Abstract
Histone modifications are regarded as the most indispensible phenomena in epigenetics. Of these modifications, lysine methylation is of the greatest complexity and importance as site- and state-specific lysine methylation exerts a plethora of effects on chromatin structure and gene transcription. Notably, paramecium bursaria chlorella viruses encode a conserved SET domain methyltransferase, termed vSET, that functions to suppress host transcription by methylating histone H3 at lysine 27 (H3K27), a mark for eukaryotic gene silencing. Unlike mammalian lysine methyltransferases (KMTs), vSET functions only as a dimer, but the underlying mechanism has remained elusive. In this study, we demonstrate that dimeric vSET operates with negative cooperativity between the two active sites and engages in H3K27 methylation one site at a time. New atomic structures of vSET in the free form and a ternary complex with S-adenosyl homocysteine and a histone H3 peptide and biochemical analyses reveal the molecular origin for the negative cooperativity and explain the substrate specificity of H3K27 methyltransferases. Our study suggests a “walking” mechanism, by which vSET acts all by itself to globally methylate host H3K27, which is accomplished by the mammalian EZH2 KMT only in the context of the Polycomb repressive complex.
Keywords: chromatin biology, crystallography, histone lysine methylation
The flexible tails of histones undergo a variety of posttranslational modifications to generate a diverse set of epigenome states to regulate gene expression in the chromatin context (1). Histone lysine methylation is unique in that one lysine residue can have one, two, or three methyl groups; such different methylation states have functionally distinctive outcomes (2). Further, methylation at different lysine sites in histones can typically result in totally different consequences. For instance, histone H3 lysine 4 (H3K4) methylation is normally associated with gene activation (3), whereas H3K9 or H3K27 methylation leads to chromatin compaction and transcriptional silencing (4, 5). The core catalytic domain of KMTs shares a conserved structural fold called the SET domain (6), so named after its founding members suppressor of variegation [Su(var)3-9], enhancer of zeste [E(z)], and trithorax.
Given the importance of lysine methylation, it was not a surprise but interesting to see that some viruses or bacteria encode SET domain proteins that target cellular proteins to aid pathogen replication (7). Specifically, we discovered that a SET domain protein, vSET, found in a mature viral particle of paramecium bursaria chlorella virus 1 (PBCV-1) (8), has explicit methyltransferase activity for host histone H3 lysine 27 (8, 9). Moreover, we demonstrated that upon PBCV-1 infection, vSET is released into the host cells and translocates to the cell nucleus where it catalyzes H3K27 methylation globally, resulting in genome-wide repression of host gene transcription (10). vSET likely exerts its activity by mimicking the function of EZH proteins of mammalian Polycomb repressive complex 2 (PRC2) that are the only KMTs known to methylate H3K27. Despite the indispensible role of PRC2 in Hox gene silencing during development (11), in maintaining embryonic stem cell identity (12), and in cancer pathophysiology (13), thus far there is no structure available for EZH proteins, nor do we understand the detailed molecular basis of their KMT specificity for H3K27 methylation. vSET serves as an attractive model to gain previously undescribed insights into the activity and specificity of EZH proteins.
vSET is one of a few known examples of viral encoded enzymes that directly modify host chromatin to benefit viral transcription and replication (14). Uniquely, vSET functions as a dimeric enzyme and is active on its own in cells, which contrasts sharply to most other KMTs that are monomeric and rely on partners to achieve optimal activity on their biological substrates (15, 16).
We sought to determine the role of vSET dimerization in its structure and activity as an independent H3K27-specific KMT, which has remained unresolved in our previous NMR structural analysis of vSET due to limited structural resolution (8, 9). In this study, we determined a high-resolution crystal structure of free vSET, which reveals a detailed network of interactions at the dimer interface. Guided by the atomic structure, we generated numerous monemeric mutant proteins and found them to be inactive, testifying to the functional importance of vSET dimer formation. Our isothermal titration calorimetry (ITC) and enzymatic activity analyses of the monemeric mutants as well as a heterodimer vSET reveal that the enzyme exhibits independence of the two active sites for the first binding event but strong negative cooperativity for the second. The single-site binding model was further strengthened by FRET studies. The atomic structure of the enzyme/substrate/cofactor ternary complex explains precisely the binding determinants of the H3K27 specificity by vSET and suggests the molecular origin of the negative cooperativity. Finally, we discuss the functional implications of these findings in a unique viral mechanism for transcriptional gene silencing on host chromatin that operates on vSET dimerization.
Results
Crystal Structure of vSET.
Overall structure.
