Abstract
Juvenile polyposis (JP) is an autosomal dominant hamartomatous polyposis syndrome where affected individuals are predisposed to colorectal and upper gastrointestinal cancer. Forty-five percent of JP patients have mutations or deletions involving the coding regions of SMAD4 and BMPR1A, but the genetic basis of other cases is unknown. We set out to identify the JP gene in a large kindred having 10 affected members without SMAD4 or BMPR1A coding sequence mutations or deletions. We found a germline deletion segregating in all affected members, mapping 119 kb upstream of the coding region of BMPR1A by multiplex ligation-dependent probe amplification and comparative genomic hybridization. To further understand the genomic structure of BMPR1A, we performed 5′ RACE from lymphoblastoid cell lines and normal colon tissue, which revealed four non-coding (NC) exons and two putative promoters. Further analysis of this deletion showed that it encompassed 12 433 bp, including one promoter and NC exon. The activities of each promoter and deletion constructs were evaluated by luciferase assays, and the stronger promoter sequence analyzed for changes in JP patients without SMAD4 or BMPR1A alterations. A total of 6 of 65 JP probands were found to have mutations affecting this promoter. All probands examined had diminished BMPR1A protein by ELISA, and all promoter mutations but one led to significantly reduced luciferase activity relative to the wild-type promoter reporter. We conclude that we have identified the promoter for BMPR1A, in which mutations may be responsible for as many as 10% of JP cases with unknown mutations.
INTRODUCTION
Juvenile polyposis (JP) is a hamartomatous polyposis syndrome that has an autosomal dominant mode of inheritance, and affects ∼1 in 100 000 individuals (1). JP predisposes patients to developing hamartomatous polyps in the upper and lower gastrointestinal (GI) tract (2), and a >50% cumulative lifetime risk of developing colorectal cancer (3), as well as gastric cancer (3–6). Germline mutations in coding regions of BMPR1A and SMAD4 have been found in 45% of patients with JP, by sequencing (7,8) and mixed ligation-dependent probe amplification (MLPA) (9–11). Therefore, the genetic basis of JP is unknown in 55% of patients, raising the question of whether additional predisposing genes or other genetic mechanisms affecting non-coding (NC) regions of BMPR1A or SMAD4 are the cause.
Germline mutations in regulatory regions of genes are more difficult to detect than in coding regions, but have been shown to be important in several heritable cancer syndromes. In hereditary non-polyposis colorectal cancer (HNPCC), mutations in the promoter regions of hMLH1 and hMSH2 have been found in affected individuals, with one segregating in a three generation family (12–14). In Cowdens syndrome, up to 10% of patients have substitutions within the PTEN promoter region, with corresponding decreases in PTEN protein levels; all of these cases were sporadic, however (15). In breast cancer, a large deletion spanning the BRCA1 promoter region, exons 1a, 1b and 2, was found in eight affected members of a breast cancer family, and a subset of familial breast cancer cases are due to deletions involving the BRCA1 promoter (16).
Understanding the regulatory regions of SMAD4 and BMPR1A is not only important for us to be able to perform more comprehensive screening of patients at risk for JP, but also because these genes have been implicated in other diseases and in the pathogenesis of human cancer. In this study, we set out to characterize the NC elements of BMPR1A because of the finding of a germline deletion in a JP family affecting a region >100 kb upstream from the coding region.
RESULTS
Genetic screening studies
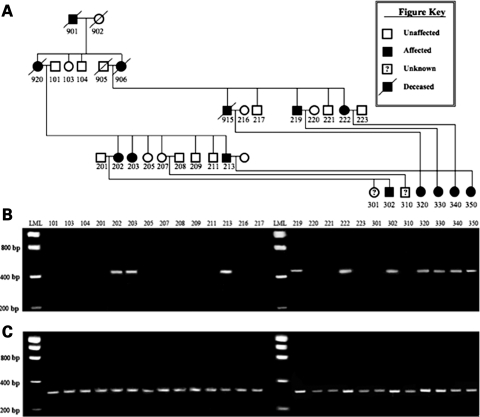
In order to search for a new JP gene, we collected DNA from a large Iowa JP family without germline mutations of BMPR1A or SMAD4 by sequencing, which consisted of 10 affected, 10 unaffected or unknown individuals and 7 spouses. We began performing a genome screen using simple tandem repeat polymorphisms, but MLPA analysis revealed a possible deletion in two probes from a NC exon of BMPR1A (Fig. 1A). We tested for this in the rest of the family, and all 10 affected members were found to have this deletion. No affected family members had upper GI polyps documented, or extraintestinal manifestations of JP, including macrocephaly, hereditary hemorrhagic telangiectasia or cardiac defects (Table 1).
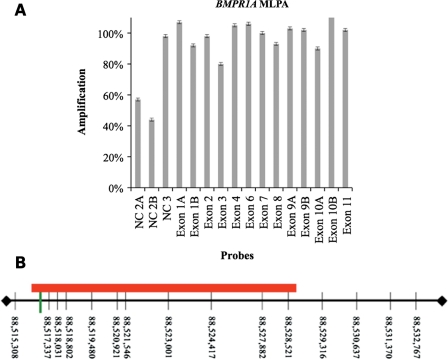
Figure 1.
