Summary
DNA methylation is an epigenetic modification associated with gene silencing. In Arabidopsis, DNA methylation is established by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), which is targeted by small interfering RNAs through a pathway termed RNA-directed DNA methylation (RdDM)[1, 2]. Recently, RdDM was shown to require intergenic noncoding (IGN) transcripts that are dependent on the Pol V polymerase. These transcripts are proposed to function as scaffolds for the recruitment of downstream RdDM proteins, including DRM2, to loci that produce both siRNAs and IGN transcripts[3]. However, the mechanism(s) through which Pol V is targeted to specific genomic loci remains largely unknown. Through affinity purification of two known RdDM components, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1)[4] and DEFECTIVE IN MERISTEM SILENCING 3 (DMS3)[5, 6], we found that they copurify with each other and with a novel protein, RNA-DIRECTED DNA METHYLATION 1 (RDM1), forming a complex we term DDR. We also found that DRD1 copurified with Pol V subunits and that, RDM1, like DRD1[3] and DMS3[7], is required for the production of Pol V-dependent transcripts. These results suggest that the DDR complex acts in RdDM at a step upstream of the recruitment or activation of Pol V.
Results and Discussion
DRD1 and DMS3 copurify with each other and a novel protein, RDM1, as well as with Pol V subunits
To better understand the roles of DRD1, a putative chromatin remodeler, and DMS3, a protein with homology to the hinge region of structural maintenance of chromosome (SMC) proteins, in RdDM, we generated transgenic Arabidopsis plants expressing epitope fusions of either DRD1 or DMS3 and used these plants to affinity purify complexes containing these proteins from flower extracts. Various epitope tagged fusions of DRD1 and DMS3, driven by their endogenous promoters, were able to complement defects in DNA methylation observed in their respective mutant backgrounds at the MEA-ISR locus (Figure 1), demonstrating that these fusion proteins are functional in vivo. Following affinity purification of DRD1-3xFlag-BLRP or DMS3-3xFlag-BLRP, co-purifying proteins were identified through mass spectrometric (MS) analyses (Table 1 and Table S1). Peptides corresponding to DRD1, DMS3, and AT3G22680 were by far the most abundant in both purifications (Table 1 and Table S1). Co-purification of At3g22680 with both DRD1 and DMS3 suggested that At3g22680 may be a novel component of the RdDM pathway. Indeed, a mutation in AT3G22680, termed RNA-DIRECTED DNA METHYLATION 1 (RDM1), was recently isolated from a genetic screen for proteins necessary for RdDM [8].
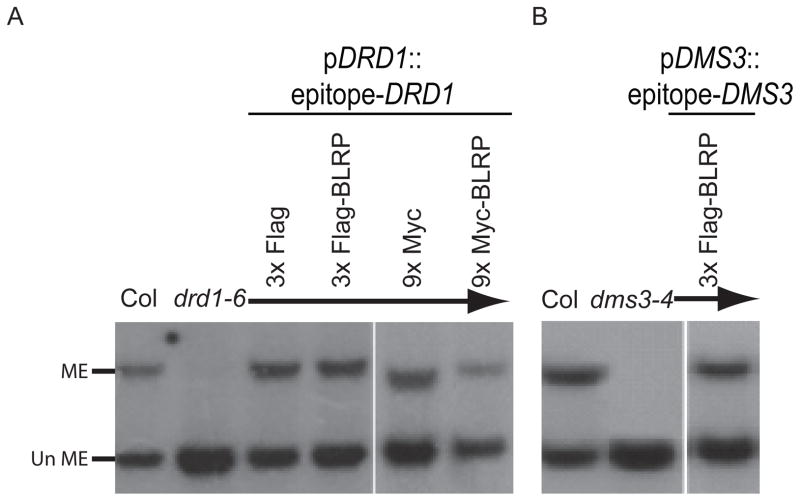
Figure 1.
