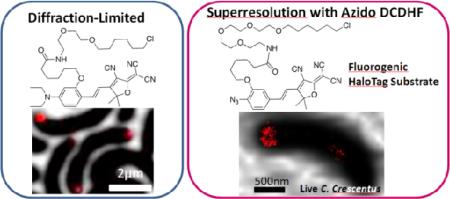
Abstract
Superresolution imaging techniques based on sequential imaging of sparse subsets of single molecules require fluorophores whose emission can be photoactivated or photoswitched. Because typical organic fluorophores can emit significantly more photons than average fluorescent proteins, organic fluorophores have a potential advantage in superresolution imaging schemes, but targeting to specific cellular proteins must be provided. We report the design and application of HaloTag-based target-specific azido DCDHFs, a class of photoactivatable push–pull fluorogens which produce bright fluorescent labels suitable for single-molecule superresolution imaging in live bacterial and fixed mammalian cells.
Recently, sequential imaging of sparse subsets of photoactivatable/photoswitchable single-molecule fluorophores has enabled optical imaging beyond the diffraction limit (DL), providing insight into the sub-diffraction world (e.g. PALM, FPALM, STORM).1–3 These single-molecule superresolution (SR) techniques have provided the impetus for development of new controllable fluorophores with large numbers of emitted photons N, because the achievable resolution scales as .4 Most previous SR experiments in living cells5 have used photocontrollable fluorescent proteins.6–9 However, despite having the advantage of being target-specific, fluorescent proteins on average provide 10-fold fewer photons before photobleaching than good organic fluorophores.10,11 Small organic fluorophores have the additional benefit of synthetic design flexibility for tuning target specificity, spectral wavelength, solubility, and other desired properties. Therefore, targeted bright organic fluorophores which are compatible with the live-cell environment and which can be turned on and/or off would be advantageous. While targeted fluorogens which activate via esterase-mediated cleavage have been previously demonstrated,12 optical control of the emitting concentration is necessary for SR.13
Many biological processes can be interrupted by fixing (i.e. killing) cells, which is required for the immunostaining used in most SR experiments that rely on organic fluorophores.16,17 While fixation can be useful, there is a need for techniques and probes that can be used for SR imaging in living cells. Single-molecule imaging in living cells using exogenous fluorophores faces the dual hurdles of cell permeability and targeted labeling.5 There are exceptions in the literature that did not use fixation and immunostaining. For instance, Heilemann et al.17 imaged mRNA in living cells using oligomers labeled with small organic fluorophores and Conley et al.18 labeled the external lysines of bacterial cells using a Cy3–Cy5 heterodimer. Very recently, fluorophores were targeted with trimethoprim and intrinsic cellular reductants enabled photoinduced blinking19. These examples demonstrated some possibilities for live-cell labeling and SR, but there is still a need for photoactivatable organic labels for SR imaging because each molecule is localized only once, while with blinking or photoswitching, each molecule can be localized a variable number of times.
Here we present a target-specific photoactivatable organic fluorophore for use inside living and fixed cells, 3, based on the commercial HaloTag targeting approach.20–22 This method requires a genetic fusion to the HaloEnzyme (HaloEnz), which forms a covalent linkage to the HaloTag substrate, thus labeling the protein of interest (i.e. a protein–HaloEnz–HaloTag–fluorophore covalent unit). Specifically, we present: (i) the basic photophysical properties of a new targeted photoactivatable probe; (ii) proof-of-principle labeling of known structures in fixed and living mammalian cells validated by co-staining with antibodies or co-transfection with fluorescent proteins; (iii) specific SR imaging of microtubules in a mammalian cell with quantification of resolution enhancement; (iv) demonstration of targeted labeling in living bacteria with diffraction-limited imaging; and finally, (v) SR imaging of poorly understood structures inside living bacteria.
As molecules with bright emission for single-molecule imaging, dicyanomethylenedihydrofuran (DCDHF) push–pull fluorophores emit millions of photons before photobleaching, and can enter living cells.15,23 Recently, we reported a photoactivatable DCDHF fluorogen based on photocaging the fluorescence by replacing the amine donor with a poorly-donating but photolabile azide, which can then be converted back to an amine using low-intensity blue light.14 Azido DCDHF fluorogens exhibit high turn-on ratios (the increase in emission from the fluorescent form compared to the dark fluorogen) of 325–1270 fold, an attractive property for SR imaging.15 (For a detailed discussion of the photophysical properties of the fluorogens, as well as the methods used to characterize them, see references 11 and 15.)
