Abstract
Genome-wide association studies of human gene expression promise to identify functional regulatory genetic variation that contributes to phenotypic diversity. However, it is unclear how useful this approach will be for the identification of disease-susceptibility variants. We generated gene expression profiles for 22 184 mRNA transcripts using RNA derived from peripheral blood CD4+ lymphocytes, and genome-wide genotype data for 516 512 autosomal markers in 200 subjects. We screened for cis-acting variants by testing variants mapping within 50 kb of expressed transcripts for association with transcript abundance using generalized linear models. Significant associations were identified for 1585 genes at a false discovery rate of 0.05 (corresponding to P-values ranging from 1 × 10−91 to 7 × 10−4). Importantly, we identified evidence of regulatory variation for 119 previously mapped disease genes, including 24 examples where the variant with the strongest evidence of disease-association demonstrates strong association with specific transcript abundance. The prevalence of cis-acting variants among disease-associated genes was 63% higher than the genome-wide rate in our data set (P = 6.41 × 10−6), and although many of the implicated loci were associated with immune-related diseases (including asthma, connective tissue disorders and inflammatory bowel disease), associations with genes implicated in non-immune-related diseases including lipid profiles, anthropomorphic measurements, cancer and neurologic disease were also observed. Genetic variants that confer inter-individual differences in gene expression represent an important subset of variants that contribute to disease susceptibility. Population-based integrative genetic approaches can help identify such variation and enhance our understanding of the genetic basis of complex traits.
INTRODUCTION
The pace of susceptibility gene mapping for common diseases has greatly accelerated with the implementation of genome-wide association studies (GWAS) in large populations. Over the past 2 years, GWAS have identified no fewer than 800 genetic loci conferring risk of more than 100 diseases or disease-related traits (1), providing new insights into the pathogenesis of these disorders and opening new avenues for therapeutic targeting. However, among the current limitations of GWAS is their reliance on ‘indirect' association testing of fixed marker sets that capture the linkage disequilibrium (LD) patterns of common genetic variation. Although in some instances, the observed pattern of genetic association and presence of an obvious functional candidate variant (typically a non-synonymous coding variant) enables precise localization of the functional locus, such efficiency is rare. More commonly, regional LD extends across multiple genes, and the disease-associated variants serve as proxies for unrecognized non-coding variation, precluding claims of specific disease gene identification. Without reliable means to recognize functional non-coding variation directly, investigators are left speculating, often with multiple relevant candidates from which to choose (2).
GWAS of human gene expression represent an innovative approach for mapping functional non-coding variation. In 2001, Jansen and Nap (3) suggested that gene transcript abundance in relevant biologic tissues (measured using microarrays) could be considered a proximal intermediate phenotype for genetic mapping studies to identify expression quantitative trait loci (eQTLs). This integrative approach has intuitive appeal, as it is increasingly evident that a substantial proportion of the genetic variation influencing complex traits is regulatory (4). Initial studies in model organisms demonstrated the feasibility of eQTL mapping and its usefulness in identifying disease-susceptibility variants. More recently, linkage and association studies of the genetics of gene expression in human population studies demonstrated that transcript expression for many genes is highly heritable (5–7). Preliminary eQTL mapping studies, primarily using immortalized lymphoblastoid cell lines (LCLs), but also in primary cell types, have identified numerous cis- and trans-acting regulatory loci (7–10). Although in several instances, this approach has facilitated the identification of novel disease-susceptibility loci (11,12), the extent to which this approach can be used for disease gene mapping remains unclear.
Here, we present results from a genome-wide survey for cis-acting regulatory variants using RNA collected from peripheral blood CD4+ lymphocytes in a cohort of young adults with asthma. Not only do we demonstrate the feasibility of eQTL mapping in primary cell types collected in the clinical setting, but also provide evidence for strong enrichment of the observed expression-associated polymorphisms for disease-susceptibility variation, highlighting the utility of eQTL mapping for the identification of putative functional variation that contributes to the pathogenesis of complex genetic traits.
RESULTS
eQTL mapping in CD4+ lymphocytes
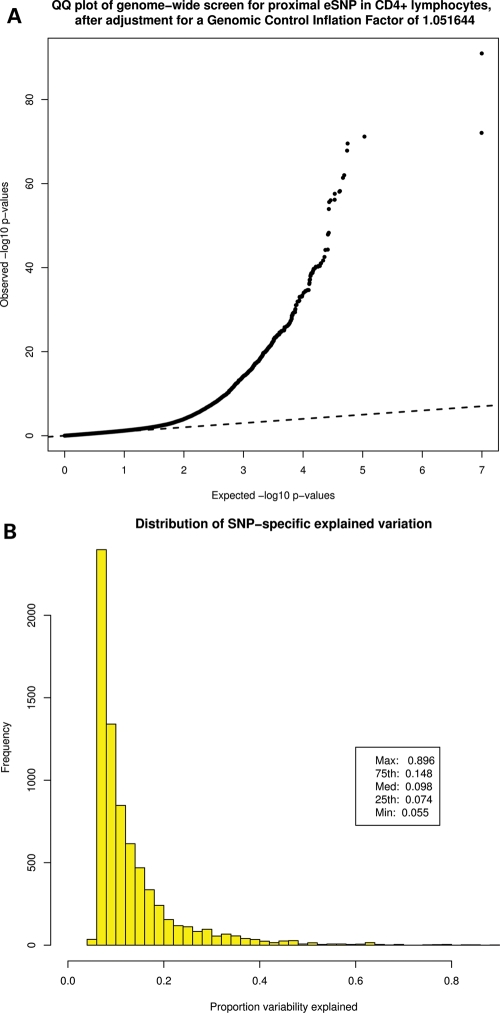
Expression data from primary peripheral blood CD4+ lymphocytes and genome-wide SNP genotype data were generated for 200 self-reported non-Hispanic white asthmatics. Of the genotyped SNPs, 258 314 mapped to within 50 kb of 19 451 transcripts with acceptable expression data (corresponding to 16 036 genes), resulting in 510 689 SNP–transcript association tests. Significant evidence for cis-acting regulatory variation was identified for 7274 SNP–transcript combinations, comprising 6706 SNPs corresponding to 1585 unique genes [9.88% of genes tested at a false discovery rate (FDR) threshold of 0.05, with corresponding P-values ranging from 0.0007 to 1 × 10−91, Fig. 1A]. The identified cis-acting expression-associated SNP (eSNP) explained a substantial proportion of the total variability in gene expression (Fig. 1B): the median of expression variability explained was 9.8% [inter-quartile range (IQR) 7.4–14.8%]. Greater than 25% of expression variability was explained by one SNP for 195 genes, and at least 50% of expression variability was explained by one SNP for 33 genes (Table 1). Although we note that the observed estimates of proportion variation explained (i.e. the eSNP-specific genetic effect size) fall between those observed in prior studies (7–10), such estimates are influenced by sample size, variance in gene expression and allele frequency distributions. It is therefore possible that given our modest sample size of 200 subjects, the observed median genetic effect size of 9.8% may be overestimated. Given our sample size and assuming 80% power to detect a genetic effect, at an FDR of 0.05 (corresponding to an eQTL P-value of 0.0007 in our data set), we calculated the expected genetic effect size to be 8.5%. Thus, our observed median effect size of 9.8% is similar to, but slightly higher than, the expected value, suggesting that our estimates are relatively good approximations of the likely underlying true distribution of magnitude of eSNP effects for common regulatory polymorphisms. However, we recognize that our sample size is underpowered to detect alleles with weak effects (i.e. variants that explain <2% of gene expression variability) and consequently that our observed distribution of genetic effects does not include such variants. A complete list of all identified eSNP is available in Supplementary Material, Table S1.