Complementation of mutants with epitope tagged DRD1 and DMS3. Analysis of DNA methylation at the MEA-ISR locus by Southern blotting after digestion of genomic DNA with the methylation-sensitive restriction enzyme, MspI. Bands representing methylation (ME) or a lack of methylation (un ME) are indicated. Digestion of genomic DNA extracted from wild-type plants of the Colombia (Col) ecotype serve as a positive control for DNA methylation levels. Panel (A) shows the loss of DNA methylation in the drd1-6 mutant alone and the restoration of DNA methylation after transformation of this mutant with a transgene carrying the indicated Flag and Myc epitope tagged fusion of the DRD1 gene under the control of its endogenous promoter (pDRD1). Panel (B) shows the loss of DNA methylation in the dms3-4 mutant alone and the restoration of DNA methylation after transformation of this mutant with a transgene carrying the indicated Flag epitope tagged fusion of the DMS3 gene under the control of its endogenous promoter (pDMS3). Complementation assays shown were conducted using tissue from T3 homozygous transgenic plant lines. BLRP, Biotin Ligase Recognition Peptide. See also Tables S2 and S3.
Table 1.
Mass Spectrometric analyses of DRD1 and DMS3 affinity purifications. Proteins co-purifying with DRD1 (upper) or DMS3 (lower) are indicated and approximate stoichiometry is shown as %DRD or %DMS3 using NSAF, normalized spectral abundance factor, values[23].
| DRD1 Purification | |||||
|---|---|---|---|---|---|
| Protein | Spectra | Unique peptides | % coverage | NSAF | % of DRD1 |
| DRD1 | 245 | 50 | 38.6 | 2.29E-03 | 1.00 |
| At3g22680 | 48 | 11 | 52.8 | 2.45E-03 | 1.07 |
| DMS3 | 115 | 30 | 48.3 | 2.28E-03 | 0.99 |
| NRPE1* | 45 | 35 | 20.2 | 1.89E-04 | 0.08 |
| NRPE2 | 20 | 10 | 9.1 | 1.42E-04 | 0.06 |
| NRPE3A | 14 | 9 | 32 | 3.65E-04 | 0.16 |
| NRPE3B | 5 | 4 | 16 | 1.30E-04 | 0.06 |
| NRPE5* | 3 | 2 | 12.6 | 1.12E-04 | 0.05 |
| NRPE7* | 2 | 2 | 11.8 | 9.34E-05 | 0.04 |
| NRPE9A | 3 | 3 | 34.2 | 2.19E-04 | 0.10 |
| DMS3 purification | |||||
| Protein | Spectra | Unique peptides | % coverage | NSAF | % of DMS3 |
| DMS3 | 2804 | 98 | 87.1 | 3.45E-02 | 1.00 |
| At3g22680 | 184 | 20 | 60.1 | 5.84E-03 | 0.1691 |
| DRD1 | 94 | 39 | 41.3 | 5.48E-04 | 0.0159 |
| NRPE1* | 4 | 4 | 3.3 | 1.05E-05 | 0.0003 |
| NRPE2 | 5 | 4 | 5 | 2.21E-05 | 0.0006 |
| NRPE3A | 4 | 4 | 13.8 | 6.49E-05 | 0.0019 |
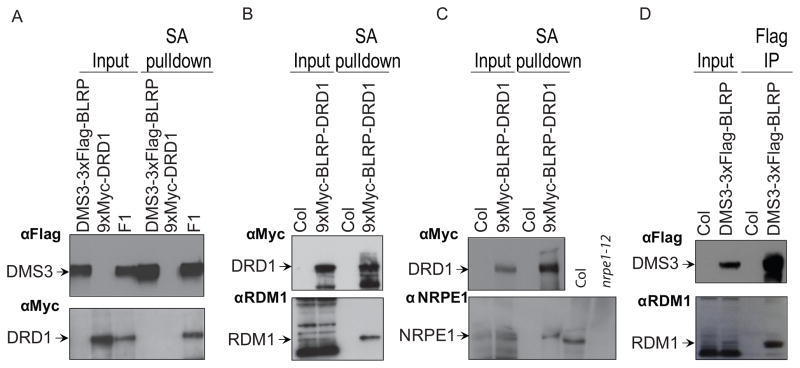
While the identities of the proteins in the DRD1 and DMS3 purifications were similar, the relative stoichiometries were quite different, suggesting these proteins may be present in more than one complex. Affinity purification of DRD1 yielded DRD1, DMS3 and RDM1 at roughly similar levels (Table 1) suggesting these three proteins may form a stable complex in vivo. Consistent with this notion, pair-wise interactions between DRD1, DMS3, and RDM1 were confirmed by co-precipitation experiments (Figure 2 and Figure S1A). Furthermore, these pair-wise interactions were found to be resistant to DNase and RNase treatment (Figure S1B,C) and were stable under high salt conditions (Figure S1C,D), suggesting the associations between these three proteins are stable and mediated by protein-protein interactions. The DRD1 purification also yielded peptides corresponding to many of the previously identified Pol V subunits, but not subunits specific to Pol II or Pol IV [9–12] (Table 1). However, the relative abundance of Pol V peptides was much lower than those of DMS3 and RDM1, which could reflect either a weak association of Pol V with DRD1 or a strong association of Pol V with a small fraction of DRD1. Nonetheless, an interaction between NRPE1, the largest subunit of Pol V, and DRD1 was confirmed by co-immunoprecipitation (Figure 2C).