MLPA and CGH result in JP family 19. (A) MLPA of the proband showing 50% decreased amplification of the probes within the second NC exon of BMPR1A. (B) aCGH array probe locations within the genomic DNA sequence upstream from BMPR1A on Chromosome 10 (vertical black lines). The red rectangle represents the deleted region and the green vertical line represents the two MLPA probes that had decreased amplification. The 5′ UTR of BMPR1A begins at 88 635 624.
Table 1.
Clinical characteristics of JP Family 19
| Proband | Age | Age at Dx | Colon polyps | Number of polyps | UGI polyps | Extraintestinal manifestations | Colon cancer | Colectomy |
|---|---|---|---|---|---|---|---|---|
| 219 | 66 | 31 | Y | >10 | − | − | − | − |
| 222 | 57 | a | N | a | − | Fibroadenoma breast | − | − |
| 202 | 55 | 50 | Y | <10 | − | Cyst on wrist | − | − |
| 203 | 57 | 39 | Y | <10 | − | Ganglion cyst | − | − |
| 213 | 60 | a | Y | a | − | − | − | − |
| 302 | 32 | 23 | Y | >10 | − | − | − | + |
| 320 | 31 | 19 | Y | >10 | − | − | − | − |
| 330 | 40 | 24 | Y | >10 | − | − | − | − |
| 340 | 32 | 24 | Y | >10 | − | − | − | − |
| 350 | 41 | 34 | Y | >10 | − | − | − | − |
aCarrier by virtue of affected offspring.
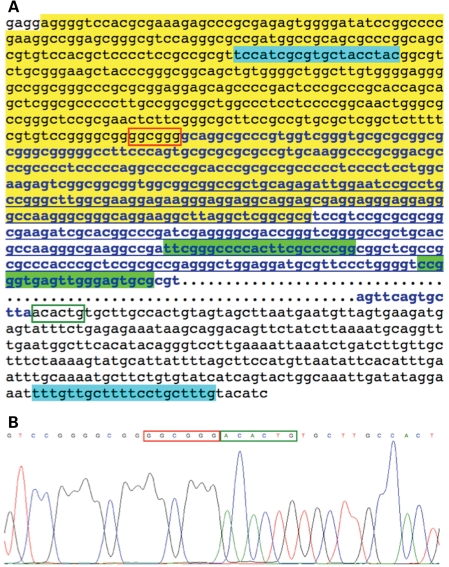
In order to further define the precise deletion, we performed CGH using DNA from an affected and unaffected member of this family. This revealed a heterozygous loss of the 10 probes between 88 515 308 and 88 529 316 on chromosome 10 (Fig. 1B). To determine the precise site of the deletion, PCR primers were chosen from just inside of the two flanking CGH probes with normal copy number. Amplification of a 515 bp product was possible in all affected members of the family (Fig. 2), despite the fact that this region spans 12 433 bp in genomic DNA. Sequencing of this product revealed that the 5′ breakpoint occurred within the second and third units of three 5 bp repeats (cgggg), and there were two 4 bp repeats (gctt) beginning 11 bp before and 2 bp after the 3′ end of the deletion (Fig. 3). Genomatix software predicted that the sequence spanning from 370 bp upstream to 150 bp downstream of the 5′ deletion breakpoint was the likely promoter region for BMPR1A. This region is located 119 379 bp upstream from the 5′UTR and first coding exon of BMPR1A.
Figure 2.
Segregation of the deletion within affected family members. (A) Pedigree of JP family 19; (B) PCR product representing the deleted segment, as demonstrated by the presence of the 515 bp product; (C) amplification products using control primers located within the deleted segment, representing the wild-type chromosome 10. LML, low-molecular weight ladder.
Figure 3.
Sequence surrounding the 12 433 bp deletion in JP family 19 upstream from BMPR1A. (A) The sequence in the box starts at base 88 515 880 and ends at 88 528 955. Bold letters in blue show the deleted region with …representing most of the nucleotides deleted. The sequence which is underlined is the second NC exon; the yellow area is the putative promoter sequence as predicted by Genomatix software; the blue highlighted areas show the primers used to sequence across the deletion; the green highlighted areas represent the two deleted MLPA probes (from Fig. 1B); the red box surrounds the six bases before the deletion, and the green open box the six bases after the end of the deletion; (B) chromatogram from the 515 bp PCR product sequence flanking the deleted segment.
5′ RACE studies
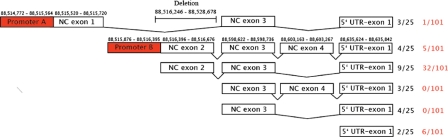
To better define putative promoter regions for BMPR1A, we performed 5′ RACE from both normal and JP patient lymphoblastoid cell lines (LCLs), and normal colon tissue. This revealed several different splicing isoforms and four different NC exons (Fig. 4). These results were compared with cDNAs and ESTs from the UCSC browser (http://genome.ucsc.edu/), and suggested two possible promoter regions for these different splice variants. Promoter B was the one with 150 bp deleted in members of JP family 19 and was also the most likely promoter for the majority of isoforms seen by 5′ RACE. Another potential promoter (promoter A) was identified just upstream of NC exon 1.