Figure 1.
(A) QQ plot of genome-wide screen for proximal eSNP in CD4+ lymphocytes. Dashed line denotes expected uniform (null) distribution. (B) Distribution of SNP-specific proportion of expression variation explained. Histogram includes 6706 SNPs with significant eQTL association findings.
Table 1.
Genes with eSNP explaining >50% of expression variability
| HUGO | SNP | Distance (kb from transcript) | MAF | Proportion variance explained | eQTL association P-value |
SNP under probe | AI | ||
|---|---|---|---|---|---|---|---|---|---|
| FDR | GC and FDR | Family-based | |||||||
| SCGB3A1 | rs2453176 | 42.7 | 0.088 | 0.896 | 1.24E − 90 | 5.39E − 86 | 3.90E − 08 | NI | |
| IPO8 | rs3910564 | 48.4 | 0.447 | 0.844 | 4.79E − 71 | 2.20E − 67 | 7.22E − 15 | Yes | Yes |
| C9orf135 | rs10521434 | 13.3 | 0.098 | 0.842 | 2.70E − 70 | 1.12E − 66 | 5.31E − 08 | NI | |
| CHURC1 | rs7143432 | −2.0 | 0.203 | 0.791 | 1.50E − 54 | 9.81E − 52 | 3.60E − 13 | Yes | |
| GYPE | rs1822841 | −15.9 | 0.263 | −0.785 | 2.41E − 60 | 3.12E − 57 | 6.70E − 11 | NI | |
| RPS23 | rs226206 | −18.8 | 0.280 | −0.776 | 1.69E − 56 | 1.39E − 53 | 1.39E − 09 | Yes | Yes |
| ANKDD1A | rs1628955 | −26.9 | 0.387 | 0.760 | 5.54E − 57 | 4.85E − 54 | 1.52E − 12 | Yes | |
| PTER | rs7909832 | 1.0 | 0.447 | −0.750 | 6.14E − 55 | 4.21E − 52 | 7.22E − 15 | No | |
| TMEM25 | rs11552421 | 28.6 | 0.138 | 0.746 | 4.71E − 55 | 3.28E − 52 | 2.17E − 06 | Yes | Yes |
| GSTM3 | rs10735234 | 1.1 | 0.443 | −0.693 | 9.67E − 47 | 2.60E − 44 | 8.75E − 14 | Yes | |
| FAM118A | rs104664 | 6.0 | 0.120 | 0.684 | 1.54E − 46 | 4.04E − 44 | 0.0002 | Yes | |
| PILRB | rs6955367 | 5.6 | 0.173 | 0.673 | 8.63E − 43 | 1.48E − 40 | 4.44E − 06 | NI | |
| FAM119B | rs10877013 | −1.3 | 0.345 | 0.659 | 5.31E − 41 | 7.40E − 39 | 2.28E − 12 | Yes | |
| LRAP | rs2161657 | 17.5 | 0.495 | −0.641 | 1.81E − 39 | 2.11E − 37 | 2.33E − 15 | NI | |
| FKSG14 | rs36133 | −33.5 | 0.370 | 0.637 | 1.04E − 38 | 1.10E − 36 | 4.93E − 10 | NI | |
| ACTA2 | rs1926196 | −41.2 | 0.498 | 0.635 | 5.01E − 37 | 4.31E − 35 | 1.49E − 13 | Yes | |
| WBSCR27 | rs4304218 | 3.9 | 0.293 | 0.627 | 1.74E − 39 | 2.02E − 37 | 7.77E − 15 | NI | |
| CPA5 | rs11761888 | 22.7 | 0.201 | −0.623 | 7.79E − 39 | 8.41E − 37 | 8.31E − 10 | Yes | No |
| SRI | rs1063964 | 13.9 | 0.296 | −0.622 | 3.27E − 38 | 3.26E − 36 | 5.51E − 09 | No | |
| USMG5 | rs11191666 | 28.9 | 0.428 | 0.621 | 7.88E − 37 | 6.64E − 35 | 5.36E − 08 | Yes | |
| MXRA7 | rs1005645 | 10.3 | 0.085 | 0.616 | 2.85E − 37 | 2.53E − 35 | 0.0005 | NI | |
| RPS6KA2 | rs9356529 | 4.6 | 0.273 | 0.582 | 1.43E − 36 | 1.16E − 34 | 8.04E − 08 | Yes | No |
| PRR17 | rs816922 | −18.5 | 0.065 | 0.579 | 4.26E − 33 | 2.34E − 31 | 6.15E − 05 | NI | |
| KCTD10 | rs9943689 | 24.8 | 0.200 | −0.571 | 1.79E − 32 | 9.13E − 31 | 1.66E − 07 | Yes | |
| NAPRT1 | rs1809148 | −2.6 | 0.140 | 0.567 | 7.30E − 33 | 3.89E − 31 | 8.94E − 07 | NI | |
| KRT1 | rs1567759 | −17.4 | 0.425 | −0.555 | 1.51E − 31 | 6.90E − 30 | 2.35E − 12 | NI | |
| LOC400566 | rs6565724 | 10.7 | 0.333 | −0.539 | 2.67E − 28 | 8.41E − 27 | 2.18E − 06 | NI | |
| C5orf35 | rs2591961 | 33.5 | 0.230 | −0.526 | 1.32E − 27 | 3.83E − 26 | 1.84E − 06 | Yes | |
| SLC25A29 | rs1059264 | 3.6 | 0.313 | 0.523 | 1.79E − 28 | 5.76E − 27 | 1.06E − 09 | Yes | |
| INPP5E | rs1127152 | −1.3 | 0.408 | −0.516 | 1.04E − 27 | 3.06E − 26 | 2.61E − 11 | Yes | |
| MRPL43 | rs2863095 | 0.8 | 0.208 | 0.513 | 1.07E − 27 | 3.15E − 26 | 1.48E − 10 | NI | |
| C1orf57 | rs3820124 | −1.6 | 0.208 | −0.511 | 2.10E − 27 | 5.96E − 26 | 5.72E − 07 | No | |
| HOXB2 | rs1042815 | 2.1 | 0.382 | −0.508 | 6.97E − 27 | 1.86E − 25 | 8.31E − 10 | NI | |
MAF, minor allele frequency. P-values derived from GLS-modeled population-based eQTL analysis are reported with FDR adjustment alone (FDR) and with both genomic control and FDR adjustment (GC and FDR). Proportion variance explained: sign indicates whether the major allele is associated with increased (+) or decreased (−) transcript abundance. AI, allelic expression observed in Verlaan et al. (52); NI, non-informative.