Figure 2.
Characterization of DDR complex components. (A–D) Streptavidin (SA) pulldown and co-immunopurification assays confirming interactions from Mass Spectrometric analyses. The BLRP tag is biotinylated in vivo allowing interaction with streptavidin. Input lanes confirm expression of the epitope fusions proteins and the endogenous NRPE1 or RDM1 proteins in the parental lines indicated above each lane. F1 represents a cross between the two parental lines. Since these F1 lines only possess a single copy of each transgene, they exhibit lower expression levels as compared to the parental lines. SA pulldown lanes show co-purification of (A) DRD1 with DMS3, (B) DRD1 with RDM1 and (C) DRD1 with NRPE1 and Flag co-immunoprecipitation lanes show (D) DMS3 with RDM1. In (C), protein extracts from Col and nrpe1-12 plants are included to confirm the identity of the co-precipitating band. For each western blot, the antibody used is indicated (upper Left). See also Figure S1.
Upon purification of DMS3, the relative abundance of DRD1 and RDM1 were significantly lower when compared to the DRD1 purification (Table 1), suggesting DMS3 may only be interacting with DRD1 and RDM1 a portion of the time. There were also fewer peptides corresponding to the subunits of the Pol V polymerase in the DMS3 purification (Table 1). Although the interaction between DMS3 and NRPE1 was not confirmed by co-immunoprecipitation analysis, presumably due to sensitivity issues, peptides corresponding to Pol V subunits were detected in two independent DMS3 purifications. Together, these findings suggest that DRD1 and DMS3 may be present in multiple complexes, one of which contains DRD1, DMS3 and RDM1, and others that contain DRD1, and possibly DMS3 to a lesser extent, as well as subunits of the Pol V polymerase.
Gel filtration profiles of DRD1, DMS3, RDM1 and NRPE1
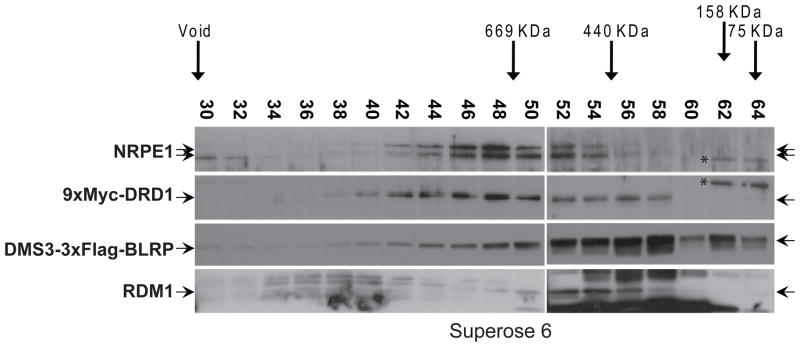
To further characterize the associations between DRD1, DMS3, RDM1 and Pol V, we generated protein extracts from F1 flowers resulting from a cross between 9xMyc-DRD1 and DMS3-3xFlag-BLRP transgenic plants and analyzed these extracts by gel filtration followed by western blotting. This analysis, like the MS analysis, supports the notion that DRD1 and DMS3 are likely present in multiple protein complexes. Using a Superose 6 column, DRD1 eluted as a broad high molecular weight peak that co-eluted with the peak of endogenous NRPE1 and a small portion of the total DMS3 protein (Figure 3). These findings are consistent with the presence of Pol V peptides in the DRD1 purification as well as with the finding that the DMS3 purification yielded fewer Pol V peptides since a smaller portion of the total DMS3 protein co-eluted with NRPE1 than is observed for DRD1. In addition to its co-elution with NRPE1, DRD1 is also present in lower molecular weight fractions, where the majority of DMS3 and RDM1 co-elute around 440KDa (Figure 3), suggesting that DRD1 associates with Pol V in a complex that is largely separate from its association with DMS3 and RDM1. This finding is also consistent with the presence of two distinct peaks of DRD1 after gel filtration using a superdex 200 column (Figures S2A), which gives better resolution of lower molecular weight complexes. Finally, DMS3 is also present in a slower eluting peak, the approximate size predicted for a DMS3 monomer (Figure 3 and Figure S2B).