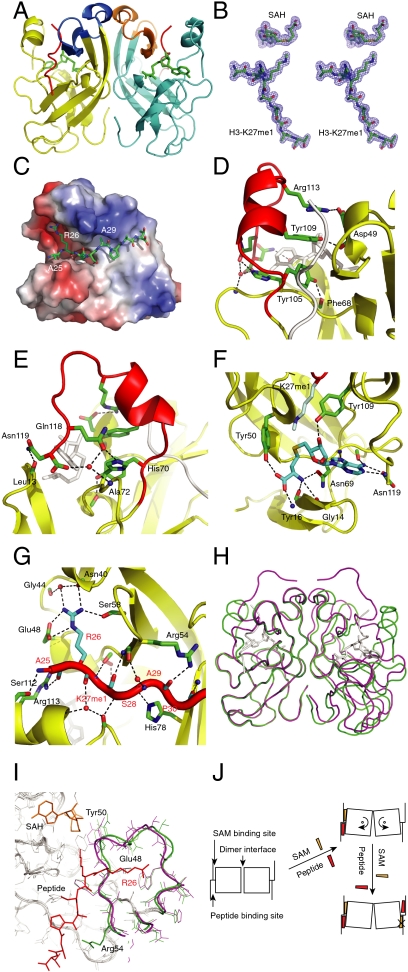
vSET crystallized in two different crystal forms. The structures were solved by single-wavelength anomalous dispersion to 1.85 Å (condition A, see Methods) and 1.60 Å (condition B), respectively (Fig. 1 and Fig. S1; see Table S1 for statistics). Condition A crystal contains one monomer per asymmetric unit that forms a dimer of C2 symmetry with another symmetry-related monomer (Fig. 1A), whereas condition B crystal contains one dimer per asymmetric unit (Fig. S1A). The two protomers of condition B structure are virtually the same with an rms deviation of 0.09 Å for the Cα atoms. Among the differences between the two structures are the conformation of the loop containing residues 9–17 (Fig. S1C) and the C-terminal tail of residues 106–119 that was seen only under condition A but not B presumably due to high mobility (Fig. S1A). The tail confirmation observed in condition A is stabilized by a few hydrogen bonds (Fig. S1D). We describe the condition A structure for the rest of study.
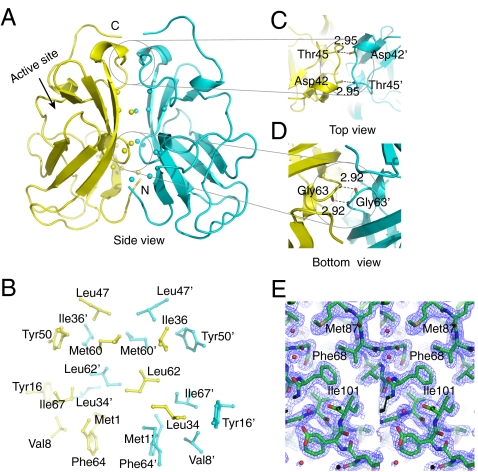
Fig. 1.
Crystal structure of vSET. (A) Cartoon representation of the overall structure of dimeric vSET, solved under crystallization condition A. The two subunits are shown in yellow and cyan, respectively. Cβ atoms of hydrophobic residues at the dimer interface are shown as spheres. (B) Hydrophobic interactions at the dimer interface. The ’ sign is used to differentiate residues from different protomers. (C) Intersubunit main chain hydrogen bonds at the top site. (D) Intersubunit main chain hydrogen bonds at the middle site. (E) Stereo view of the 2Fo - Fc electron density map of the condition A structure contoured at 1σ.
The overall fold of each protomer of either structure is similar to SET domain structures solved previously (17, 18, 19, 20, 21), which consists of three β-sheets positioned around a pseudoknot structure at the C terminus (9, 22). One major difference in the crystal structure is a long continuous β-strand around the dimer interface that connects two β-sheets, which is made up by two connecting β-strands in the solution (8). The two structures obtained under different conditions are similar with an rms deviation of 0.61 Å for the Cα atoms (Fig. S1C).
Dimer interface.
The dimer interface consists of many hydrophobic residues (Fig. 1 A and B). There are also main chain hydrogen bonds across the dimer interface at two discrete locations. The top site contains two symmetry-related hydrogen bonds between the carbonyl of Asp42 of one protomer and the amide of Thr45′ of the other (′ denotes residues of the protomer in cyan) (Fig. 1C). At the middle site, the carbonyl group of Gly63 interacts with the amide group of the corresponding Gly63′ in the other protomer, thus forming an eight-membered ring due to C2 symmetry (Fig. 1D). The combination of hydrophobic and polar interactions creates a stable dimer interface burying over 1,800 Å2 of solvent accessible surface area upon dimer formation.
Dimer Interface Residue Mutants of vSET.
Oligomeric state of mutants.