A series of statistical and technical validation procedures, including family-based testing and comparisons with results from previously published allelic imbalance and eQTL mapping studies, confirmed a large majority of identified eSNP. First, to assess whether population stratification could explain our results, even after adjustment for ancestry using the EIGENSTRAT program (13), we repeated the association testing incorporating parental genotype information in family-based association testing. The availability of parental genotype data for 154 of the 200 probands enabled confirmatory family-based association testing using PBAT (version 3.5) (14,15). A robust empirical variance estimator was used to calculate the variance in each family-based association test, which estimates correlation among family members when calculating the genetic variance–covariance matrix. Despite reduced statistical power resulting from the nearly 25% reduction in sample size, nearly three-fourths (74.9%) of the population-based associations were also significant using family-based association testing (P ≤ 0.05). The remaining 25.1% of the significant population-based tests not associated using family-based methods had lower minor allele frequency (mean of 0.277 versus 0.304 for those that were significant, P = 6.6 × 10−15) and were consequently tested in fewer informative families [means (standard deviations) of 79.3 (19.7) versus 84.4 (22.0), P < 10−16], suggesting that failure to associate using family-based methods was largely related to reduced statistical power and that observed associations identified by the population-based methods were not due to occult population stratification.
We next compared our results with a recent genome-wide survey for allelic expression (AE) in Epstein–Barr virus (EBV)-transformed lymphoblastic cell lines of Caucasian origin (16). This approach detects only cis-acting variation and provides an orthogonal test for heritable gene expression changes. The AE mapping was carried out in CEU-HapMap LCLs using three or more consecutive expressed marker SNPs as a trait (AE windows); ∼33 000 informative AE windows with local SNPs phased in HapMap Phase 2 data (rel. 22) were included, resulting in ∼6.6 million AE association tests. Of these, ∼200 000 SNP–AE window pairs (∼3% of tested pairs) showed association at P < 10−5 level, corresponding to permutation significance of 0.001, which corrects for multiple testing in each window. For details, see Ge et al. (16). We limited our analysis to variants assessed by both methods. Information was available for 3283 of the 7274 identified CD4+ eQTL associations, corresponding to 673 genes. Evidence for significant AE (P < 10−5) was confirmed for 818 transcript–SNP pairs (24.9%) in 217 genes (32.2%) when only 3 (0.06%) transcript–SNP pairs were expected to overlap by chance (P < 10−6). Given that prior observations suggest that only 50% of eQTL associations overlap with AE signals when both are performed in the same tissue (16), the degree of observed overlap between our eQTL associations in CD4+ lymphocytes with the AE findings in LCLs is considerable. Of particular note, 15 of the 19 informative variants with the strongest evidence of eQTL association (Table 1) demonstrated replication by AE mapping.
We also compared our results with two similar studies performed using EBV-transformed immortalized LCLs (8,9,12). The studies differ with respect to ascertainment strategies, sample size, expression and genotyping platforms, and methods of statistical analysis. However, considerable overlap in replicated associations was noted across studies: 18.8 and 39.9% of genes found to have cis-acting variants in the GeneVar and Dixon studies, respectively, were also noted in Childhood Asthma Management Program (CAMP). Although more genes with evidence of cis-acting regulatory variation were identified in the current analysis (45 and 296% more than the GeneVar and Dixon data sets, respectively), this is likely a function of between-study differences in sample size and defined significance thresholds rather than cell type.
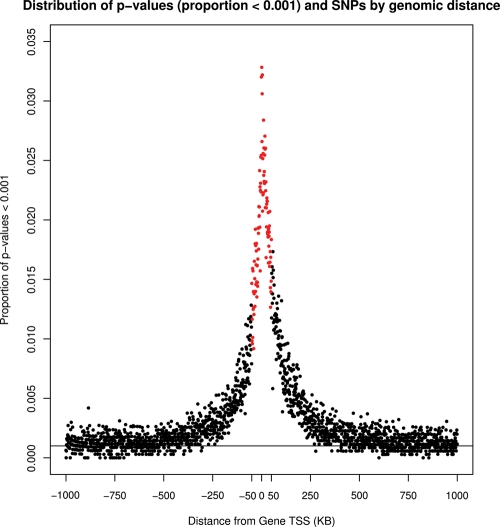
We examined the physical distribution of eSNP in our data set in relation to genomic distance from transcript (Fig. 2). Similar to prior observations (17,18), enrichment of expression-associated variation increased exponentially with increasing proximity to transcript, with a >30-fold increase over expectation under the null (horizontal line at P = 0.001 in Fig. 2) for variants within 1 kb of transcript. We also note persistent enrichment at 50 kb from transcript (generally 10–15-fold over null expectation), suggesting that more distal regulatory variation is notable in some genes. To further explore the extent of more distal regulatory variation, we extended our association testing to variants mapping as far as 1 Mb from transcript and found significant residual enrichment as far as 500 kb from transcript (P < 10−5), suggesting the existence of more remote regulatory variation in a subset of genes.
Figure 2.
The proportion of expression–trait association results with population-based P-values <0.001 are plotted against distance from transcript boundaries for SNP within 1 Mb of transcript (6.86 million association tests). SNP distances were rounded up to the nearest kilobase, resulting in 2000 bins. A lowess curve with smoothing span of 0.1 is plotted in solid black. The line at 0.001 on the ordinate reflects the proportion of SNP that would be expected under the null hypothesis of no association. The red data points denote the 50 kb window considered for our primary association studies.
eQTL mapping of disease-associated variation
A catalog of putative regulatory variants could facilitate mapping the genetic determinants of complex traits. We compared our results with a catalog of 285 published GWAS that includes results for 1544 SNPs associated with 198 traits (19). We found strong enrichment for cis-acting regulatory variation among the disease-associated genes: of the 783 genes with SNP–disease associations (as designated in the Catalog of Published GWAS, abstracted from primary GWAS publications) for which eQTL data were available in our data set, evidence of cis-acting regulatory variation was observed for 119 (15.2%), 1.63 times more frequent as expected by chance (95% confidence interval 1.32–2.00, Fisher's exact P = 6.41 × 10−6; see Supplementary Material, Table S2). The degree of eQTL enrichment was similar across strata of minor allele frequencies, with fold enrichment of 1.59 (P = 0.001), 1.69 (P = 0.0005), 1.61 (P = 0.001) and 1.78 (P = 0.0003) for minor allele frequency bins of 0.05–0.20, 0.21–0.30, 0.31–40 and 0.41–0.50, respectively, suggesting that the observed enrichment of eSNP cannot be explained by confounding by allele frequency. We note that the degree of significance of this fold enrichment may be slightly overestimated due to some non-independence of transcripts in our eQTL data set, due to their co-expression in CD4+ lymphocytes.