Figure 3.
Gel filtration of co-purifying proteins. The elution profiles of NRPE1, RDM1, 9xMyc-DRD1, and DMS3-3xFlag-BLRP on a Superose6 column were detected using antibodies against endogenous NRPE1, endogenous RDM1 and either the Myc or Flag epitope, respectively. Fraction numbers and sizing standards are indicated. In fractions 62 and 64 nonspecific background bands are marked by an asterisk (*). See also Figure S2.
Together the elution profiles of these proteins are in general agreement with the co-precipitation data and the MS analyses, demonstrating that a portion of DRD1, DMS3, and RDM1 co-elute as a complex around 440KDa and that DRD1, and DMS3 to a lesser extent, co-elute with NRPE1 in higher molecular weight associations. However, the stoichiometry of the complex containing DRD1, DMS3 and RDM1 appears to differ between the MS and gel filtration techniques. This difference could be attributed to the different sample preparation procedures used for the two techniques, with only the most stable interactions withstanding the more lengthy affinity purification procedure.
RDM1 is required for the production of Pol V-dependent transcripts and DNA methylation
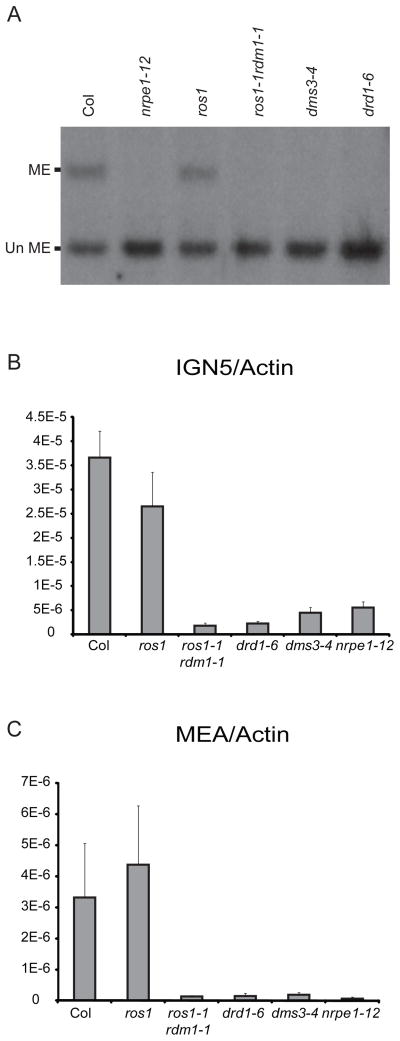
Co-purification of peptides corresponding to RDM1 with DRD1 and DMS3 suggests this gene may also be required for RdDM. RDM1 encodes an ~18KDa protein of unknown function and a crystal structure of RDM1 revealed this protein has a unique fold[13]. To assess the role of this protein in RdDM, we analyzed the level of DNA methylation at the MEA-ISR locus by Southern blotting in a ros1-1 rdm1-1 mutant background that was isolated from a ros1 suppressor screen[8]. DNA methylation was lost in the ros1-1 rdm1-1 mutant to a similar extent as observed for nrpe1-12, drd1-6 and dms3-4, demonstrating that RDM1 is required for RdDM at this locus (Figure 4A). Extensive analysis of DNA methylation at several other loci in several rdm1 alleles, including an allele in a wildtype background, showed similarly strong losses of methylation demonstrating a general role of RDM1 in RNA-directed DNA methylation[8].
