in vivo. J. Biol. Chem. 1983;258:1942–1949. [PubMed] [Google Scholar]
- 21.Towne C.F., York I.A., Neijssen J., Karow M.L., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Neefjes J.J., Rock K.L. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J. Immunol. 2008;180:1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 22.Kita H., Carmichael J., Swartz J., Muro S., Wyttenbach A., Matsubara K., Rubinsztein D.C., Kato K. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum. Mol. Genet. 2002;11:2279–2287. doi: 10.1093/hmg/11.19.2279. doi:10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 23.Raspe M., Gillis J., Krol H., Krom S., Bosch K., van Veen H., Reits E. Mimicking proteasomal release of polyglutamine peptides initiates aggregation and toxicity. J. Cell Sci. 2009;122:3262–3271. doi: 10.1242/jcs.045567. doi:10.1242/jcs.045567. [DOI] [PubMed] [Google Scholar]
- 24.Steffan J.S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B.L., Kazantsev A., Schmidt E., Zhu Y.Z., Greenwald M., et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. doi:10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 25.Jackson G.R., Salecker I., Dong X., Yao X., Arnheim N., Faber P.W., MacDonald M.E., Zipursky S.L. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. doi:10.1016/S0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 26.Franceschini N., Kirschfeld K. Pseudopupil phenomena in the compound eye of drosophila. Kybernetik. 1971;9:159–182. doi: 10.1007/BF02215177. doi:10.1007/BF02215177. [DOI] [PubMed] [Google Scholar]
- 27.Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. doi:10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 28.Kabuta T., Suzuki Y., Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J. Biol. Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. doi:10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 29.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. doi:10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. doi:10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 31.Sengupta S., Horowitz P.M., Karsten S.L., Jackson G.R., Geschwind D.H., Fu Y., Berry R.W., Binder L.I. Degradation of tau protein by puromycin-sensitive aminopeptidase in vitro. Biochemistry. 2006;45:15111–15119. doi: 10.1021/bi061830d. doi:10.1021/bi061830d. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 33.Ravikumar B., Berger Z., Vacher C., O'Kane C.J., Rubinsztein D.C. Rapamycin pre-treatment protects against apoptosis. Hum. Mol. Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. doi:10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. doi:10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 35.Taylor A. Aminopeptidases: structure and function. FASEB J. 1993;7:290–298. doi: 10.1096/fasebj.7.2.8440407. [DOI] [PubMed] [Google Scholar]
- 36.Kakuta H., Koiso Y., Nagasawa K., Hashimoto Y. Fluorescent bioprobes for visualization of puromycin-sensitive aminopeptidase in living cells. Bioorg. Med. Chem. Lett. 2003;13:83–86. doi: 10.1016/s0960-894x(02)00845-4. doi:10.1016/S0960-894X(02)00845-4. [DOI] [PubMed] [Google Scholar]
- 37.Kakuta H., Koiso Y., Takahashi H., Nagasawa K., Hashimoto Y. Novel specific puromycin-sensitive aminopeptidase inhibitors: 3-(2,6-diethylphenyl)2,4(1H, 3H)-quinazolinedione and N-(2,6-diethylphenyl)2-amino-4H-3,1-benzoxazin-4-one. Heterocycles. 2001;55:1433–1438. doi:10.3987/COM-01-9247. [Google Scholar]
- 38.Komoda M., Kakuta H., Takahashi H., Fujimoto Y., Kadoya S., Kato F., Hashimoto Y. Specific inhibitor of puromycin-sensitive aminopeptidase with a homophthalimide skeleton: identification of the target molecule and a structure-activity relationship study. Bioorg. Med. Chem. 2001;9:121–131. doi: 10.1016/s0968-0896(00)00231-5. doi:10.1016/S0968-0896(00)00231-5. [DOI] [PubMed] [Google Scholar]
- 39.McDermott J.R., Mantle D., Lauffart B., Kidd A.M. Purification and characterization of a neuropeptide-degrading aminopeptidase from human brain. J. Neurochem. 1985;45:752–759. doi: 10.1111/j.1471-4159.1985.tb04056.x. doi:10.1111/j.1471-4159.1985.tb04056.x. [DOI] [PubMed] [Google Scholar]
- 40.Mantle D., Lauffart B., Perry E.K., Perry R.H. Comparison of major cortical aminopeptidase activity in normal brain and brain from patients with Alzheimer's disease. J. Neurol. Sci. 1989;89:227–234. doi: 10.1016/0022-510x(89)90024-5. doi:10.1016/0022-510X(89)90024-5. [DOI] [PubMed] [Google Scholar]
- 41.Gonzales T., Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 1996;18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. doi:10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 42.Kanazawa T., Taneike I., Akaishi R., Yoshizawa F., Furuya N., Fujimura S., Kadowaki M. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J. Biol. Chem. 2004;279:8452–8459. doi: 10.1074/jbc.M306337200. doi:10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 43.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. doi:10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 44.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. doi:10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narain Y., Wyttenbach A., Rankin J., Furlong R.A., Rubinsztein D.C. A molecular investigation of true dominance in Huntington's disease. J. Med. Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin D.M., Goodman C.S. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. doi:10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 47.Schulz C., Perezgasga L., Fuller M.T. Genetic analysis of dPsa, the Drosophila orthologue of puromycin-sensitive aminopeptidase, suggests redundancy of aminopeptidases. Dev. Genes. Evol. 2001;211:581–588. doi: 10.1007/s00427-001-0194-z. doi:10.1534/genetics.104.026658. [DOI] [PubMed] [Google Scholar]
- 48.Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N., Johnson G., et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. doi:10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.