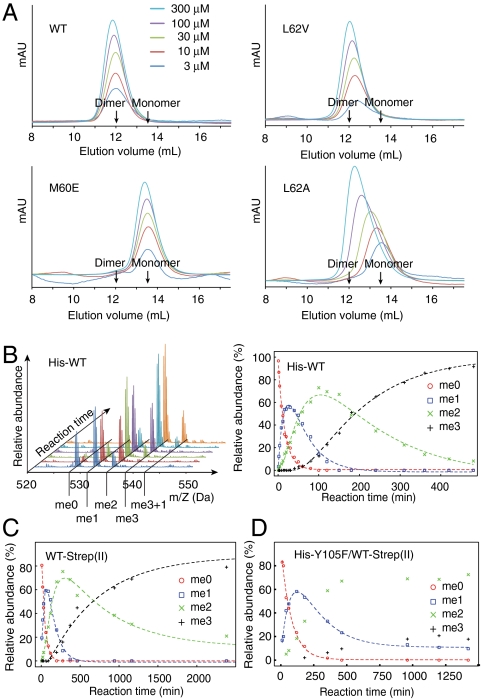
To determine whether dimer formation is critical for the activity of vSET, we mutated residues at the dimer interface including Ile36, Met60, and Leu62. We checked the elution profile of wild-type and mutant proteins at loading concentrations from 3 to 300 μM to assess the oligomeric state as a function of protein concentration. Wild-type vSET is a dimer at all concentrations tested (Fig. 2A, Upper Left). Conservative mutations such as L62V and I36V shift the elution curves slightly to the right (Fig. 2A, Upper Right), indicating that they are still dimers. Mutation of Met60 or Leu62 to a charged amino acid, such as M60E, M60D, and L62K, rendered the protein monomeric even at a loading concentration of 300 μM (Fig. 2A, Lower Left). Alanine mutations of the interfacial residues I36A, M60A, L62A, as well as I36K, produced proteins that behave monomeric at low concentration and become dimeric at high concentration (Fig. 2A, Lower Right).
Fig. 2.
Oligomeric state and methyltransferase activity of vSET. (A) Analytical size exclusion chromatography elution profiles of vSET and its mutants. The elution profiles of selected mutants representing different behaviors are shown. The elution volumes of pure monomer and dimer are shown based on protein standards. The y axes are not drawn to scale. (B) Mass spectrometry results of wild-type vSET reaction at different time points. Peaks are labeled that correspond to unmodified peptide (me0) as well as mono-, di-, or trimethylated peptide (me1, me2, or me3). The me3 + 1 peaks represent low level and nonspecific activity of vSET. The relative abundance of each species is plotted against reaction time (Right). (C) Reaction time course for vSET-Strep(II) homodimer. (D) Reaction time course for His-Y105F/WT-Strep(II) heterodimer. Only k1 and k2 are obtained.
Activity of monomeric mutants.
The activity of wild-type and mutant vSET was measured by in vitro methyltransferase assay using histone H3 peptide substrate (residues 13–33). The reaction mixtures were analyzed by mass spectrometry so that at each reaction time point relative abundances were known of the original unmodified substrate and the products with different numbers of methyl group added (Fig. 2B, Left). The relative abundance was plotted against reaction time for each species. Curve fitting was performed assuming a three-step irreversible consecutive reaction model (Fig. 2B, Right). Three pseudo first-order rate constants for three methylation steps were obtained through this analysis, k1, k2, and k3, respectively. The rate constants for wild-type vSET are listed in Table 1. The activities of monomeric mutants were found to be much lower than those of the wild type; only k1 was obtained by analyzing samples taken at two time points, 5 or 29.3 h of the reaction (Table 2). Mutants L62D, M60D, and M60E did not produce any trace amount of monomethylated product even after 29.3 h of reaction, whereas the activities of I36A, I36K, and L62A were reduced by more than a thousandfold (Table 2). Even the most active monomeric mutant L62K has only 0.35% of wild-type activity. Therefore, we concluded that dimer formation of vSET is indispensible for its enzymatic activity.
Table 1.
H3K27 methylation activity of vSET with different tags and the heterodimer
| Protein | k1, min-1 | k2, min-1 | k3, min-1 |
| His-vSET | 0.062 | 0.021 | 0.0066 |
| vSET-Strep(II) | 0.026 | 0.0099 | 0.0015 |
| His-Y105F/WT-Strep(II) | 0.013 | 0.0054 | – |
Table 2.