Figure 4.
Clones derived from 5′ RACE experiments. The exact positions of the NC exons, the 5′UTR and first coding exon, and the two promoters are located above the corresponding rectangles [from February 2009 Genome Reference Consortium (GRCh37), UCSC hg19 assembly]. The deletion found in family 19 is shown in the figure. The numbers in black at right represent the number of 5′ RACE clones identified and the numbers in red are the ESTs described in the UCSC genome browser (44 containing sequence 5′ to the coding exons and 57 with coding exons only).
Luciferase expression vectors and luminometry
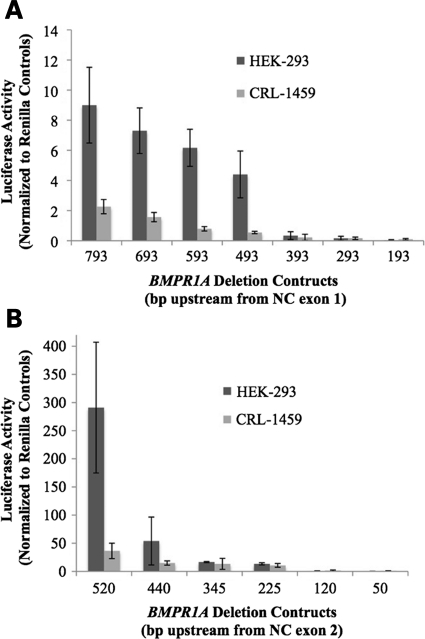
The candidate promoter sequences were cloned into luciferase vectors and transfected into human embryonic kidney cells (HEK-293) and colonic fibroblast cells (CRL-1459) to test their activity. Progressively shorter deletion constructs were also made to determine the regions of greatest promoter activity. Promoter A was found to have minimal activity, with a mean of eight normalized light units (lu) in HEK-293 and 2 lu in CRL-1459 (Fig. 5A). In contrast, promoter B was much stronger, with 291 lu in HEK-293 and 36 lu in CRL-1459. Deletion constructs showed a sharp drop in luciferase activity with loss of the 5′ 80 bp, and gradually decreasing to negligible activity in the terminal 120 bp construct (Fig. 5B).
Figure 5.
Luciferase expression from BMPR1A promoter deletion constructs. Results of luciferase assays (with standard deviations) from the various deletion constructs of promoters A (A) and B (B) from the two cell lines HEK-293 and CRL-1459; the size of each is the number of bases 5′ to the TSS. The 520 bp construct of promoter B did not have significant increase in the luciferase activity when NC exon 3 was added to the construct (data not shown).
Molecular screening for genetic alterations within the promoter region
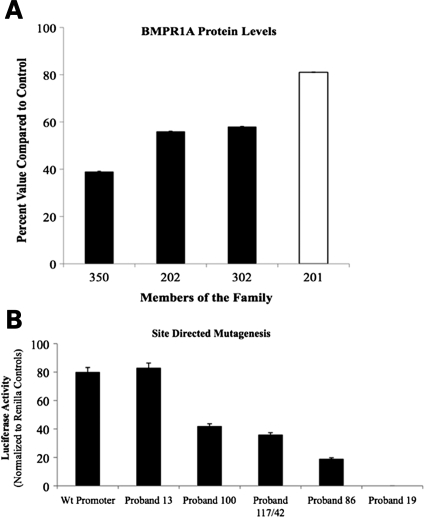
BMPR1A protein concentrations derived from LCLs from three affected and one unaffected member of family 19 with partial deletion of promoter B revealed that affected members had levels 40–60% of that seen in normal controls (Fig. 6A). Promoter B was then sequenced in 100 normal controls, and 64 additional JP probands who had previously not been found to have mutations in coding exons of BMPR1A or SMAD4, nor deletions by MLPA. Thirty-one of these probands had previously been sequenced for ENG and PTEN and no disease-causing mutations were identified (17). Two common polymorphisms were found (−434G/A and −61C/T; the most 3′ base in the promoter is position −1) within promoter B in 39 of 64 JP probands and 62 of 100 controls. Four different substitutions were found in five JP probands that were not found in controls (Table 2), and BMPR1A protein levels were reduced in LCLs from these patients (LCLs were available for three of the five probands with substitutions).
Figure 6.
BMPR1A protein levels in family 19 and luciferase activity for different alterations in the BMPR1A promoter. (A) Results of ELISA normalized to controls for BMPR1A with the corresponding standard deviations from four members of kindred 19; #350, 202 and 302 have the deletion and are affected (black), whereas 201 does not and is unaffected (white). (B) Luciferase activity in HEK-293 cells of genetic alterations in the BMPR1A promoter found in JP probands, created by SDM of the wild-type promoter construct.
Table 2.