Figure 4.
DNA methylation and IGN transcript defects in a ros1 rdm1 mutant. (A) Southern blot analysis as described in Figure 1 using DNA from wild type Col plants or from the indicated mutant plants. (B and C) Quantitative Reverse-Transcriptase PCR analysis of the abundance of Pol V-dependent transcripts corresponding to the (B) IGN5 and (C) MEA-ISR loci in the indicated genetic backgrounds after normalization to the level of an ACTIN transcript. Error bars represent the standard deviation among at least three biological replicas.
Since RDM1 co-purified with components of RdDM known to affect the accumulation of IGN transcripts[3, 7], we tested the hypothesis that RDM1 would also be required for wild-type levels of such transcripts. Using quantitative Reverse-Transcriptase PCR the levels of the IGN5 transcript were assessed in a ros1-1 rdm1-1 mutant and found to be reduced to a similar level as observed in drd1-6, dms3-4, and nrpe1-12 mutants (Figure 4B). A previously unidentified Pol V-dependent transcript corresponding to the MEA-ISR locus was also found to depend on DRD1, DMS3, NRPE1 and RDM1 (Figure 4C). Thus, all the major proteins co-purifying with DRD1 and DMS3 are required for the accumulation of Pol V-dependent IGN transcripts.
Our findings demonstrate that in addition to other associations, DRD1, DMS3, and RDM1 form a complex that we term DRD1-DMS3-RDM1 (DDR) and that RDM1, like DRD1 and DMS3, is required for the accumulation of Pol V-dependent transcripts. Furthermore, we show that DRD1 associates with many subunits of the Pol V complex. Together, these findings provide further insight into the mechanism through which intergenic transcripts are produced by Pol V. Since Pol V subunits copurify with DRD1, and DMS3 to a lesser extent, and since both DRD1 and DMS3 are required for the association of the NRPE1 subunit of Pol V with chromatin[3, 7], it is tempting to speculate that the DDR complex assists in the recruitment or activation of Pol V, after which DRD1, which contains a chromatin remodeling domain, may be important for the initiation or elongation of IGN transcripts by remodeling chromatin ahead of the Pol V polymerase.
Materials and Methods
Generation of Gateway entry clones
Genomic fragments containing the promoter and genomic DNA corresponding to either the DRD1 or DMS3 locus were amplified from the F16F14 BAC (ABRC) or genomic DNA isolated from the Col ecotype, respectively, by PCR using the following primers (Table S2): JP4003 and JP4004 for DRD1 and JP5446 and JP5447 for DMS3. PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen) per manufacturer instructions. For DRD1 and DMS3, carboxy-terminal 3xFlag and 3xFlag-BLRP tags (Table S3) were inserted into a 3’ Asc I site in the pENTR/D-TOPO vector. For DRD1, amino-terminal 9xMyc and 9xMyc-BLRP tags were inserted into a Nco I restriction site engineered into the DRD1 genomic sequence upstream of the start codon through quickchange site directed mutagenesis (Stratagene) per manufactures instructions using the following primers: JP4430 and JP4431.
Generation of Gateway destination clones and selection of transgenic Arabidopsis plants
The described pENTR/D constructs were digested with the Mlu I restriction enzyme and then recombined into a modified gateway destination vector based on the pEarleyGate vectors[14], as described in[15], which contains the BirA gene under the control of an ACTIN promoter and a gene conferring resistance to the BASTA herbicide, per manufacturer instructions (Invitrogen). BirA recognizes a lysine residue in the BLRP tag and catalyzes the addition of a biotin moiety onto this residue which is recognized by streptavidin. These DNA constructs were then transformed into the AGLO strain of Agrobacterium by electroporation. Arabidopsis plants of carrying the drd1-6 mutant allele or the dms3-4 mutant allele were transformed with DRD1 or DMS3 epitope tagged constructs, respectively, as described in[16]. Transformed plants were BASTA selected and scored for single inserts of the transgene by segregation analysis. Protein expression and biotinylation were assessed by western blotting using antibodies against the Flag or Myc epitope or with streptavidin.