H3K27 methylation activity of vSET and its mutants
| vSET | Conversion after 5 h, % | Conversion after 29.3 h, % | k1, min-1 | k1(mutant)/k1(wild type) |
| Wild type | – | – | 0.062 | 1 |
| L62D | 0 | 0 | 0 | 0 |
| M60D | 0 | 0 | 0 | 0 |
| M60E | 0 | 0 | 0 | 0 |
| I36A | 0 | 1.3 | 7.4E-6 | 0.00014 |
| I36K | 4.3 | 8.8 | 3.3E-5 | 0.00063 |
| L62A | 1.4 | 3.8 | 1.7E-5 | 0.00033 |
| L62K | 6.4 | 27.6 | 1.8E-4 | 0.0035 |
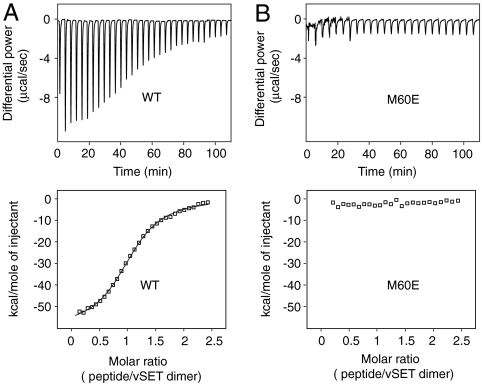
We ruled out a possibility that the extremely low activities of these mutants were caused by misfolding, as the solubility and stability of these mutants under normal buffer condition are similar to those of the wild type. This was further supported by the observation that 1D proton NMR spectrum of I36K shows no significant differences from that of wild type (Fig. S2), thus confirming that the overall fold of vSET is maintained in I36K. So what could cause the loss of activity after dimer disruption? We postulated that dimerization maintains the two active sites in an optimal conformation for catalysis and that vSET monomeric mutations allosterically alter the optimal active site conformation for cofactor and substrate binding required for a productive catalysis. We performed isothermal titration calorimetry measurements for some of the mutants to test this hypothesis. Although wild-type vSET showed nice binding (Fig. 3A), mutant M60E showed almost no binding to H3K27 peptide substrate in the presence of cofactor SAH (S-adenosyl homocysteine) at the protein concentration of 300 μM (Fig. 3B). Similar results were obtained for L62K (Fig. S3A).
Fig. 3.
Isothermal titration calorimetry analysis of vSET. (A) ITC data for wild-type vSET. The solid line at the lower panel represents the fit of the calorimetric data to a single binding site model. The vSET dimer and SAH were kept at 0.05 mM and 1 mM, respectively, whereas the H3 peptide used in the titration ranged from 0 to 0.12 mM. The curve fitting gave a peptide to protein dimer ratio of 1.1 for binding. The association constant is 2.0E5 M-1 and the ΔH is -60 kJ mol-1. (B) ITC data for vSET monomeric mutant M60E. The vSET mutant and cofactor SAH concentrations were kept at 0.3 mM and 1 mM, respectively, whereas the histone H3 peptide was added in incrementally from 0 to 0.72 mM, as indicated.
Activity of a Heterodimer.
To test whether the two active sites in vSET dimer are interdependent, we generated a heterodimer of one wild-type and one dead mutant. Y105F mutation was chosen as it was shown to abolish the enzymatic activity (8). Y105F mutant behaves as a dimer in solution as supported by size exclusion chromatography, but shows abrogated peptide binding as confirmed by ITC (Fig. S3B). pETDuet-1 vector from Novagen was utilized for the coexpression of N-terminal His-tagged Y105F and C-terminal Strep(II)-tagged wild-type vSET (Fig. S4A). A two-step purification procedure with Strep-Tactin and Ni-NTA affinity chromatography columns afforded a purified heterodimer with a nearly equal amount of the wild-type and the Y105F mutant (Fig. S4B).
Because the C-terminal residues of vSET are involved in both cofactor and substrate binding (9), we determined as a control the activity of the C-terminal Strep(II) tagged wild-type vSET homodimer (Fig. 2C) to be k1 (0.026 min-1) and k2 (0.0099 min-1), which are 42% and 47% of the rate constants of the C-terminal untagged vSET, respectively (Table 1). We then measured the heterodimer activity and obtained k1 and k2 of 0.013 and 0.0054 min-1, respectively (Fig. 2D). Notably, rate constants for the first two steps are almost exactly half of those of the WT-Strep(II) homodimer (Table 1), arguing that the activity of the wild-type active site in the heterodimer was not affected because the concentration of functional active site was reduced by half in the heterodimer at a given protein concentration. In essence, even if one active site in the vSET dimer is knocked out, the other site is still functional.
Peptide Binding Stoichiometry of vSET Dimer.
As described above, vSET binds to the H3K27 peptide in the presence of saturating concentration of cofactor SAH with a dissociation constant (Kd) of 5 μM (Fig. 3A). Astonishingly, we found the binding stoichiometry of H3 peptide to vSET dimer to be 1.1, which means that each dimer can bind only one peptide molecule. This finding along with the data of the enzymatic activity of the monomeric mutants and heterodimer vSET strongly suggests that peptide binding at one site triggers some conformation change that propagates through the dimer interface to the other active site so that binding affinity at the other site becomes much lower.