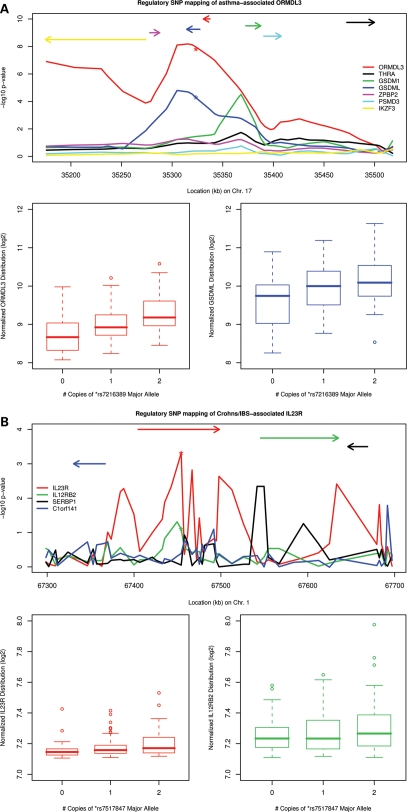
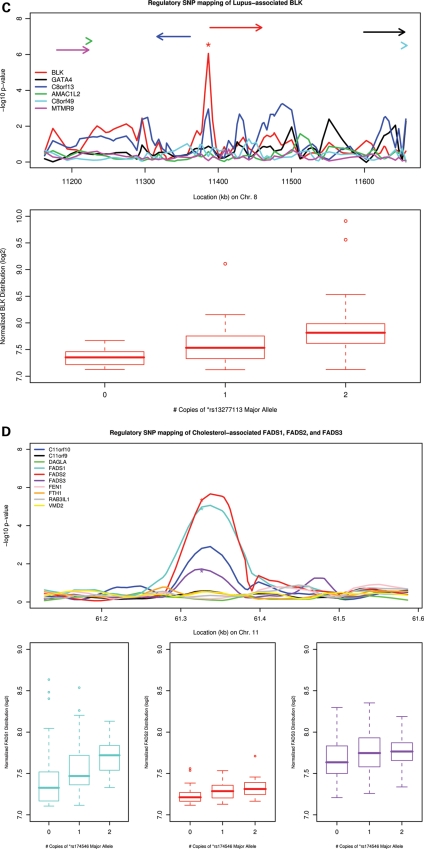
Of 119 GWAS disease-associated genes harboring cis-acting eQTL, we found 24 examples in which the variant most strongly associated with clinical phenotype also exhibits strong association with gene expression (Table 2). Regulatory function has been demonstrated previously for several of these variants, including multiple variants on chromosome 17q and ORMDL3/GSDML in asthma (12) and Crohn's disease (20) (Fig. 3A); rs7517847 and interleukin 23 receptor expression in inflammatory bowel disease (Fig. 3B) (21,22); and rs13277113 with BLK in systemic lupus erythematosus (Fig. 3C) (23). For example, a GWAS identified variants on chromosome 17q (including rs7216389) that confer increased susceptibility to asthma and were strongly associated with ORMDL3 expression in LCLs (12). We confirmed these findings in the CD4+ lymphocyte data set: rs7216389 rare-allele count was associated with ORMDL3 expression in a dose-dependent manner, explaining 16.5% of expression variability (P = 1.6 × 10−8). In our CD4+ lymphocyte data set, stronger evidence was demonstrated for variants immediately upstream of ORMDL3, including rs4795405, which is located 4.6 kb upstream of the ORMDL3 transcription start site, explaining 20.2% of ORMDL3 expression variability (P = 3.0 × 10−10). We note that the mechanism for the genetic co-regulation of ORMDL3 and GSDML was recently defined (24), resulting from regional gene regulation by allele-specific binding of insulator CTCF at SNP rs12936231, in strong LD with rs4795405 (D′ = 1.0, r2 = 0.69) (24). This polymorphism was strongly associated with asthma in three independent populations (combined P = 8.74 × 10−7), including the CAMP asthmatic probands (family-based association study, P = 0.007) (24).
Table 2.
CD4+ lymphocyte eSNP associated with complex genetic traits
| SNP | Published association studies |
CD4+ eSNP associations |
Dominanta eSNP association in CD4+ cells |
Relationship between trait eSNP and dominant eSNP |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Trait–GWAS association P-value | PubMed ID | Symbol | eQTL P-value | % variance explained | SNP | P-value | LD (r2) | Disease-eSNP P-value, conditioned on dominant eSNP | |
| Immune-related | ||||||||||
| rs3764021 | Type 1 diabetes | 5.00E−08 | 17554300 | CLECL1 | 1.04E−09 | −0.182 | rs10466829 | 6.09E−11 | 0.926 | 0.853 |
| rs4763879 | 2.00E−11 | 19430480 | 2.43E−05 | −0.094 | rs10466829 | 6.09E−11 | 0.709 | 0.596 | ||
| rs1701704 | Type 1 diabetes | 9.00E−10 | 18198356 | SUOX | 2.02E−06 | 0.118 | rs773107 | 7.59E−07 | 0.949 | 0.781 |
| rs2290400 | Type 1 diabetes | 6.00E−13 | 19430480 | GSDML | 4.07E−06 | 0.108 | Same SNP | NA | NA | NA |
| ORMDL3 | 3.05E−09 | 0.182 | rs4795405 | 3.00E−10 | 0.848 | 0.357 | ||||
| rs7216389 | Asthma | 9.00E−11 | 17611496 | ORMDL3 | 1.63E−08 | 0.165 | rs4795405 | 3.00E−10 | 0.886 | 0.868 |
| GSDML | 5.30E−05 | 0.083 | rs2290400 | 4.09E−06 | 0.968 | 0.33 | ||||
| rs2872507 | Crohn's disease | 5.00E−09 | 18587394 | ORMDL3 | 7.98E−09 | 0.172 | rs4795405 | 3.00E−10 | 0.87 | 0.574 |
| GSDML | 3.20E−05 | 0.088 | rs2290400 | 4.09E−06 | 0.894 | 0.593 | ||||
| rs2188962 | Crohn's disease | 2.00E−18 | 18587394 | SLC22A5 | 2.42E−13 | −0.261 | Same SNP | NA | NA | NA |
| rs7517847 | Crohn's disease | 3.00E−12 | 17435756 | IL23R | 4.23E−04 | 0.066 | Same SNP | NA | NA | NA |
| IBD | 4.00E−13 | 17068223 | Same SNP | NA | NA | NA | ||||
| rs3890745 | Rheumatoid arthritis | 1.00E−07 | 18794853 | MMEL1 | 1.33E−06 | 0.098 | Same SNP | NA | NA | NA |
| rs13277113 | SLE | 1.00E−10 | 18204098 | BLK | 2.83E−07 | 0.136 | rs2736340 | 1.52E−08 | 0.977 | 0.363 |
| rs1420101 | Plasma eosinophil count | 5.00E−14 | 19198610 | IL18R1 | 6.88E−04 | 0.064 | rs12998521 | 1.23E−04 | 0.945 | 0.475 |
| rs2066808 | Psoriasis | 1.00E−09 | 19169254 | TMEM4 | 4.23E−04 | 0.071 | Same SNP | NA | NA | NA |
| Metabolic | ||||||||||
| rs10889353 | Triglycerides | 3.00E−07 | 19060906 | DOCK7 | 1.57E−05 | −0.099 | rs2031373 | 6.62E−06 | 0.61 | 0.031 |
| LDL and total cholesterol | 4.00E−12 | 19060911 | ||||||||
| rs1167998 | Triglycerides | 2.00E−12 | 19060911 | 1.97E−05 | −0.098 | rs2031373 | 6.62E−06 | 0.613 | 0.037 | |
| rs174546 | LDL cholesterol | 1.00E−07 | 19060910 | FADS1 | 1.32E−05 | 0.093 | rs968567 | 3.70E−08 | 0.681 | 0.31 |