Compared with the original DCDHF-V-P-azide,14 3 exhibits similar spectral changes upon optical pumping of the aryl azide (Figure 1) but a higher photoconversion quantum yield (ΦP, see Table 1). Presumably the oxygen on the aryl azide stabilizes the intermediate nitrene, making photoconversion more favorable.14,15,24 The photoconversion of 3 was so sensitive such that an additional activating blue laser was not necessary; instead, the diffuse ambient light (e.g. the blue light emitted from a nearby computer monitor in an otherwise dark room) was sufficient to activate sparse sets of the fluorogen. The thermal activation rate and activation by the 594 nm imaging laser were significantly lower, as measured in complete darkness in a covered sample (see SI). Because the fluorogen's sensitivity to photoconversion is so high, it was possible to set the level of the ambient light such that the bleaching rate from 594 nm pumping was similar to the activation rate. For instance, with room lights off and a nearby computer monitor on, we maintained a steady-state concentration of isolated emitters. This reduced the complexity of the experiment by allowing the use of only one laser.17 Moreover, because no blue or UV laser was necessary for activation, photodamage to the imaging sample was greatly reduced. (The drawback to this high sensitivity is that it increases the difficulty of preventing photoactivation before imaging. We successfully minimized preactivation by performing all preparations in complete darkness or under dim red lights only.)
Figure 1.
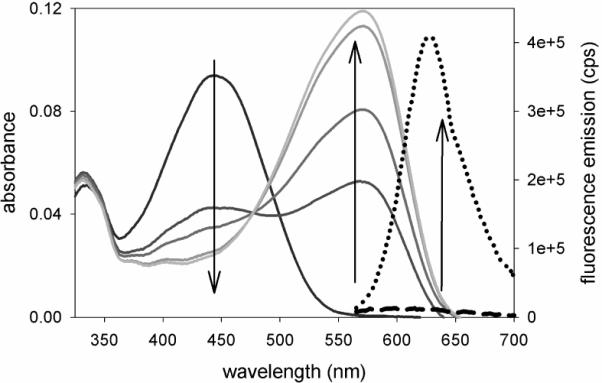
The absorption of 1 in ethanol decreases during irradiation with 1.1 mW/cm2 at 385 nm shown at 15 s, 30 s, 90 s, and 150 s (left down arrow). Concurrently, the absorption of amine photoproduct 2 grows proportionally (center up arrow). The bright fluorescence from 2 when pumped at 594 nm is the dotted curve; the heavy dashed curve is dim fluorescence from the original sample of 1 (the small emission signal is most likely from the pre-activated amine contaminants in the azide sample). Preactivation can be minimized by keeping the samples in complete darkness. Compound 3 has the same photophysical properties as 1.
Table 1.
Photophysical/photochemical parameters. DCDHF-V-P-azide is the earlier underivatized azido-DCDHF from ref 9 for comparison.
| λabs,azide (nm)[a] | λabs,amine/λfl,amine (nm)[b] | Yield[c] | Φ p[d] | |
|---|---|---|---|---|
| DCDHF-V-P-azide | 424 | 570/613 | 65% | 0.0059 |
| 1 (and 3) | 443 | 572/627 | ~50% | 0.095 |
| 4 | – | 598/629 | – | – |
Peak absorbance for azido fluorogen.
Absorbance and fluorescence peak wavelengths of the amino fluorophore.
Overall chemical reaction yield to the primary fluorescent amine.
Photoconversion quantum yield of azido fluorogens to any product (i.e. the probability of photoconverting after absorbing one photon). For reference, the value of ΦP for mEos is on the order of 10−5 (see reference 11 for more detailed comparisons). For all DCDHFs, the fluorescence quantum yield of the photoactivated form varies greatly depending on the precise nanoenvironment, typically <0.1% in buffer to >30% when rigidized (e.g. in the membrane or when bound to proteins).23 For measurement details, see SI. Compound 3 has the same photophysical properties as 1.
We verified the specificity of 3 in mammalian culture. Wild-type HeLa cells and HeLa cells transfected to express HaloEnz–α-tubulin were stained live with 3, fixed and immunostained with Alexa488-mAb to α-tubulin. Fluorescence images after photoactivation clearly demonstrate that 3 is only retained in the cells that expressed–HaloEnz-α-tubulin (Fig. 2A–G). Next, live CHO cells were co-transfected with HaloEnz-α-tubulin and α-tubulin–eGFP, and the labeling by 4 was shown to co-localize well with the eGFP labeling (Fig. 2H–I).