GA of the BMPR1A promoter found in JP probands
| Proband | GA | RBSsa | Proteinb | Luciferasec |
|---|---|---|---|---|
| 19 | Del −150 to −1 | 2 MZF-1, TF2B, 2 SP-1 | 39% | 0 |
| 86 | −224 T/T>A/T | ZF5F | Not done | 24 |
| 100 | −306 G/G>C/G | E2F | 43% | 53 |
| 117 | −328 G/G>T/G | MZF-1 | 27% | 45 |
| 42 | −328 G/G>T/G | MZF-1 | Not done | 45 |
| 13 | −386 G/G>A/G | None identified | 31% | 104 |
GA, genetic alterations.
aPutative RBSs at the site of each substitution.
bBMPR1A protein levels relative to controls for the four JP probands evaluated.
cPercent of luciferase activity of SDM clone relative to the wild-type promoter construct.
Site-directed mutagenesis of transcription factor regulatory-binding sites
To confirm that the 150 bp deletion seen in our JP family or the substitutions found in other JP probands would have functional significance, the luciferase activity of constructs with these changes were compared with the full-length promoter sequence. Deletion of the terminal 150 bp led to complete loss of promoter activity, while the proband 86 (−482 A>T) construct had only 24% the activity of the wild-type promoter, the proband 100 (−320 G>C) 52%, and the probands 42 and 117 (−328 G>T) construct had 46% activity (Fig. 6B). In contrast, the proband 13 (−386 G>A) construct had similar luciferase activity to the wild-type promoter, while BMPR1A protein levels were reduced to 31% of controls.
In silico promoter analysis
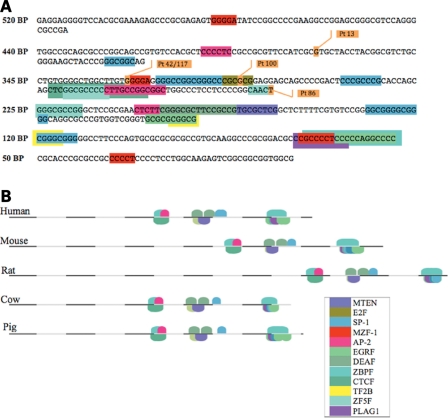
Gene2Promoter software predicted the 520 bp region just upstream from NC exon 2 to be a promoter for BMPR1A. This region had 78 CpG sites, and MatInspector software predicted many potential transcription factor regulatory-binding sites (RBSs) (Fig. 7A). This was evaluated by RegionMiner for orthologous sequences within 12 different species (Macaca mulatta, Pan troglodytes, Mus musculus, Rattus norvegicus, Equus caballus, Canis familiaris, Bos taurus, Sus scrofa, Monodelphis domestica, Ornithorhnuchus anatinus, Danio rerio and Gallus gallus) and 4 species were found to have levels of conservation ranging between 47.2 and 66.8% (Table 3). Sequences from these four species were then analyzed alongside the human BMPR1A promoter B sequence using FrameWorker, and two 10-element models were identified. These models had conserved transcription factors (EGRF, DEAF, AP2F, ZBPF, ZF5F, CTCF, SP1F and MTEN) across the five species. Both ZBPF and EGRF-binding sites were seen within the last 150 bp of promoter B (Fig. 7B), which would be lost in the deletion affecting JP family 19.
Figure 7.
(A) Map of the 520 nucleotides that make up BMPR1A promoter B. The various deletion constructs that were cloned into luciferase vectors are shown, and the putative transcription factor-binding sites are depicted. JP patient (Pt) substitutions are also shown. (B) Cross-species comparison of the BMPR1A promoter reveals two 10-element models that are conserved (key in inset for both A + B).
Table 3.
Percentage of conservation between species for the BMPR1A promoter B region
| Species | Percent conservation |
|---|---|
| M. musculus | 55.5 |
| R. norvegicus | 47.2 |
| B. taurus | 64.6 |
| S. scrofa | 66.8 |
DISCUSSION
By current sequencing methods and MLPA screening, ∼40–45% of JP patients have genetic alterations detected in BMPR1A or SMAD4 (9). The failure to detect mutations in over half of the JP patients is a serious obstacle for the screening of patients at risk, and our further understanding of the molecular basis of JP. JP kindred 19 clearly shows that deletion of the promoter and NC exon 2 of BMPR1A causes aberrant BMP signaling, with decreased levels of BMPR1A protein in affected members, and elimination of luciferase activity of this promoter construct.
Two candidate promoter regions were selected for study due to their high GC content, proximity to transcription start sites (TSS), the presence of regulatory sequences (such as a TATA-box, SP1 and AP2 sites), and gene structure. There were six different splice variants found, with only one containing NC exon 1, two with NC exon 2, five with NC exon 3, two with NC exon 4 and one without NC exons (Fig. 4). Isoforms with NC exon 1 splicing into NC exon 3 and NC exon 2 into 3 were found, but not 1 into 2, suggesting that there might be separate promoter regions upstream of NC exons 1 and 2. These regions were cloned into luciferase vectors, as were the sequences immediately upstream from NC exon 3 and the 5′UTR. No luciferase activity was found in the latter two regions (data not shown), while promoter A (upstream of NC exon 1) showed some weak activity and promoter B much greater activity in HEK-293 and CRL-1459 cells (Fig. 5).