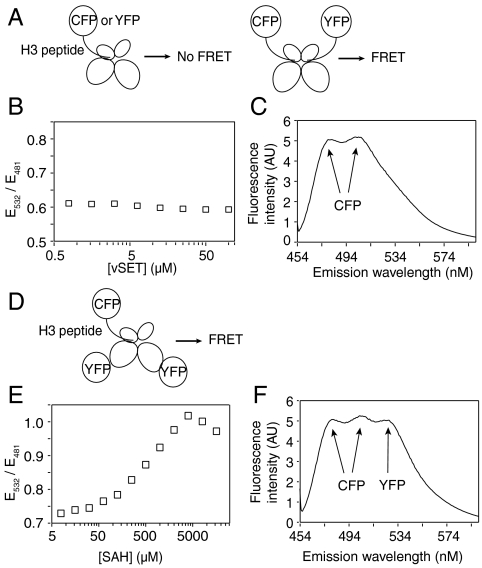
To validate this single-site binding model, we performed a fluorescence resonance energy transfer study as follows: If a histone H3 peptide substrate could bind both active sites of vSET dimer simultaneously, we would detect a FRET signal between two vSET-bound H3 peptides (residues 24–35), one fused to cyan fluorescence protein (H3-CFP) and the other to yellow fluorescence protein (H3-YFP) (Fig. 4A). However, we observed virtually no change of the ratio of emission at 532 nm to that at 481 nm (an indicator of FRET signal) for the mixture as a function of vSET concentration from 0.78–100 uM while keeping the concentrations of H3-CFP and H3-YFP constant (Fig. 4B). One representative emission spectrum of the mixture shows no prominent peak at 532 nm, which is almost identical to that of the mixture of H3-CFP and H3-YFP without vSET (Fig. 4C). These two lines of evidence argue that H3-CFP and H3-YFP could not bind to vSET at the same time, consistent with the single-site occupancy model. To generate direct evidence for this model, we mixed vSET-YFP and H3-CFP and observed a large increase of E532/E481 from 0.73 to 1.02 as a function of SAH concentration from 7.8 uM–4 mM (Fig. 4 D and E). A peak at 532 nm, as a FRET signal, was clearly seen at high concentration of SAH (Fig. 4F). Notably, as SAH concentration passed 4 mM, a decrease of FRET signal was observed (Fig. 4E). This could be explained by ligand binding to the second site at very high concentration, which would weaken the binding of peptide at the first site. The antagonistic effect of the second binding event is also in line with our hypothesis that only one active site of vSET dimer is suitable for stable binding at any given moment under physiological conditions. To understand the molecular basis for this negative cooperativity between these two sites and to find out how substrate specificity is achieved, we solved the crystal structure of a ternary complex of vSET.
Fig. 4.
FRET analysis of vSET binding to histone H3. (A) Schematic diagrams showing hypothetical FRET outcome resulting from one-site binding (Left) vs. two-site binding (Right). (B) E532/E481 as a function of vSET concentration for a vSET/H3-CFP/H3-YFP system. Each sample has 1 μM H3-CFP, 1 μM H3-YFP, and 0.5 mM SAH in a 20 mM Tris buffer of pH 8, containing 300 mM NaCl and 0.01% triton X-100. The excitation wavelength is 434 nm. (C) Emission spectrum of one sample from B with 12.5 μM of vSET. (D) Illustration of H3-CFP’s single-site binding to vSET-YFP that would lead to a robust FRET signal. (E) E532/E481 as a function of SAH concentration for a vSET-YFP/H3-CFP system. Each sample contains 1.6 μM vSET-YFP and 0.8 μM H3-CFP. (F) Emission spectrum of one representative of E with 1 mM of SAH.
Crystal Structure of vSET/SAH/H3 Peptide Ternary Complex.
Overall structure.
The crystal structure of the ternary complex of vSET, SAH, and H3K27me1 peptide (resides 25–32) was solved to 1.78-Å resolution by molecular replacement (Fig. 5 A and B). There are three monomers in the asymmetric unit, two of which form a dimer. The other monomer forms another dimer through symmetry operation. The rms deviation is 0.29 Å between the Cα atoms of these two dimers so that each is representative of both. We focus on the dimer in the asymmetric unit for the rest of the paper. The overall fold of vSET in the ternary complex is similar to that of free protein with an rms deviation of 0.87 Å (Fig. 5 A and Fig. S5A). The rms deviation between the two protomers in the ternary complex dimer is 0.20 Å, so the ternary complex still maintains the C2 symmetry. Peptide and SAH molecules are found at both active sites in the dimer. This seeming discrepancy with the ITC data could be explained by the much higher concentration of all components needed for crystallization. Like all other SET domain complex structures (21), the cofactor and peptide are found to bind two opposite sides of the vSET surface that are connected through a narrow pore (Fig. 5C and Fig. S5B).