Southern blotting
Complementation of the epitope tagged DRD1 and DMS3 proteins, as well as the effect of a mutation in the RDM1 gene on DNA methylation, were assessed by Southern blotting using a probe specific of the MEA-ISR locus as previously described[15].
Affinity purification
Approximately 8g of flower tissue collected from 3xFlag and 3xFlag- BLRP-DRD1 or 3xFlag-BLRP-DMS3 transgenic T4 plants, or from Col plants as a negative control, were ground to a fine powder with a mortar and pestle in liquid nitrogen, and suspended in 45ml of lysis buffer (LB: 50mM Tris pH7.6, 150mM NaCL, 5mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5mM DTT, 1μg/μL pepstatin, 1mM PMSF and 1 protease inhibitor cocktail tablet (Roche, 14696200)). The tissue was further homogenized by douncing and then centrifuged in an SS34 rotor for 25 minutes at 12,500 rpm. 125μL of Dynabeads (M-270 Epoxy, Invitrogen, 143.01) that had been conjugated with Flag antibody (Sigma F 3165) according to manufacturer instructions were added to the supernatant for the DRD1 purification and 600μL of 50% slurry Flag agarose beads for the DMS3 purification. After incubation at 4°C with rotation for 2.5 hours, the Flag beads were washed twice for 5 minutes with 40ml of LB and then 5 times for 5 minutes with 1mL of LB. Proteins were then eluted from the Flag beads by competition with 150μL of 100μg/mL of 3xFlag peptide (Sigma, F 4799) five times at room temperature.
Mass spectrometry
The eluted protein complexes were precipitated by the addition of trichloroacetic acid and then digested by the sequential addition of lys-C and trypsin proteases as previously described[17]. The digested peptide samples were then fractionated online using sequential strong-cation exchange and reversed-phase chromatography and eluted directly into a LTQ-Orbitrap mass spectrometer (Thermofisher) where MS/MS spectra were collected[18, 19]. Data analysis was performed using the SEQUEST and DTASelect2 algorithms and peptide identifications were filtered using a false positive rate of less than 5% as estimated using a decoy database strategy[20–22]. Normalized Spectral Abundance Factor (NSAF) values were calculated as described in Florens et al[23].
SA pulldowns and co-immunoprecipitation analysis
0.5g of flower tissue from the indicated plant lines was ground in liquid nitrogen with 2.5ml of LB and spun in microfuge tubes for 10 minutes at 4°C at 13,200rpm. The supernatants were incubated with 100μL of streptavidin agarose (50% slurry Upstate, 16–126) or with M2 Flag agarose (50% slurry, Sigma A2220) for 2.5 hours at 4°C with rotation. After washing the beads 5 times with 1mL of LB for 5 minutes each, the beads were resuspended in 50μL of SDS-PAGE loading buffer and boiled for 5 minutes. 30μL of input and bead eluate were run on 8% (Figure 2A) or 4–12% (Figure 2B–D) SDS-PAGE gels and the various proteins were detected by western blotting. Flag Westerns were carried out using the ANTI-FLAG M2 Monoclonal Antibody-Peroxidase Conjugate (Sigma A 8592) at a dilution of 1:5000. Myc Westerns used the c-Myc 9E10 mouse monoclonal antibody (Santa Cruz Biotechnology, sc-40) at a dilution of 1:5000 as the primary antibody and goat anit-mouse IgG horseradish peroxidase (Thermo scientific, 31430) was used at a dilution of 1:5000 as the secondary antibody. For NRPE1, an antibody to the endogenous protein initially described in[24] was used at a dilution of 1:1000 as the primary antibody and goat anti-rabbit IgG horseradish peroxidase (Thermo scientific, 31460) was used at a dilution of 1:5000 as the secondary antibody. For RDM1, an antibody to the endogenous protein was used at a dilution of 1:3000 as the primary antibody and goat anit-rabbit IgG horseradish peroxidase (Thermo scientific, 31460) was used at a dilution of 1:25000 as the secondary antibody. All westerns were developed using ECL Plus Western Blotting Detection System (GE healthcare RPN2132).