The Gene2Promoter prediction software program identified several potential BMPR1A promoters; the highest quality prediction was that for promoter B and the next was promoter A. On the basis of this analysis and our luciferase data, promoter B appears to be the most important promoter for BMPR1A. However, we cannot rule out the possibility that promoter A (or others) plays a role in different tissues, during various stages of development, or in different contexts influenced by the abundance and combinations of specific transcription factors. Further evaluation of promoter B using MatInspector (http://www.genomatix.de/products/MatInspector/) and Promoter Scan 1.7 (http://www-bimas.cit.nih.gov/molbio/proscan/) identified several potential binding sites that may regulate transcription of BMPR1A (Fig. 7). One important transcription factor may be the myeloid zinc finger 1 factor (MZF1), for when this site was lost in the 440 bp deletion construct, there was a 59–81% drop in luciferase activity (in CRL-1459 and HEK-293, respectively) relative to the full-length 520 bp construct. Between the 440 and 225 bp constructs (which has 70–95% loss of activity relative to the 520 bp construct) there are four SP-1 sites, one E2F site, one AP-2 site and one additional MZF1-binding site. The 120 bp construct had >95% reduction in promoter activity relative to the 520 bp construct, and there is one SP-1 site, one AP-2, an RNA PolII transcription 2B-binding site (TF2B) and a core promoter motif 10 (MTEN) element located in the sequence between the 225 and 120 bp deletion constructs. Since the greatest decrease in luciferase activity was seen between the 520 and 225 bp constructs, and this region contains two MZF1-binding sites, two AP-2 sites, one E2F site, one ZF5F site and four SP-1-binding sites, we hypothesize that these are important regulatory elements for BMPR1A transcription. Also of interest was the fact that JP patient substitutions were found within MZF1, E2F and ZF5F sites. Finally, since in silico analysis determines potential binding sites for transcription factors based upon degrees of homology to various consensus sequences, further refinement of important regulatory sites can be assessed by cross-species comparison. This 520 bp region was conserved across several species, with 47% orthology to the brown rat and 68% to the wild boar. The 10 element transcription factor models were remarkably similar across these species, thereby confirming their potential relevance (Fig. 7B).
Deletions of BMPR1A have previously been reported in JP patients, but these have been very large, usually involving the entire gene. Delnatte et al. (18) described four patients with JP having early age of onset (1–18 months), both upper and lower GI polyps, and macrocephaly who all had contiguous deletions of both PTEN and BMPR1A. Salviati et al. (19) reported one patient with a 12 Mb deletion on chromosome 10q23 (encompassing 87 genes, including PTEN and BMPR1A), with lower GI polyps (but no upper GI), onset at age 5, and no macrocephaly. Menko et al. presented four additional patients with deletions encompassing PTEN, BMPR1A and at least eight other genes. All these patients had macrocephaly, facial dysmorphism and psychomotor retardation, three had juvenile polyps (presenting at ages 2–4 years) and the other had metastatic rectal cancer diagnosed at age 24. They concluded that the clinical phenotype seen with contiguous deletion of these two genes was variable, did not always fit the criteria for JP of infancy, and was not necessarily related to the size of the deletion (20).
Others have found deletions of BMPR1A by MLPA. Aretz et al. (11) found one JP patient with deletions of all probes of PTEN and BMPR1A (but did not describe the phenotype), one with loss of two BMPR1A NC exon probes and 2 exon 1 probes (age of onset 2 years, 11 total colonic polyps), and the final with loss of exon 1 probes (onset at age 8, total of >30 colonic polyps). van Hattem et al. (10) found one patient with deletion of BMPR1A exons 10 and 11, and two patients had deletions of all probes for both PTEN and BMPR1A, but no clinical information was provided. We previously described a patient with deletions of all MLPA probes for PTEN and BMPR1A (9). This individual was diagnosed with rectal bleeding at age 20 and had a partial colectomy at age 21, later had a gastrectomy for gastric polyps at age 54, and then developed rectal cancer at age 55 and had a total proctocolectomy for a T1N0M0 lesion. He had head circumference in the 95th percentile (55.7 cm), asymmetric mouth opening, inguinal hernia repairs in infancy and childhood, sensorineural hearing loss and had had trichoepitheliomas and dermatofibromas removed in the past. His parents and two siblings were phenotypically normal, without known polyps or rectal bleeding, and one of his sons had polyps and a colectomy performed at age 24.
Members of our JP family with a 12.4 kb deletion segregating in 10 affected individuals from two generations did not have upper GI polyps, macrocephaly or an early age of onset (Table 1). This deletion does not involve any other genes on 10q23, besides the NC region of BMPR1A. More specifically, this deletion results in loss of the last 150 bases of promoter B, which by luciferase experiments caused a >95% drop in activity relative to the 520 bp construct (Fig. 6B). This loss of activity is perhaps due to the loss of an SP-1 site from positions −113 to −120 or another from positions −150 to −161, which has the last base at position −150 within the large deletion, or the TF2B-binding site (from −119 to −124), or 2 MZF1 sites (Fig. 7A).