Fig. 5.
Crystal structure of vSET/SAH/H3-K27me1 ternary complex. (A) Cartoon representation of the ternary structure. One protomer is shown in yellow, and the other in cyan. SAH is shown as sticks in green. H3-K27me1 peptide is shown in red. Residues 39–57 of the two subunits that undergo rotation upon complex formation (see H) are depicted in marine and orange. (B) Stereo view of the 2Fo - Fc electron density map around the cofactor and H3 peptide contoured at 1σ. (C) Electrostatic surface potential representation of vSET. H3 peptide is shown as sticks. (D and E) Two different views of the polar interactions between the vSET C-terminal tail and the rest of the protein in the complex. The tail is shown in red. (F) Hydrogen bonds involved in cofactor recognition. Side chains of vSET residues, cofactor and peptide are color-coded by atom type with carbon shown in green, cyan, and light cyan, respectively. See Fig. S4E for another view. (G) Interactions between the H3-K27me1 peptide and vSET. Another angle is shown in Fig. S4F. (H) Superimposition of the complex (green) and condition A free form (magenta) structures using one protomer from each structure. (I) Close-up view of the left protomers in G. vSET residues 39–57 of the complex and the free form are highlighted in green and magenta, respectively. (J) A simplified model that explains the negative cooperativity between the two sites. vSET residues 39–57 are depicted as squares. S-adenosyl methionine and peptide are shown as orange and red objects, respectively.
Ordering of the C-terminal tail upon complex formation.
Despite the overall structural similarity between the complex and free form, the C-terminal tail undergoes a dramatic conformational change upon complex formation adopting a one-and-a-half turn α-helix and a type I β-turn around Pro114 (Fig. 5A vs. Fig. 1A). The tail is anchored to the rest of the protein through a myriad of polar interactions (Fig. 5 D and E). Arg113 forms a salt bridge with Asp49, the main chain carbonyl of which forms a hydrogen bond with Tyr109 (Fig. 5D). Mutation of the residue in Drosophila E(z) that corresponds to Asp49 of vSET was shown to abrogate its activity (23). Side chains of Arg113 and Tyr109 stack, contributing to the stability of the α-helix of the tail. Tyr105 is hydrogen-bonded to carbonyl of Phe68 with a short distance of 2.61 Å. These hydrogen bonds explain the importance of Tyr105 and Tyr109, mutation of either of which abrogates the methyltransferase activity of vSET (8, 9). The Tyr105 hydrogen bond also explains why even the conservative phenylalanine mutation eliminates binding as observed in our ITC study. The very last two residues of vSET are also important for the positioning of the tail. Gln118 contacts His70 and is also a member of a water-mediated hydrogen-bonding network involving the C-terminal carboxylate of Asn119 and the amide of Ala72 (Fig. 5E). The side-chain amide of Asn119 in addition interacts with the carbonyl of Leu13. The conformation of the tail is further stabilized by hydrophobic interactions of Trp110 and its surrounding residues (Fig. S5C). It is interesting to see that through this combination of interactions vSET adopts the conformation of its tail suitable for the catalysis, which is achieved by zinc sulfur cluster in some other SET KMTs (24) (Fig. S5D). Just as substrate and cofactor binding is a prerequisite for the tail to adopt this conformation, this conformation in turn is also essential for their binding (seen below).
Cofactor binding.
The SAH cofactor fits snugly in vSET upon complex formation with its adenine ring inserted deeply into a pocket (Fig. S5B). One face of the adenine is covered by the aforementioned Trp110 (Fig. S5C), whereas the other face interacts with Leu13. Mutation of corresponding latter residue in Drosophila E(z) leads to its loss of methyltransferase activity (23). All hydrogen-bonding capabilities of the amino acid moiety of SAH are satisfied (Fig. 5F and Fig. S5E). The extracyclic amino group of adenine interacts with both the carbonyl of His70 and the terminal carboxylate of Asn119. The involvement of Tyr109 and Asn119 in cofactor binding partially explains that the emergence of the new conformation of the C-terminal tail happens only after ligand and/or peptide binding. The fact that the C-terminal carboxylate of vSET is involved both in the ordering of the tail (Fig. 5E) and in cofactor binding (Fig. 5F) explains why adding a C-terminal tag decreases its activity to about half of its original value (Table 1).
Substrate specificity.