| FADS2 | 4.58E−06 | 0.11 | rs968567 | 1.80E−16 | 0.681 | 0.248 | ||||
| rs10838738 | Body mass index | 5.00E−09 | 19079261 | C1QTNF4 | 7.36E−08 | −0.151 | Same SNP | NA | NA | NA |
| Miscellaneous | ||||||||||
| rs6899976 | Height | 6.00E−06 | 18391951 | L3MBTL3 | 9.74E−21 | 0.373 | rs6569648 | 5.90E−23 | −0.825 | 0.006 |
| rs210138 | Testicular germ cell tumor | 1.00E−13 | 19483681 | FLJ43752 | 3.73E−06 | 0.111 | rs375555 | 2.45E−06 | −0.866 | 0.25 |
| rs8034191 | Lung cancer | 5.00E−20 | 18385738 | IREB2 | 2.87E−04 | 0.07 | Same SNP | NA | NA | NA |
| 3.00E−18 | 18385676 | |||||||||
| COPD | 1.00E−08 | 18780872 | ||||||||
| 1.00E−10 | 19300482 | |||||||||
| rs2290416 | ADHD | 9.00E−06 | 18821565 | NAPRT1 | 2.97E−04 | 0.068 | rs1809148 | 3.88E−35 | 0.071 | 9.85E−07 |
| rs3799977 | ADHD | 5.00E−06 | 18839057 | SUPT3H | 5.87E−13 | −0.25 | rs9472409 | 3.83E−15 | −0.852 | 0.216 |
| rs420259 | Bipolar disorder | 6.00E−08 | 17554300 | DCTN5 | 6.41E−07 | 0.127 | rs34514 | 7.20E−13 | −0.645 | 0.477 |
| rs4654748 | Vitamin B6 | 8.00E−18 | 19303062 | NBPF3 | 4.35E−07 | −0.136 | rs1780324 | 2.27E−10 | 0.818 | 0.968 |
| rs1780324 | Alkaline phosphatase | 7.00E−15 | 18940312 | NBPF3 | 2.26E−10 | −0.203 | Same SNP | NA | NA | NA |
| rs657152 | Alkaline phosphatase | 2.00E−30 | 18940312 | ABO | 5.56E−04 | −0.064 | rs11244079 | 4.96E−06 | 0.337 | 0.028 |
| rs505922 | TNF-α levels | 7.00E−40 | 18464913 | 4.26E−05 | −0.088 | rs11244079 | 4.96E−06 | 0.352 | 0.005 | |
| Venous thromboembolism | 4.00E−15 | 19278955 | ||||||||
| rs7112513 | Soluble transferrin receptor | 6.00E−09 | 18464913 | TAGLN | 2.47E−04 | 0.068 | rs236919 | 6.08E−06 | −0.432 | 0.049 |
| rs10919071 | QT interval | 1.00E−15 | 19305409 | ATP1B1 | 3.51E−24 | 0.458 | Same SNP | NA | NA | NA |
LD, linkage disequilibrium; IBD, inflammatory bowel disease; ADHD, attention deficit hyperactivity disorder; SLE, systemic lupus erythematosus; COPD, chronic obstructive pulmonary disease; sign of % variance explained denotes whether major allele is associated with increased (+) or decreased (−) transcript abundance.
aFor the purposes of this table and accompanying analysis, the dominant eSNP is defined as the eSNP with the lowest GLS association P-value at the target gene.
Figure 3.
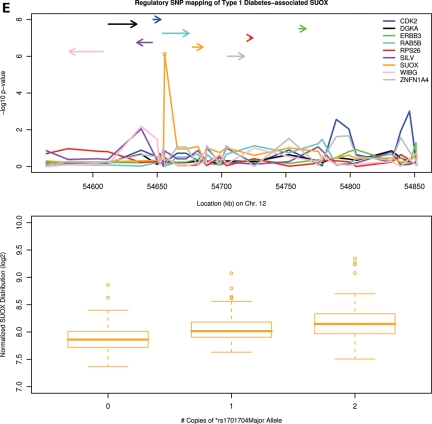
Examples of disease-associated eQTL findings in peripheral blood CD4+ lymphocytes. For each of five panels (A–E), upper figure displays the –log10P-values of population-based tests of association as a function of physical distance. Line colors correspond to results for individual genes (defined in legend), with relative position and strand orientation of genes depicted as arrows. Lower figure displays box plots of transcript intensity (log2) as a function of disease-associated SNP genotype, the position of which is denoted by (*) in the upper figure. (A) Asthma, Crohn's and type I diabetes-associated ORMDL3/GSDML; (B) Crohn's disease/inflammatory bowel disease-associated IL23R; (C) lupus-associated BLK locus; (D) lipid-associated FADS1, FADS2, and FADS3; and (E) type I diabetes-associated SUOX.
Although a substantial proportion of the disease-associated variants for which we have identified regulatory function relate to T-cell-associated diseases [including asthma, autoimmune disease, type I diabetes (T1D) and inflammatory bowel disease], regulatory variation for other disease classes, including serum lipid levels, anthropomorphic measures, cancer and neuropsychiatric disorders, were also noted. For example, strong associations of DOCK7 and FADS1 expression levels with lipid-associated risk alleles that have been previously demonstrated using liver-derived RNA samples were also noted in our CD4+ lymphocyte data set (Fig. 3D). Similarly, a recently identified determinant of height (25) (rs6899976) was strongly associated with the expression of L3MBTL3 in CD4+ lymphocytes, explaining 37.3% of the variance of L3MBTL3 expression (P = 9.7 × 10−21). No other neighboring gene's expression was associated with rs6899976 genotype, despite similar ranges of expression. L3MBTL3 is expressed in osteoblasts and embryonic bone, harbors multiple vitamin D response element-binding sites and is a target of down-regulation by 1,25-hydroxyvitamin D (26), all supporting L3MBTL3 as a plausible determinant of height.