Besides affecting this promoter, the deletion also included the contiguous NC exon 2 and 12 kb of downstream intron. The role of BMPR1A NC exons is more difficult to define, and further study into their activity and molecular structure will help to unravel the role they play in transcription, translation, cellular localization, mRNA stability or interaction with other molecules involved in BMP signaling. Differential expression of the BMPR1A NC exons may be tissue specific, and may be an important influence on the variability and timing of gene and protein expression. Whether the loss of NC exon 2 in family 19 plays a role in the development of JP in addition to promoter B remains to be seen.
There are several examples of inherited GI cancer syndromes that result from genetic alterations of promoter regions of their predisposing genes. Up to 10% of patients with Cowden syndrome have substitutions within the PTEN promoter region with a corresponding 50% decrease in PTEN protein levels. Furthermore, these substitutions lead to increased phosphorylation of Akt, which is believed to lead to increased activity of the pro-proliferative PI3K/Akt pathway, possibly leading to the genesis of cancer in patients with Cowden's sydnrome (15). All of these cases were sporadic, however, and inheritance of these mutations within families was not demonstrated.
In HNPCC, Shin et al. described hMLH1 and hMSH2 core promoter mutations in 96 cases or suspected cases, and found germline changes in the hMSH2 promoter that segregated within affected members of 2 families. One was an A insertion at −80 in an affected mother and son but not the unaffected daughter, and the other was a G-C transversion at −225 in the proband of a family with multiple cancers. The former change reduced the activity of a luciferase reporter vector by 82%, and was within an E1A-F consensus sequence (12). Yan et al. screened 37 suspected HNPCC cases for changes in the promoters of hMSH2, hMLH1 and hMSH6, since mutations could not be found in the coding regions. They found a deletion of GT at −78–9 in the promoter of hMSH2 segregating in three affected members of an HNPCC family. This mutation led to an 87% reduction in luciferase activity, and also appeared to affect E1A-F binding (14). Green et al. found a C to T heterozygous mutation at position −42 upstream of the translation start site in the promoter region of hMLH1 in two affected members of an HNPCC family and another individual with adenomatous polyps. This change affected a putative Myb-binding site, and led to reduced luciferase activity relative to the wild-type sequence (13).
Consequently, there is precedent for promoter mutations causing autosomal dominant diseases, although these are relatively uncommon. However, greater understanding of NC regions of DNA will likely lead to many more such discoveries in the future. The specific example described here of a 12 kb deletion 119 kb upstream from the coding region of BMPR1A segregating in all 10 affected members of a JP family is particularly striking. The potential significance of alterations of the promoter was confirmed by the finding that 5 of 64 additional JP probands without coding region mutations of BMPR1A or SMAD4 had single base pair substitutions associated with reduced BMPR1A protein levels and reduced luciferase activity. Therefore, we conclude that BMPR1A promoter mutations cause JP, and may be responsible for up to 10% of genetically undefined JP cases.
MATERIALS AND METHODS
DNA screening and sequencing
All JP patient blood samples were collected under a protocol approved by the Institutional Review Board at the University of Iowa. Genomic DNA was extracted from peripheral blood by salting out (21) or the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN, USA), and from LCLs by the Qiagen AllPrep DNA mini kit (Qiagen, Valencia, CA, USA). DNA was amplified using primers flanking each intron-exon boundary of the 11 coding exons of BMPR1A and SMAD4, and products were purified using Qiagen spin columns (Qiagen). PCR products were sequenced by dideoxy cycle sequencing followed by electrophoresis through an ABI model 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA).
Individuals with no identifiable mutations by sequencing were screened by MLPA for exonic deletions (MRC Holland, Amsterdam). The large deletion identified in a JP proband by the MLPA method at the 5′ end of BMPR1A was further characterized using an oligonucleotide array comparative genomic hybridization (aCGH) chip (HG18_WG_CGH_5 of 8 array; NimbleGen Systems, Madison, WI, USA), which has probes spaced approximately every 700 bp. Primers were designed using the Primer3 program (http://frodo.wi.mit.edu/) (22) to make probes flanking the deletion as determined by the aCGH chip.
Protein analysis
Sandwich ELISA was performed in the four members of family 19 to determine relative protein levels of BMPR1A using capture antibodies BMPR1A-MAB2406 (R&D Systems, Minneapolis, MN, USA), and for actin as a control using Actin-600-401-886 (Rockland Immunochemials, Gilbertsville, PA, USA). The detection antibodies used were BMPR1A-ab59947-100 (Abcam, Cambride, MA, USA) and Actin-A5316-Sigma (Sigma). Quantification of protein was performed by measuring optical density as read at 450 nm in a Spectra Max Plus 384 (Molecular Devices, Sunnyvale, CA, USA) after addition of secondary antibody conjugated with HRP (Anti-Rabbit-HRP-sc-2030 or Anti-Mouse-HRP-sc-3697; SantaCruz Biotechnology, Santa Cruz, CA, USA).
5′ RACE
cDNA was created using gene-specific primers from RNA extracted from a control LCL and fresh colon tissue, using the Invitrogen 5′ RACE kit (Invitrogen). Clones with inserts were sequenced to determine different splice variants, potential TSS and the most abundant mRNA isoforms.