Salt stability, DNase and RNase treatment
Co-immunoprecipitation and pulldown assays testing the salt stability of the protein associations were conducted as above with the following alterations: 1.5g of the indicated tissue was ground in 7.5 ml of LB, centrifuged as above, and incubated with 300 μL of either streptavidin agarose (50% slurry Upstate, 16–126) (Figure S1D) or M2 Flag agarose beads (50% slurry, Sigma A2220) (Figure S1E) for 2.5 hours at 4°C with rotation. The beads were then washed once with 10ml of LB and then distributed evenly between three microfuge tubes. One aliquot of beads was washed an additional 5 times with 1mL of LB for 5 minutes each, another aliquot with LB supplemented with NaCl to a final concentration of 300mM and another with LB supplemented with NaCl to a final concentration of 500mM. The beads were then resuspended in 50μL of SDS-PAGE loading buffer and boiled for 5 minutes. 10μL or 12μL of input and bead eluate were run on 4–12% SDS-PAGE gels (Figure S1D,E), respectively, and the various proteins were detected by western blotting as above. Co-immunoprecipitation and pulldown assays testing stability of the protein associations upon DNAse and RNAse treatment were conducted with the following alterations: 1.5g of the indicated tissue was ground in 7.5 ml of LB, then split into three 15mL conical tubes. 30μL of TURBO DNase (Ambion #AM2239) was added to one tube, 30μLof RNAse, DNase-free (Roche #11 119 915 001) was added to another, and 30μL of buffer was added to the third tube. Tubes were rotated at 4°C for 30 minutes and 250μL of each extract were removed to assess the DNase and RNase efficiency after phenol:chlorophorm extraction and isopropanol precipitation (data not shown). The remaining extract was centrifuged as indicated above and incubated with 100 μL of either streptavidin agarose (50% slurry Upstate, 16–126) (Figure S1B) or M2 Flag agarose beads (50% slurry, Sigma A2220) (Figure S1C) for 2.5 hours at 4°C with rotation. The beads were then washed 5 times with 1mL of LB for 5 minutes each and then resuspended in 50μL of SDS-PAGE loading buffer and boiled for 5 minutes. 10μL of input and bead eluates were run on 4–12% SDS-PAGE gels (Figure S1B,C) and the various proteins were detected by western blotting as above.
Gel filtration
0.3g of flower tissue from the indicated plant lines were ground in liquid nitrogen with 1.8ml of LB and spun in microfuge tubes for 10 minutes at 4°C at 13,200rpm. The supernatants were transferred to new tubes and spun again for 10 minutes at 4°C at 13,200rpm. The supernatants were then filtered through a .2 micron filter and 500μLs were loaded onto either a Superdex 200 10/300GL column (GE healthcare, 17-5175-01) column or a Superose 6 10/300 GL column (GE Healthcare, 17-5172-01) and 250μL fractions were collected. For the Superose 6 column, 45μL of every other fraction were run on a 4–12% SDS-PAGE and probed for NRPE1, RDM1 and 9xMyc-DRD1 using the antibodies and dilutions outlined above. For DMS3-3xFlag-BLRP, 10μL of the same fractions were run on an 8% SDS-PAGE gel and detected using the Flag antibody described above. For the Superdex 200 columns, DRD1-3xFlag-BLRP was detected in 45μL from every other fraction and DMS3-3xFlag-BLRP was detected in 10μL of each fraction using the Flag antibody. Each column was calibrated prior to use with the Gel Filtration Calibration kit HMW (GE Healthcare, 28-4038-42).