Figure 2.
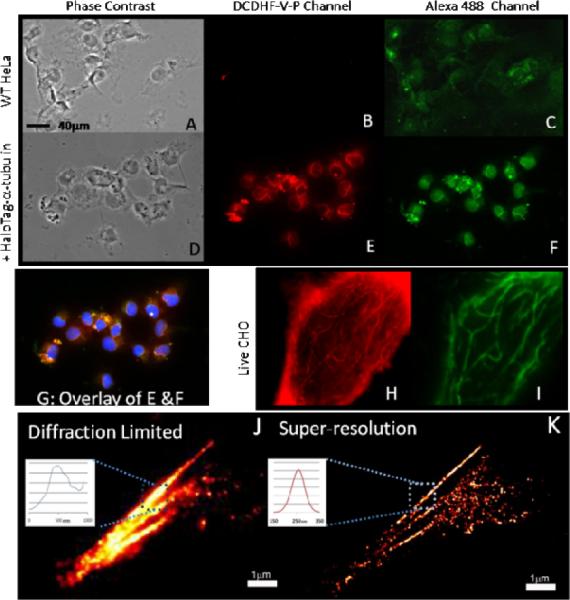
Evidence that the HaloTag-targeted fluorogen correctly labels specific proteins and enables SR imaging in mammalian cells. (A) Phase image of fixed WT HeLa cells. (B) The cells in A imaged in the DCDHFV-P channel. (C) The cells in A imaged in the Alexa488 channel. (D) Phase image of fixed HeLa expressing HaloEnz-α-tubulin labeled with 3. (E) The cells in D imaged in the DCDHF-V-P channel. (F) The cells in D imaged in Alexa488 channel. (G) Overlay of E, F, and additional blue DAPI channel to show nuclei. (H) Live CHO cells co-transfected to express both HaloEnz–α-tubulin and α-tubulin–eGFP labeled with 3 and imaged in DCDHF-V-P channel. (I) Cells from H imaged in the EGFP channel. (The higher background in H may be the result of nonspecific binding and imperfect washing of untargeted fluorophores.) (J) Fixed BS-C-1 cells expressing HaloEnz-α-tubulin labeled with 3 imaged using conventional diffraction-limited imaging. Indicated microtubule measures 450±40nm FWHM. (K) Same cell as J with SR imaging. Indicated microtubule measures 85±15nm FWHM. See SI for sample preparation procedures.
Most importantly, BS-C-1 cells were transfected with HaloEnz-α-tubulin, fixed, stained with 3 and washed before SR imaging by PALM. Comparing DL and SR fluorescence images (Fig. 2J,K), the microtubule structure is clearly imaged with resolution beyond the optical diffraction limit. After corroborating the utility of the DCDHF fluorophores for labeling known structures in mammalian cells, we then moved our attention to cells with protein organization that is not fully understood.
Bacteria are tiny (about a thousand bacteria could fit within one HeLa cell), and the details of protein localization in prokaryotes are poorly understood, yet essential for function and phenotype.25 DL imaging of labeled proteins in bacteria only produces diffuse blobs, complicating meaningful interpretation. For these reasons, living bacterial studies benefit greatly from SR imaging.11,26 We used HaloTag-DCDHFs to highlight protein localization patterns in live Caulobacter crescentus bacteria,26–29 which demonstrates the biologically interesting ability to divide asymmetrically. Elucidating the mechanisms of asymmetric cell division and intracellular organization requires understanding how cytoskeletal proteins localize through the life cycle of the cell.25 In this Communication, a polar protein PopZ,27 and mid-plane proteins FtsZ28 and AmiC29 were expressed as HaloEnz fusions. PopZ, FtsZ, and AmiC have distinct roles: PopZ anchors the chromosomal origin at the “swarmer” pole; FtsZ and AmiC are recruited to the mid-plane and are components of the cell division machinery.25 DL imaging using HaloTag targeting of the (nonphotoactivatable) fluorophore 4 shows correct PopZ localization at cell poles and FtsZ at the cellular division plane as expected (Figure 3), confirming that this HaloTag labeling system does not significantly interfere in phenotype.
Figure 3.