The H3K27me1 peptide binds to vSET in an extended conformation (Fig. 5 A, C, and G). One outstanding feature is the recognition of H3R26 through exhaustive hydrogen-bonding/salt bridge network. H3R26 has a divalent salt bridge with Glu48 using Nε and Nη2 atoms (Fig. 5G). The same Nη2 forms water-mediated hydrogen bonds with the carbonyls of Gly44 and Asn40. Nη1 of H3R26 is contacted by both the carbonyl of Asn40 and the side chain of Ser58. All other polar interactions between the peptide and vSET involve only the main chain atoms of residues 25 to 30 of the peptide. The amide and carbonyl of H3K27me1 interact with the carbonyl of Leu51 and amide of Ser53, respectively. The same groups of Leu51 and Ser53 have been found to interact with the tail for the protein alone under crystallization condition A (Fig. S1D).
To investigate whether side chains other than those of H3R26 and H3K27 contribute to vSET specificity for H3K27 methylation, we looked at the surface complementarity between the enzyme and the substrate. H3R26 packs against a flat surface with one tall wall on one side and a small bump on the other (Fig. 5C). The methyl group of H3A25 is pointed to a wall. Any residue with large side chain at this position would induce congestion. Another critical residue H3A29 is found to point to a shallow concave surface. Replacement of A29 with any nonglycine amino acid would cause steric clashes between the substrate and vSET. This explains vSET’s specificity for H3K27 but not H3K9. Although the H3K9 region also contains ARKS sequence as found around H3K27, it is followed by threonine instead of alanine. This threonine would clash with the protein surface if a H3K9 peptide were to bind. Indeed, the methyltransferase activity of vSET was found to be 20-fold lower when a H3K9 peptide was used as a substrate (8). Taken together, the preferred substrate sequence for vSET can be described as #RKXA where # represents an amino acid with small side chain and X represents any amino acid.
Structural Basis for the Negative Cooperativity.
Aligning the complex and the free structures using just one subunit from each structure reveals previously unseen conformation differences at the other subunit. (Fig. 5H vs. Fig. S5A). So substrate and cofactor binding does seem to affect dimer organization. Moreover, significant conformational changes are seen around resides 39 to 57 (Fig. 5A, residues in marine and orange) for the aligning subunits (Fig. 5I), which corresponds to some rotational motion from the top view. Upon complex formation, Tyr50 is pulled away and makes room for cofactor binding. Arg54 also becomes ordered and the chain around this residue gets closer to the peptide substrate. These seemingly minor but large-scale conformational changes may explain the negative cooperativity between the two active sites (Fig. 5J). In the free state, the original cofactor pocket is slightly smaller than needed for tight binding, whereas the peptide pocket is slightly larger. Because interprotomer main chain hydrogen bonds involve residues 42 and 45 (Fig. 1C), it is conceivable that binding of cofactor and peptide could trigger local conformational adjustments in the region of residues 39 to 57, which could cause similar rotational movement in the other protomer at the opposite direction. This ligand binding-induced trans protomer motion would render the cofactor-binding pocket even smaller and the peptide pocket even larger, making them unfit for productive ligand and substrate binding at the second site simultaneously as the first site at their physiological concentrations.
Discussion
The current study reports the atomic resolution structures of an H3K27-specific lysine methyltransferase. The ternary complex structure illustrates how H3K27 recognition is achieved. R26 of histone H3 is recognized by five ionic/hydrogen-bonding interactions (Fig. 5G). H3A29, on the other hand, is identified through the existence of a shallow pocket around its side chain on the protein surface (Fig. 5C). But can we apply the same principles to the important mammalian EZH KMTs that have the same specificity? Sequence alignment of vSET with human EZH1 and EZH2, and Drosophila E(z) shows a sequence identity of 26% and a similarity of 53% (Fig. S6). The residues that are responsible for creating the shallow pocket around peptide H3A29 are Arg54, Leu80, and Leu84 in vSET, which are replaced by Phe, Met, and Asp, respectively, in E(z) proteins (Fig. S6, residues in red). One common feature about these residues (the original and the replacements) is their bulkiness. So this critical recognition mode that distinguishes H3K27-specific KMTs from H3K9 KMTs may also exist in E(z) family KMTs. In vSET, Glu48 forms a salt bridge with H3R26. It is substituted by Asp in E(z) proteins (Fig. S6, green residues), so bonding possibility remains. The other H3R26-contacting residue in vSET, Ser58, appears to be changed to Asn. Collectively, it seems that whereas the precise conformation of active site residues in the EZH KMTs required for productive catalysis is dependent on and controlled by their interactions with other component proteins in the PRC2 complex, E(z) family SET domain KMTs likely use similar strategies to recognize R26 and A29 of histone H3.