We assessed whether the disease-associated eSNP was also the SNP most strongly associated with target gene expression (i.e. is the disease-associated eSNP the dominant eSNP of the target gene). In approximately one-third of the cases (10 of 31 instances), the disease-associated eSNP was the dominant eSNP for the target gene. Of the remaining variants, LD between the disease-associated eSNP and the dominant eSNP was very high (median 0.82, IQR 0.61–0.89), with only four instances where r2 was <0.60. To evaluate the implications of this, we repeated the eQTL association tests for the disease-associated eSNP, conditioned on the most dominant eSNP. In all but a few instances, this adjustment resulted in loss of evidence for eSNP association of the disease-associated SNP. These data suggest that in the vast majority of the cases, the disease-associated variants are tightly linked to the dominant regulatory variants underlying the variability in transcript abundance. Although few, there were several instances where evidence for eSNP association of the disease-associated variant persisted even after conditioning on the dominant eSNP, including rs505922 at the ABO locus (conditioned P-value 0.005), rs6899976 at L3MBTL3 (P = 0.006) and rs2290416 at NAPRT1 (P = 9.85 × 10−7). These results suggest that in these three instances, the target genes are controlled by at least two loci (the primary dominant eSNP and the disease-associated eSNP). It is possible in these cases that the primary eSNP may therefore independently contribute to disease susceptibility.
In addition to the 24 instances in which we found that a disease-associated SNP was an eSNP, we identified evidence for regulatory variation in 95 other disease-susceptibility genes (Supplemental Material, Table S2). Measures of LD in the HapMap samples of Western European ancestry were available for 72 eSNP/disease-associated SNP pairs. We found that in one-third of the cases (24 of 72), D′ between the disease- and expression-associated variants was 1, suggesting that the identified disease associations are likely due to a regulatory effect marked by a shared haplotype; although indirect association due to bystander effect of neighboring causative markers (27) could also explain these patterns. Among the remaining 48 cases, pair-wise LD between the disease and expression-associated variants was low (median r2 0.031, IQR 0.006–0.181), suggesting that in these cases, the GWAS-identified disease associations are independent from our observed expression associations and that testing of these novel regulatory variants for evidence of disease association may reveal heretofore unknown allelic heterogeneity at these disease-susceptibility loci.
SNP-under-probe effects
Interference of probe hybridization due to polymorphism in the target transcript sequence can bias expression association studies (28). Alignment of probe sequences with dbSNP (build 129) revealed a non-significant trend for excess SNP-under-probe effects, as 7.4% (123 of 1662) of expression-associated transcripts harbor at least one known polymorphism, compared with 6.2% (1105/17 789) among the remaining Illumina HumanRef8 v2 target sequences not associated with cis-acting variants (Fisher's exact test, P = 0.06). We note that repeating the cis-acting expression association studies after removal of probes with known sequence variation did not change our results (i.e. the P-value distributions were similar, resulting in similar FDR cut-offs). Moreover, we note that several eSNP-associated genes for which the Illumina probes have known polymorphism (i.e. IPO8, RPS23 and TMEM25; Table 1) demonstrated confirmed AE (a method immune to SNP-probe effects), suggesting that the observed expression association for these variants may not be due to SNP-under-probe effects. Similar to observations by others (29), these results suggest that though SNP-under-probe effects are present, they do not present a significant problem in the interpretation of our results.
DISCUSSION
Identification of functional non-coding genetic polymorphisms is an ongoing challenge in human genetics. Unlike coding variation, differentiating functional variants from among the millions of common human polymorphisms is hampered by the lack of accurate predictive algorithms and limited availability of functional sequence annotation. As we and others have already demonstrated, association mapping of regulatory polymorphisms in human populations is feasible with relatively small sample sizes and can facilitate the identification of disease-susceptibility loci (30). The enrichment for eQTL-associated variants within the catalog of GWAS studies observed in our analysis is very similar to that recently observed by Nicolae et al. (30) using LCL-derived eQTL data. These and future studies, in conjunction with complementary approaches like AE mapping, should facilitate annotation of regulatory sequence variation and help accelerate identification of functional disease-susceptibility variation.
Our analyses provide several insights regarding the role of regulatory variation in common disease. For several instances in which the mechanism of SNP–disease association was not discernible from the initial GWAS of disease susceptibility (due to the proximity of the variant to more than one plausible candidate), expression of only one gene was associated with disease-susceptibility genotype, implicating the expressed candidate over other neighboring loci in disease pathogenesis. For example, rs1701704 on chromosome 12q13 is strongly associated with T1D in three populations (P = 9.13 × 10−10) (3). Although rs1701704 resides within a 250 kb haplotype block that includes 13 genes (several of which are plausible biologic candidates for T1D), eQTL mapping revealed that the T1D-associated risk allele was associated with increased transcript abundance of only one gene—sulfite oxidase (SUOX)—explaining 11.8% of the variation in its expression (P = 0.0004, Fig. 3E). rs1701704 was not significantly associated with expression of any of the other candidates in this region (despite similar variances in expression for most), suggesting a functional role for SUOX over others loci in this region in T1D pathogenesis. Other notable examples include the association of the T1D-associated variant rs11052552 (31) with expression of C-type lectin-like 1 (CLECL1, P = 4.11 × 10−5) but not C-type lectin domain family 2D (CLEC2D, min P = 0.24); peripheral blood eosinophil level-associated rs1420101 (32) with expression of IL18R1 (P = 0.0489) but not IL1RL1 (min P = 0.74); and the height-associated rs6899976 with L3MBT3 but not SAMD3 (P = 0.20).
In several instances (including rs3890745 with MMEL1 and TNFRSF14 in rheumatoid arthritis and chromosome 17 variants with ORMDL3 and GSDML in asthma and Crohn's), the disease-associated variant appears to influence expression of two or more neighboring genes, with a similar proportion of expression explained for both genes. In these cases, it is not possible to discern which gene is the more likely to influence disease pathogenesis, and it is intriguing to speculate that it is in fact altered expression of both genes which affects disease susceptibility. Conversely, we also note several examples of confirmed eSNPs that are associated with susceptibility to more than one clinical trait (i.e. genetic pleiotropy) (12,20,33,34). Co-segregation of inflammatory bowel disease with rheumatoid arthritis (35,36) and asthma with Crohn's disease (36,37) suggests shared molecular determinants among these pairs of conditions. Our observation that identified susceptibility loci for these traits appear to regulate specific genes implies that the determinants of these pleiotropic disease associations operate downstream of the variants’ direct influences on gene expression and may be due to interactions with other susceptibility loci.