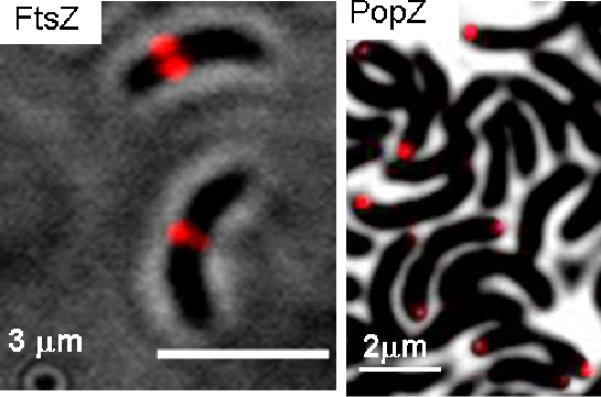
Diffraction-limited imaging of 4 inside live C. crescentus cells expressing fusion proteins to FtsZ and PopZ. These proteins localize as expected,25 indicating that the HaloTag–DCDHF labeling does not disrupt typical cellular behavior.
SR images produced by photoactivation of fluorogen 3 display the expected localization patterns, but also reveal additional detail unseen in the DL images of Figure 3. For PopZ at the cell pole, the protein forms an asymmetric cap-like structure with a curvature that hugs the shape of the bacterial membrane (Figure 4A–C). Also, in the case of AmiC, the protein localizes to the cellular mid-plane, as expected (Figure 4D–E). The SR images of AmiC may reveal a tighter organization than seen in DL microscopy, but further study is necessary before any definitive statement can be made. In either case, these SR images provide new detail not available in DL images.
Figure 4.
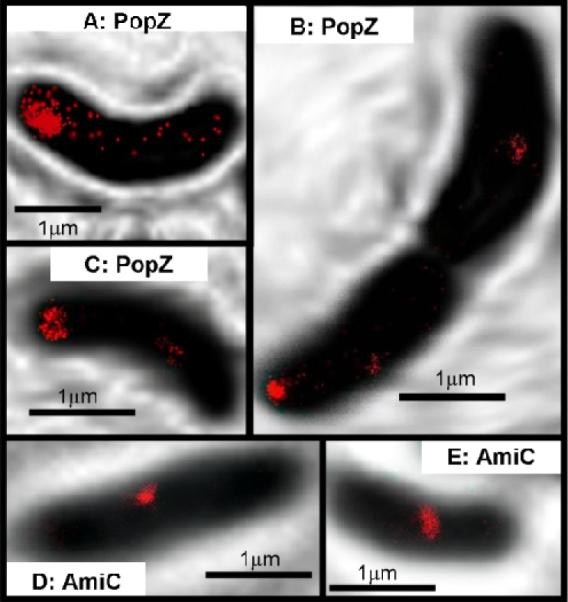
SR imaging of protein fusions inside live C. crescentus cells using 3. (A–C) PopZ forms a polymeric network at the poles of the cells. Compared to the DL images in Figure 3, these SR images reveal distinct shapes of the PopZ structure, including the cap-like network in C. (D–E) AmiC is recruited to the division plane early in the cell cycle. These SR images indicate that AmiC may form a structure that hugs the membrane. For details of imaging and image processing, see SI. The SR images are extracted from localizations over 75 seconds with a mean localization precision of 32 ± 12 nm.
The target-specific DCDHF single-molecule fluorogen presented here represents the first successful installation of target-specificity to small organic photoactivatable fluorogens for single-molecule SR imaging. Compared to existing schemes, the photoactivation of the fluorogen does not require other additives (e.g. thiols for Cy5,30 or redox chemicals17) nor activation by UV light, and thus can be used inside living cells. SR imaging has been directly demonstrated for fixed mammalian and live bacterial cells; additional effort to improve washout for live mammalian cells is an important topic for future work. This and future photoactivatable fluorogens should be helpful tools for SR imaging in the complex environment within the living cell.
Supplementary Material
Scheme 1.
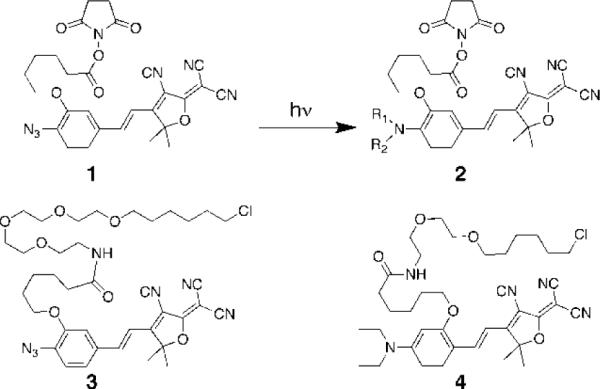
Photochemical activation produces 2 from 1. A mixture of photoproducts is produced,14,15 but the primary amine with R1=R2=H is the significant product (see Table 1 for reaction yield of primary amine). HaloTag versions of 1 and a separate fluorophore are also shown (3 and 4).