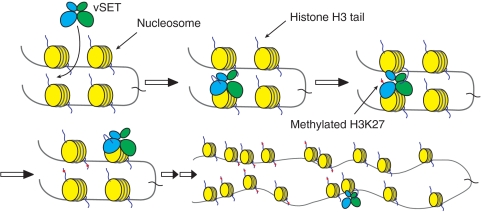
Our combined structural, biophysical and enzymatic study illuminates how vSET functions as a dimer, and uniquely operates H3K27 methylation catalysis one active site at a time. Our results lead us to the following in vivo model (Fig. 6). When in its free form, each subunit of the vSET dimer has equal chance of finding its substrate and cofactor due to the C2 symmetry. However, ligand occupancy of one active site triggers conformational change around residues 39 to 57, which propagates across the dimer interface so that the other active site is not suitable for simultaneous binding. Binding at the second site could occur after the methylation reaction and release of cofactor and H3 at the first site; the second binding could also facilitate ligand dissociation at the first site. The above sequence of events could repeat numerous times on chromatin only at the expense of methyl donor cofactor, S-adenosyl methionine, which would result in the deposition of H3K27 trimethylation marks over large regions of chromosome. In this proposed mechanism, vSET remains associated with chromatin all the time while methylating H3K27 by simply “walking” on chromatin. This mechanism would endow this viral enzyme an efficiency much higher than that by constant dissociation and reassociation were it monomeric. This would thus make it possible for vSET to execute genome-wide methylation of histone H3K27 to suppress global host gene transcription. Similar walking mechanisms are best known in motor proteins such as myosins and kinesins (25), which are driven by ATP hydrolysis. But in vSET, cofactor conversion is used concurrently for the viral enzyme to catalyze target histone H3K27 methylation and feed itself the host substrate, a unique function that vSET owes all to its dimerization.
Fig. 6.
A walking model for vSET. Each site in the dimer has an equal chance for the initial binding. Once that happens, it triggers some conformational change around the first binding site that propagates across the dimer interface to the other active site making it unfit for simultaneous binding. After the reaction takes place at the first site, binding occurs at the second site with concurrent release of cofactor and peptide at the first site. The aforementioned events could be repeated many times to allow efficient deposit of H3K27 methylation marks along long stretches or clustered regions of chromatin. The methylated H3K27 is shown as red dots that represent a mixture of products at different methylation stages. The movement of vSET along chromatin may be a random walk.
Methods
The experimental procedures are briefly described here. A detailed description is provided in SI Methods. vSET proteins were obtained from BL21(DE3) cells. The protein used for structural studies was the full-length vSET without any tag, whereas the protein used for activity and binding studies contained an N-terminal His tag by default. Crystal structures were solved first by single-wavelength anomalous dispersion using selenomethionine-labeled protein and subsequently by molecular replacement. Similar conditions were used for methyltransferase assay as described previously (6). Isothermal titration calorimetric measurements were performed with an Omega instrument (Microcal).
Supplementary Material
Acknowledgments.
We acknowledge the staff at the X6A and X4C beamlines of the National Synchrotron Light Sources at the Brookhaven National Laboratory for facilitating X-ray data collection. We also thank X. Q. Wang and A. N. Plotnikov for technical advice and helpful discussion. The work was in part supported by the grants from the National Institutes of Health (GM073207 and DA028776) (to M.-M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors for free form and ternary complex vSET have been deposited in Protein Data Bank, www.pdb.org (PDB ID codes 3KMJ, 3KMA, and 3KMT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009911107/-/DCSupplemental.
References
- 1.Jenuwein T, Allis C. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama J, Rice J, Strahl B, Allis C, Grewal S. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L, Iyer L. Provenance of SET-domain histone methyltransferases through duplication of a simple structural unit. Cell Cycle. 2003;2:369–376. [PubMed] [Google Scholar]
- 7.Qian C, Zhou M. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manzur K, et al. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat Struct Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 9.Qian C, et al. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol. 2006;359:86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Mujtaba S, et al. Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol. 2008;10:1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 12.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. [PubMed] [Google Scholar]
- 14.Wei H, Zhou MM. Viral-encoded enzymes that target host chromatin functions. Biochim Biophys Acta. 2009;1799:296–301. doi: 10.1016/j.bbagrm.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 16.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J, Zhang X, Cheng X, Grewal SI, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 18.Wilson J, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 19.Trievel R, Beach B, Dirk L, Houtz R, Hurley J. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs SA, et al. The active site of the SET domain is constructed on a knot. Nat Struct Biol. 2002;9:833–838. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 21.Xiao B, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi P, et al. Dominant alleles identify SET domain residues required for histone methyltransferase of Polycomb repressive complex 2. J Biol Chem. 2008;283:27757–27766. doi: 10.1074/jbc.M804442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 25.Cross RA. Molecular motors: Walking talking heads. Curr Biol. 1999;9:R854–856. doi: 10.1016/s0960-9822(00)80045-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.