Luciferase analysis
The putative promoter regions were evaluated using the Gene2Promoter program from Genomatix (http://www.genomatix.de) and Promoter Scan 1.7 (http://www-bimas.cit.nih.gov/molbio/proscan/). Deletion constructs from the putative promoters were then amplified from genomic DNA by PCR using successively shorter sequences upstream from each putative TSS. The primers used to amplify the deletion constructs had a MluI restriction endonuclease site incorporated into the 5′ end and BglII at the 3′ end. The PCR products were then ligated into the pGL3 luciferase basic reporter vector (Promega, Madison, WI, USA).
Five micrograms of pGL3 constructs was transfected using Transfast (Promega) in a 1:2 ratio into a normal human CRL-1459 and the HEK-293 cell lines (American Type Culture Collection, Manassas, VA, USA), and co-transfected with 1 μg of control Renilla luciferase vector with CMV promoter (pRL-CMV; Promega). Cells were incubated with MEM with 10% serum and penicillin/streptomycin/amphotericin for 72 h. Triplicate reactions were performed for each construct.
Cells were lysed and firefly luciferase activity was measured at 562 nm for 10 s (using the Dual-Luciferase Reporter Assay kit, Promega) with the TD 20/20 luminometer (Turner BioSystems, Sunnyvale, CA, USA). Renilla luciferase activity was then measured at 480 nm for 10 s. Firefly luciferase activity for each construct was determined by subtracting the background firefly luciferase activity from the control pGL3 basic vector and normalizing to the Renilla luciferase activity.
Promoter evaluation
MatInspector software (www.genomatix.de/products/MatInspector) was used to identify potential RBSs from each promoter sequence, RegionMiner to search for orthologous sequences in different species, and FrameWorker to identify different transcription factor-binding modules (www.genomatix.de) (23–26). The promoter B region was screened by sequencing in 64 JP probands that did not have any coding mutations in both SMAD4 and BMPR1A or large exonic deletions by MLPA, and 100 normal controls. Site-directed mutagenesis (SDM) of potential RBSs was performed using the QuikChange II kit to evaluate the luciferase activity of mutations in the promoter (Stratagene, La Jolla, CA, USA).
FUNDING
This work was supported by NIH 1R01CA136884-01, the Roy J. Carver Charitable Trust and the Susser family.
ACKNOWLEDGEMENTS
We are grateful to all the patients who donated samples for this study and to the physicians and genetic counselors who follow them.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Burt R.W., Bishop D.T., Lynch H.T., Rozen P., Winawer S.J. Risk and surveillance of individuals with heritable factors for colorectal cancer. Bull. World Health Organ. 1993;68:655–664. [PMC free article] [PubMed] [Google Scholar]
- 2.Jass J.R. Pathology of polyposis syndromes with special reference to juvenile polyposis. In: Utsunomiya J., Lynch H.T., editors. Hereditary Colorectal Cancer. Tokyo, Japan: Springer; 1990. pp. 343–350. [Google Scholar]
- 3.Giardiello F.M., Hamilton S.R., Kern S.E., Offerhaus G.J.A., Green P.A., Celano P., Krush A.J., Booker S.V. Colorectal neoplasia in juvenile polyposis or juvenile polyps. Arch. Dis. Child. 1991;66:971–975. doi: 10.1136/adc.66.8.971. doi:10.1136/adc.66.8.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn M.C., Pricolo V.E., DeLuca F.G., Bland K.I. Malignant potential in intestinal juvenile polyposis syndromes. Ann. Surg. Oncol. 1995;2:386–391. doi: 10.1007/BF02306370. doi:10.1007/BF02306370. [DOI] [PubMed] [Google Scholar]
- 5.Howe J.R., Mitros F.A., Summers R.W. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann. Surg. Oncol. 1998;5:751–756. doi: 10.1007/BF02303487. doi:10.1007/BF02303487. [DOI] [PubMed] [Google Scholar]
- 6.Friedl W., Uhlhaas S., Schulman K., Stolte M., Loff S., Back W., Mangold E., Stern M., Knaebel H-P., Sutter C., et al. Juvenile polyposis: massive gastric polyposis is more common in MADH4 mutation carriers than in BMPR1A mutation carriers. Hum. Genet. 2002;111:108–111. doi: 10.1007/s00439-002-0748-9. doi:10.1007/s00439-002-0748-9. [DOI] [PubMed] [Google Scholar]