A primary distinction of our study from many others is our focus on a primary cell type (CD4+ lymphocytes) harvested directly from study subjects in the clinical setting. Although our samples were collected using a standardized protocol, and batching of samples during hybridization was randomized to avoid center-specific biases in our analyses, we anticipated substantial between-sample variability that could compromise our ability to detect SNP-specific genetic effects, considering that sample collection took place over an 18-month period at four clinical centers across the USA. However, we observed associations of substantial genetic effect on par with prior in vitro studies in LCLs, suggesting that eQTL mapping studies using clinical samples are robust to these unavoidable experimental influences. Successful mapping of eQTLs in peripheral blood mononuclear cells (11), adipose tissue (29) and cortical tissue (18) support this notion.
Although immortalized LCLs are a convenient, renewable source of study materials, recent evidence of substantial tissue-dependent differences in the patterns of regulatory variation suggests that the genetics of gene expression for the purposes of disease gene identification should be studied in disease-specific cell types (38). Comparisons of our results with the eQTL and AE studies, all conducted in LCLs, support these sentiments, in that although we saw considerable overlap for many loci (32.2% overlap with AE, 18.8% with GeneVar eQTLs and 39.9% with Dixon eQTLs), the majority of identified variants appear to be uniquely (or at least more easily) observed in the primary CD4+ lymphocytes. These observations provide further impetus for the development of large-scale integrative genome data sets in diverse cell types.
In summary, using a population-based integrative genomics genetic mapping approach, we have identified common genetic variants that influence the expression of 1585 genes in CD4+ lymphocytes. These polymorphisms represent an important subset of total genetic variation that can be prioritized for association testing of common traits, particularly those with an immune basis. Similar studies across various cell types and tissues could facilitate annotation of all regulatory variation relevant in health and disease.
MATERIALS AND METHODS
Study population and sample collection
The CAMP was a 4.5-year multicenter clinical trial of childhood asthmatics designed to evaluate the long-term efficacy and safety of inhaled asthma medications (39). Nine hundred sixty-three of the 1041 trial participants and 1518 parents provided DNA samples for genetic studies of asthma. The trial was followed by two 4-year observation studies—CAMP Continuation Study (CAMPCS) 1 and 2. RNA was obtained from peripheral blood CD4+ lymphocytes collected during year 3 or 4 CAMPCS/2 clinic visits at four CAMP study centers (Baltimore, Boston, Denver and St Louis). We isolated CD4+ lymphocytes using anti-CD4+ microbeads by column separation (Miltenyi Biotec, Auburn, CA, USA) (40) and extracted total RNA using the RNAeasy Mini Protocol (QIAGEN, Valencia, CA, USA) (41). High-quality RNA was available for 378 CAMP participants, of whom 200 were of self-reported non-Hispanic white ancestry and had available genotype data. Eighteen of the 200 subjects were siblings (i.e. nine sibling pairs). The remaining 78 subjects were of diverse ethnic backgrounds (including African-Americans, Hispanics and other). Owing to known between-population differences in gene expression and eQTL results (42–44), and because the largest group of subjects (African-Americans, n = 49) was too small and underpowered for separate eQTL studies, we restricted our analysis to the non-Hispanic white subset only. Approval was obtained from the Institutional Review Boards of Brigham and Women's Hospital (Boston, MA, USA) and each of the CAMP participating institutions. Informed consent was obtained from study participants if they were over the age of 18 years. Otherwise, informed consent was obtained from parents of participating children, and the child's assent was obtained prior to study enrollment.
Expression profiling
Expression profiles were generated with Illumina HumanRef8 v2 BeadChip arrays (Illumina, San Diego, CA, USA). Raw expression intensities generated using BeadStudio (v3.1.7) were processed with background adjustment with RMA convolution using the lumi package (45,46) and normalized using VSN (47). Two thousand two hundred eighty of 22 184 transcripts were not considered for analysis because they did not uniquely map or were located on sex chromosomes. The microarray data are available through the Gene Expression Omnibus repository (GEO, at http://www.ncbi.nlm.nih.gov/geo/, accession number GSE22324).
Genotyping
DNA was available for 200 subjects of self-reported white ancestry, as well as 292 of their parents (146 complete nuclear families). Genotyping of families was performed by Illumina on the Infinium II HumanHap550 Genotyping BeadChip. Forty-seven additional singletons were genotyped on the Human610W-Quad platform, with excellent genotype concordance rates among four subjects genotyped on both platforms (average 99.99%, minimum 99.89%). Association studies were limited to the set of overlapping markers present on both platforms passing quality control (QC). The merged data set comprised 516 512 autosomal SNPs. QC evaluations and data cleaning were performed using PLINK (48). Passing subjects all had a completion rate higher than 96.5% (average 99.8%). Markers were excluded (16 419 and 9022 from the 550 and 610 K platforms, respectively) for the following reasons: (i) probe sequences did not map uniquely to the hg18 genome build, (ii) poor genotype cluster separation, (iii) –log10(P-value) for Hardy–Weinberg equilibrium ≥8, (iv) marker completion rate <95%, (v) monomorphic markers or (vi) Mendelian error count ≥5. Unlike some other researchers (8), we did not apply non-specific gene filtering, as we have found that transcripts with lower overall intensities and/or narrower intensity distributions still displayed informative differential expression by genotype.
Statistical analysis
Population-based analyses were conducted using generalized least squares (GLS) models with the nlme R package, adjusting for age and sex. To control for potential population stratification, the model was further adjusted for four significant principal components derived from the genotype data with EIGENSTRAT (49). Data were managed using an smlSet from the Bioconductor package GGtools (50). The GLS model covariance matrix was modified to accommodate correlation among the few related probands (nine sibling pairs) in the data set, enabling accurate genetic effect size estimation. Specifically, the correlation structure that models within-family correlation was fixed to reflect the expected number of alleles shared by siblings. We used this approach rather than estimating the correlation empirically because we felt that the number of sibling pairs was too small (n = 9) to reliably estimate the correlation. We adjusted the test statistics by the genomic inflation factor λ, which was estimated to be 1.051644 from the distribution of test statistics from the 1 Mb cis-eQTL screen. We employed the FDR procedure (51) with a threshold of 0.05 to adjust for multiple comparisons. Comparisons of our results with those from prior GWAS studies were performed using a catalog of 285 published GWAS available at www.genome.gov/gwastudies (19). Estimates of LD were obtained using the HapMap samples of Western European ancestry (rel. 21). Fold enrichment of eSNP in the Catalog of Published GWAS (19) was assessed using the tabular data (accessed December 15, 2008) as abstracted from primary GWAS publications; significance was evaluated by Fisher's exact tests.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Heart, Lung and Blood Institute, National Institutes of Health (NIH/NHLBI, grant numbers R01 HL086601 and RC2 HL101543). The CAMP Genetics Ancillary Study is supported by the NIH/NHLBI (grant numbers U01 HL075419, U01 HL65899, P01 HL083069 and T32 HL07427) and by the National Institutes of Health (NIH) and National Center for Research Resources (NCRR) Colorado CTSA (grant number 1 UL1 RR025780). T.P. is a recipient of a Canada Research Chair and supported by the CIHR and Genome Canada/Quebec.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. Special thanks to Anne Plunkett, Teresa Concordia, Debbie Bull, Denise Rodgers and D. Sundstrom for their assistance with sample collection; to Huiqing Yin-DeClue, PhD, Michael McLane and Chris Allaire for their assistance with T cell isolations and RNA preparation; and to Ankur Patel for his assistance running the microarrays. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subject protections.