Detection of Pol-V dependent transcripts by reverse transcriptase PCR (RT-PCR)
RNA was isolated from approximately 0.2g of flowers or seedlings by grinding in liquid nitrogen and 1ml of TRIzol Reagent (Invitrogen). RNA was then extracted using 0.2ml of chloroform and precipitated with 0.5 ml isopropanol. The RNA pellet was washed with 1ml 75% ethanol and resuspended in 88μl DEPC-treated H2O. 10μl of 10X Turbo buffer and 2μl Turbo DNase (Ambion) were added and samples incubated for 2 hours at 37°C. RNA was then cleaned up using RNAeasy Mini Kit (Qiagen). Purified RNA was then eluted with 62μl DEPC-treated H2O, to which 7μl of 10X Turbo buffer and 1μl of Turbo DNase was added. Samples were incubated for another 2 hours at 37°C and DNase was removed with DNAse inactivation beads.
Absence of DNA contamination was determined by PCR with no reverse transcriptase added to the reaction. RT-PCR was performed as follows: 1μg RNA was mixed with 2μl of dNTPs (2.5 mM each) and 1μl 12.5 uM Primer 1 in a final volume of 11μl. This was heated to 65°C for 5 minutes and cooled on ice for 1 minute. 14μl of a mix containing 2.5μl Platinum Taq buffer (minus MgCl2), 2μl 50 mM MgCl2, 1μl 0.1 M DTT, 0.3μl RNaseOUT, 0.3μl Platinum Taq (Invitrogen), 0.3μl SuperScriptIII (Invitrogen) and 0.25μl 10 μM Taqman probe was added to each sample and incubation continued for 30 minutes at 55°C, followed by 15 minutes at 70°C. After the addition of Primer 2, the qPCR was started (2 minutes 95°C; 40 cycles of 15 seconds at 95°C, 1 minute at 60°C). Quantities were determined from a standard curve and results are shown normalized to ACTIN. At least three biological replicas were done and standard errors determined. Primers: ACTIN. Primer 1: JP2453, Primer 2: JP2452, Probe: TTTTCCCTAGTTGAGATGGGAATT IGN5. Primer 1: JP6606, Primer 2: JP6607, Probe: TGACCACGGTTAAATGGCGGG. MEA-ISR. Primer 1: JP3734, Primer 2: JP3734, Probe: TTGGGCCGAATAACAGCAAGTCC.
Highlights.
DRD1 and DMS3 copurify with each other and a novel protein, RDM1
DRD1, DMS3, and RDM1 form a stable complex in vivo
DRD1 also copurified with many previously identified Pol V subunits
RDM1, like DRD1 and DMS3, is required for Pol V-dependent transcript accumulation
Supplementary Material
Acknowledgments
We thank T. LaGrange for providing the NRPE1 antibody and members of the Jacobsen laboratory for helpful discussion. Jacobsen lab research was supported by US National Institutes of Health grant GM60398. I.A. was supported by a postdoctoral fellowship from the Ministerio de Educacion y Ciencia. J.A.L. was supported the National Institutes of Health National Research Service Award 5F32GM820453. Wohlschlegel lab research was supported by University of California, Los Angeles Jonsson Cancer Center. S.E.J. is an investigator of the Howard Hughes Medical Institute. S.E.J., J.A.W., J.A.L., and I.A designed the experiments; J.A.L, I.A., L.M.J, and A.A.V. performed the experiments; J.K.Z. provided the ros1-1 rdm1-1 mutant allele and the RDM1 antibody; J.A.L. wrote the paper.
References
- 1.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 6.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, Lui HL, Daxinger L, Pontes O, He XJ, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, Pontier D, Lagrange T, Jin H, Matzke AJ, Matzke M, Pikaard C, Zhu J. A novel methyl DNA binding protein interacts with AGO4 and RNA polymerase II and functions in RNA-direced DNA methylation in Arabidopsis. Nature. doi: 10.1038/nature09025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 10.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 2009;106:941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allard ST, Bingman CA, Johnson KA, Wesenberg GE, Bitto E, Jeon WB, Phillips GN., Jr Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:647–650. doi: 10.1107/S1744309105019743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 17.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR., 3rd Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: Single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. International Journal of Mass Spectrometry. 2002;219:245–251. [Google Scholar]
- 18.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 19.Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 20.Eng J, McCormack A, Yates J. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 22.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.