- 7.Howe J.R., Roth S., Ringold J.C., Summers R.W., Jarvinen H.J., Sistonen P., Tomlinson I.P.M., Houlston R., Bevan S., Mitros F.A., et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. doi:10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 8.Howe J.R., Bair J.L., Sayed M.G., Anderson M.E., Mitros F.A., Petersen G.M., Velculescu V.E., Traverso G., Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 2001;28:184–187. doi: 10.1038/88919. doi:10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 9.Calva-Cerqueira D., Chinnathambi S., Pechman B., Bair J., Larsen-Haidle J., Howe J. The rate of germline mutations and large deletions of SMAD4 and BMPR1A in juvenile polyposis. Clin. Genet. 2008;75:79–85. doi: 10.1111/j.1399-0004.2008.01091.x. doi:10.1111/j.1399-0004.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 10.van Hattem W.A., Brosens L.A., de Leng W.W., Morsink F.H., Lens S., Carvalho R., Giardiello F.M., Offerhaus G.J. Large genomic deletions of Smad4, Bmpr1a And Pten in juvenile polyposis. Gut. 2008;57:623–627. doi: 10.1136/gut.2007.142927. doi:10.1136/gut.2007.142927. [DOI] [PubMed] [Google Scholar]
- 11.Aretz S., Stienen D., Uhlhaas S., Stolte M., Entius M.M., Loff S., Back W., Kaufmann A., Keller K.M., Blaas S.H., et al. High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J. Med. Genet. 2007;44:702–709. doi: 10.1136/jmg.2007.052506. doi:10.1136/jmg.2007.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin K.H., Shin J.H., Kim J.H., Park J.G. Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res. 2002;62:38–42. [PubMed] [Google Scholar]
- 13.Green R.C., Green A.G., Simms M., Pater A., Robb J.D., Green J.S. Germline hMLH1 promoter mutation in a Newfoundland HNPCC kindred. Clin. Genet. 2003;64:220–227. doi: 10.1034/j.1399-0004.2003.t01-1-00110.x. doi:10.1034/j.1399-0004.2003.t01-1-00110.x. [DOI] [PubMed] [Google Scholar]
- 14.Yan H., Jin H., Xue G., Mei Q., Ding F., Hao L., Sun S.H. Germline hMSH2 promoter mutation in a Chinese HNPCC kindred: evidence for dual role of LOH. Clin. Genet. 2007;72:556–561. doi: 10.1111/j.1399-0004.2007.00911.x. doi:10.1111/j.1399-0004.2007.00911.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X.P., Waite K.A., Pilarski R., Hampel H., Fernandez M.J., Bos C., Dasouki M., Feldman G.L., Greenberg L.A., Ivanovich J., et al. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am. J. Hum. Genet. 2003;73:404–411. doi: 10.1086/377109. doi:10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swensen J., Hoffman M., Skolnick M.H., Neuhausen S.L. Identification of a 14 kb deletion involving the promoter region of BRCA1 in a breast cancer family. Hum. Mol. Genet. 1997;6:1513–1517. doi: 10.1093/hmg/6.9.1513. doi:10.1093/hmg/6.9.1513. [DOI] [PubMed] [Google Scholar]
- 17.Howe J.R., Larsen Haidle J., Lal G., Bair J., Song C., Pechman B., Chinnathambi S., Lynch H.T. ENG Mutations in MADH4/BMPR1A Mutation Negative Patients with Juvenile Polyposis. Clin. Genet. 2007;71:91–92. doi: 10.1111/j.1399-0004.2007.00734.x. doi:10.1111/j.1399-0004.2007.00734.x. [DOI] [PubMed] [Google Scholar]
- 18.Delnatte C., Sanlaville D., Mougenot J.-F., Vermeesch J.-R., Houdayer C., de Blois M.C., Genevieve D., Goulet O., Fryns J.-P., Jaubert F., et al. Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am. J. Hum. Genet. 2006;78:1066–1074. doi: 10.1086/504301. doi:10.1086/504301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salviati L., Patricelli M., Guariso G., Sturniolo G.C., Alaggio R., Bernardi F., Zuffardi O., Tenconi R. Deletion of PTEN and BMPR1A on chromosome 10q23 is not always associated with juvenile polyposis of infancy. Am. J. Hum. Genet. 2006;79:593–596. doi: 10.1086/507151. doi:10.1086/507151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menko F.H., Kneepkens C.M.F., de Leeuw N., Peeters E.A.J., Van Maldergem L., Kamsteeg E.J., Davidson R., Rozendaal L., Lasham C.A., Peeters-Scholte C.M.P., et al. Variable phenotypes associated with 10q23 microdeletions involving the PTEN and BMPR1A genes. Clin. Genet. 2008;74:145–154. doi: 10.1111/j.1399-0004.2008.01026.x. doi:10.1111/j.1399-0004.2008.01026.x. [DOI] [PubMed] [Google Scholar]
- 21.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. doi:10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. doi:10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 24.Tarling E., Salter A., Bennett A. Transcriptional regulation of human SREBP-1c (sterol-regulatory-element-binding protein-1c): a key regulator of lipogenesis. Biochem. Soc. Trans. 2004;32:107–109. doi: 10.1042/bst0320107. doi:10.1042/BST0320107. [DOI] [PubMed] [Google Scholar]
- 25.Tasheva E.S., Klocke B., Conrad G.W. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol. Vis. 2004;10:758–772. [PubMed] [Google Scholar]
- 26.Werner T., Nelson P.J. Joining high-throughput technology with in silico modelling advances genome-wide screening towards targeted discovery. Brief. Funct. Genomic. Proteomic. 2006;5:32–36. doi: 10.1093/bfgp/ell010. doi:10.1093/bfgp/ell010. [DOI] [PubMed] [Google Scholar]