Acknowledgements
This work was supported in part by Grant No. R01-GM086196 from the National Institute of General Medical Sciences. We thank S. Pfeiffer for BS-C-1 cells.
Footnotes
Supporting Information Available: Procedures, chemical synthesis, analysis, additional figures, and complete Ref. 20. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott–Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- (2).Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rust MJ, Bates M, Zhuang X. Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lord SJ, Lee HD, Moerner WE. Anal. Chem. 2010;82(6):2192–2203. doi: 10.1021/ac9024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dickson RM, Norris DJ, Tzeng YL, Moerner WE. Science. 1996;274:966–969. doi: 10.1126/science.274.5289.966. [DOI] [PubMed] [Google Scholar]
- (7).Ando R, Mizuno H, Miyawaki A. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- (8).Patterson GH, Lippincott–Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- (9).Hess ST, Gould TJ, Gudheti MV, Maas SA, Mills KD, Zimmerberg J. Proc. Nat. Acad. Sci. (USA) 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schmidt T, Kubitscheck U, Rohler D, Nienhaus U. Single Mol. 2002;3:327. [Google Scholar]
- (11).Thompson MA, Biteen JS, Lord SJ, Conley NR, Moerner WE. Methods Enzym. 2010;475:27–59. doi: 10.1016/S0076-6879(10)75002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Watkins RW, Lavis LD, Kung VM, Los GV, Raines RT. Org. Biomol. Chem. 2009;7:3969–3975. doi: 10.1039/b907664f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Maurel D, Banala S, Laroche T, Johnsson K. ACS Chem. Biol. 2010;5:507–516. doi: 10.1021/cb1000229. [DOI] [PubMed] [Google Scholar]
- (14).Lord SJ, Conley NR, Lee HD, Samuel R, Liu N, Twieg RJ, Moerner WE. J. Am. Chem.. Soc. 2008;130:9204–9205. doi: 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lord SJ, Lee HL, Samuel R, Weber R, Liu N, Conley NR, Thompson MA, Twieg RJ, Moerner WE. J Phys Chem. B. 2009 doi: 10.1021/jp907080r. ASAP. ( http://dx.doi.org/10.1021/jp907080r) [DOI] [PMC free article] [PubMed]
- (16).Folling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, Jakobs S, Eggeling C, Hell SW. Nat. Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- (17).Heilemann M, van de Linde S, Mukherjee A, Sauer M. Angew. Chem., Int. Ed. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- (18).Conley NR, Biteen JS, Moerner WE. J. Phys. Chem. B. 2008;112:11878–11880. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Nat. Meth. 2010;7:717–721. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- (20).Los GV, et al. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- (21).O'Hare HM, Johnsson K, Gautier A. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- (22).So M-K, Yao H, Rao J. Biochem. Biophys. Res.Comm. 2008;374:419–423. doi: 10.1016/j.bbrc.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lord SJ, Conley NR, Lee HD, Nishimura SY, Pomerantz AK, Willets KA, Lu Z, Wang H, Liu N, Samuel R, Weber R, Semyonov A, He M, Twieg RJ, Moerner WE. ChemPhysChem. 2009;10(1):55–65. doi: 10.1002/cphc.200800581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Schriven EFV, editor. Azides and Nitrenes: Reactivity and Utility. Academic Press; Orlando, FL: 1984. [Google Scholar]
- (25).Shapiro L, McAdams HH, Losick R. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Biteen J, Moerner WE. Cold Spr. Harb. Persp. Biol. 2010;2(3):a000448. doi: 10.1101/cshperspect.a000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bowman G, Comolli L, Gaietta G, Fero M, Hong S-H, Jones Y, Lee J, Downing K, Ellisman M, McAdams H, Shapiro L. Mol. Micro. 2010;76(1):173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McAdams H, Shapiro L. FEBS Lett. 2009;583:3984–3991. doi: 10.1016/j.febslet.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bernhardt TG, de Boer PA. Mol. Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dempsey G, Bates M, Kowtoniuk WE, Liu DR, Tsien RY, Zhuang X. J. Am. Chem. Soc. 2009;131:18192–18193. doi: 10.1021/ja904588g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.