Conflict of Interest statement. All authors attest that they do not have any financial interests or connections, direct or indirect, that might raise the question of bias in the work reported or the conclusions, implications or opinions stated. A.M. is currently an employee at Merck Pharmaceuticals. Her work presented here was performed prior to her employment at Merck Pharmaceuticals and is unrelated to her current activities.
REFERENCES
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakonarson H., Qu H.Q., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–1146. doi: 10.2337/db07-1305. doi:10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 3.Jansen R.C., Nap J.P. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. doi:10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 4.Jais P.H. How frequent is altered gene expression among susceptibility genes to human complex disorders? Genet. Med. 2005;7:83–96. doi: 10.1097/01.gim.0000153665.55420.c3. doi:10.1097/01.GIM.0000153665.55420.C3. [DOI] [PubMed] [Google Scholar]
- 5.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. doi:10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 6.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. doi:10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 7.Brem R.B., Yvert G., Clinton R., Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. doi:10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 8.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. doi:10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 9.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. doi:10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan S., Huang R.S., Zhang W., Bleibel W.K., Roe C.A., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. doi:10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. doi:10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 12.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. doi:10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 13.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Gen. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1086/381563. [DOI] [PubMed] [Google Scholar]
- 14.Lange C., DeMeo D., Silverman E.K., Weiss S.T., Laird N.M. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. doi:10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Steen K., McQueen M.B., Herbert A., Raby B., Lyon H., Demeo D.L., Murphy A., Su J., Datta S., Rosenow C., et al. Genomic screening and replication using the same data set in family-based association testing. Nat. Genet. 2005;37:683–691. doi: 10.1038/ng1582. doi:10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 16.Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C., Gagne V., et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. doi:10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 17.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. doi:10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. doi:10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff L.A., Junkins H.A., Mehta J.P., Manolio T.A. A Catalog of Published Genome-Wide Association Studies. 2008. www.genome.gov/gwastudies. (accessed December 15, 2008) [Google Scholar]
- 20.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. doi:10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. doi:10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. doi:10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V., et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. doi:10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 24.Verlaan D.J., Berlivet S., Hunninghake G.M., Madore A.M., Lariviere M., Moussette S., Grundberg E., Kwan T., Ouimet M., Ge B., et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am. J. Hum. Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. doi:10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. doi:10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 26.Wang T.T., Tavera-Mendoza L.E., Laperriere D., Libby E., MacLeod N.B., Nagai Y., Bourdeau V., Konstorum A., Lallemant B., Zhang R., et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol. Endocrinol. 2005;19:2685–2695. doi: 10.1210/me.2005-0106. doi:10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 27.Reiner A.P., Barber M.J., Guan Y., Ridker P.M., Lange L.A., Chasman D.I., Walston J.D., Cooper G.M., Jenny N.S., Rieder M.J., et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. doi:10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberts R., Terpstra P., Li Y., Breitling R., Nap J.P., Jansen R.C. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. doi:10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi:10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 30.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. doi:10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudbjartsson D.F., Bjornsdottir U.S., Halapi E., Helgadottir A., Sulem P., Jonsdottir G.M., Thorleifsson G., Helgadottir H., Steinthorsdottir V., Stefansson H., et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. doi:10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 33.Kugathasan S., Baldassano R.N., Bradfield J.P., Sleiman P.M., Imielinski M., Guthery S.L., Cucchiara S., Kim C.E., Frackelton E.C., Annaiah K., et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. doi:10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plenge R.M., Seielstad M., Padyukov L., Lee A.T., Remmers E.F., Ding B., Liew A., Khalili H., Chandrasekaran A., Davies L.R., et al. TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N. Engl. J. Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. doi:10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen R., Robinson D., Jr, Paramore C., Fraeman K., Renahan K., Bala M. Autoimmune disease concomitance among inflammatory bowel disease patients in the United States, 2001–2002. Inflamm. Bowel. Dis. 2008;14:738–743. doi: 10.1002/ibd.20406. doi:10.1002/ibd.20406. [DOI] [PubMed] [Google Scholar]
- 36.Weng X., Liu L., Barcellos L.F., Allison J.E., Herrinton L.J. Clustering of inflammatory bowel disease with immune mediated diseases among members of a northern California-managed care organization. Am. J. Gastroenterol. 2007;102:1429–1435. doi: 10.1111/j.1572-0241.2007.01215.x. doi:10.1111/j.1572-0241.2007.01215.x. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein C.N., Wajda A., Blanchard J.F. The clustering of other chronic inflammatory diseases in inflammatory bowel disease: a population-based study. Gastroenterology. 2005;129:827–836. doi: 10.1053/j.gastro.2005.06.021. doi:10.1053/j.gastro.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 38.Hovatta I., Zapala M.A., Broide R.S., Schadt E.E., Libiger O., Schork N.J., Lockhart D.J., Barlow C. DNA variation and brain region-specific expression profiles exhibit different relationships between inbred mouse strains: implications for eQTL mapping studies. Genome Biol. 2007;8:R25. doi: 10.1186/gb-2007-8-2-r25. doi:10.1186/gb-2007-8-2-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N. Engl. J. Med. 2000;343:1054–1063. doi: 10.1056/NEJM200010123431501. doi:10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 40.Zorn E., Miklos D.B., Floyd B.H., Mattes-Ritz A., Guo L., Soiffer R.J., Antin J.H., Ritz J. Minor histocompatibility antigen DBY elicits a coordinated B and T cell response after allogeneic stem cell transplantation. J. Exp. Med. 2004;199:1133–1142. doi: 10.1084/jem.20031560. doi:10.1084/jem.20031560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu L., Tseng S., Horner R.M., Tam C., Loda M., Rollins B.J. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. doi:10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 42.Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., Cheung V.G. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. doi:10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storey J.D., Madeoy J., Strout J.L., Wurfel M., Ronald J., Akey J.M. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007;80:502–509. doi: 10.1086/512017. doi:10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Duan S., Kistner E.O., Bleibel W.K., Huang R.S., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. doi:10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. doi:10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 46.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. doi:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 47.Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl. 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Carey V.J., Davis A.R., Lawrence M.F., Gentleman R., Raby B.A. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25:1447–1448. doi: 10.1093/bioinformatics/btp169. doi:10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. doi:10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 52.Verlaan D.J., Ge B., Grundberg E., Hoberman R., Lam K.C., Koka V., Dias J., Gurd S., Martin N.W., Mallmin H., et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–127. doi: 10.1101/gr.084798.108. doi:10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.