Abstract
Many proteins that respond to DNA damage are recruited to DNA lesions. We used a proteomics approach that coupled isotopic labeling with chromatin fractionation and mass spectrometry to uncover proteins that associate with damaged DNA, many of which are involved in DNA repair or nucleolar function. We show that polycomb group members are recruited by poly(ADP ribose) polymerase (PARP) to DNA lesions following UV laser microirradiation. Loss of polycomb components results in IR sensitivity of mammalian cells and Caenorhabditis elegans. PARP also recruits two components of the repressive nucleosome remodeling and deacetylase (NuRD) complex, chromodomain helicase DNA-binding protein 4 (CHD4) and metastasis associated 1 (MTA1), to DNA lesions. PARP plays a role in removing nascent RNA and elongating RNA polymerase II from sites of DNA damage. We propose that PARP sets up a transient repressive chromatin structure at sites of DNA damage to block transcription and facilitate DNA repair.
Keywords: B lymphoma Mo-MLV insertion region 1 homolog (BMI1), damage signaling, MEL-18, polycomb repressive complex 1, polycomb repressive complex 2
The cellular response to DNA damage is initiated by the sensing of structural alterations in DNA that culminates in the activation of phosphoinositide-3-kinase–related protein kinases (PIKKs) that include the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinases (1). With the help of mediators, ATM and ATR subsequently signal downstream to activate effector kinases checkpoint 1 (CHK1) and checkpoint 2 (CHK2), leading to transcriptional induction, cell-cycle arrest, DNA repair, senescence, or apoptosis. This DNA damage response induces the sequential recruitment of an extensive network of proteins to the sites of damage. For example, in response to double-strand breaks (DSBs), ATM phosphorylates histone H2AX adjacent to the break to initiate a H2AX-dependent concentration of proteins involved in the DNA damage response, such as mediator of DNA damage checkpoint protein 1 (MDC1), which recruits additional molecules of the ATM kinase. This recruitment effectively initiates a positive feedback loop that promotes the spread of γH2AX-flanking DSBs (2). Phosphorylation of MDC1 by ATM creates a motif that is recognized by the ubiquitin ligase ring finger 8 (RNF8) (3–6) that, with the help of ring finger 168 (RNF168), catalyzes the formation of lysine 63 (K63)-linked polyubiquitin chains that ultimately recruit the breast cancer 1 (BRCA1) A complex containing receptor-associated protein 80 (RAP80), Abraxas, BRCA1, new component of the BRCA1 A complex (NBA1), and BRCA1/BRCA2-containing complex, subunit 3 (BRCC36) (3–10) as well as p53 binding protein 1 (53BP1) and RAD18 homolog (RAD18) (3–8, 11).
Several factors, such as Nijmegen breakage syndrome 1 (NBS1), 53BP1, and BRCA1, are recruited to the sites of damage in an H2AX-independent manner (12). However, these interactions appear to be more transient and may play a role as an initial response to DNA damage that is distinct from the extended association of factors via γH2AX. Several additional pathways also have been shown to direct the recruitment of various proteins to sites of DNA damage. For example, monoubiquitination of Fanconi anemia complementation group D2 (FANCD2) and Fanconi anemia complementation group I (FANCI) by the Fanconi anemia core complex directs their binding to DNA interstrand crosslinks (13–16). Separately, the poly(ADP ribose) polymerase (PARP) family of proteins also has been implicated in the recruitment of DNA repair factors to lesions. PARP1, the best-characterized family member in mammalian cells, is involved in single-strand break repair (SSBR) and an alternative nonhomologous end-joining (aNHEJ) pathway (17). The binding of PARP1 to DNA breaks stimulates its enzymatic activity and leads to the rapid assembly of poly(ADP ribose) (PAR) chains adjacent to the lesion. This assembly leads to the recruitment of X-ray repair cross-complementing group 1 (XRCC1) and ligase III (LIG3), as well as polynucleotide kinase (PNK), polymerase-β (POLB), flap endonuclease-1 (FEN1), and PARP2, to repair the break.
The physical recruitment to sites of damage is a characteristic hallmark shared by many players involved in the DNA damage response. We therefore sought to discover additional factors involved in the DNA damage response based on their ability to be recruited to chromatin after DNA damage by combining chromatin fractionation with quantitative mass spectrometry.
Results
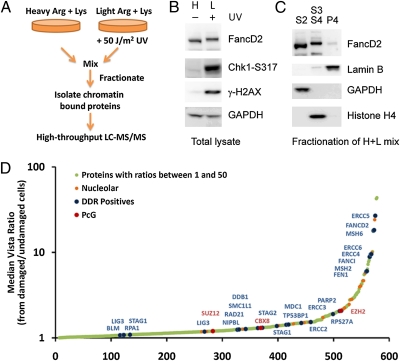
To survey the chromatin-associated proteome to identify factors that are selectively enriched in the chromatin fraction after DNA damage, we combined stable isotope labeling in cell culture (SILAC) (18), chromatin fractionation, and high-throughput identification of proteins by mass spectrometry to compare the chromatin-associated proteome from damaged and undamaged cells (Fig. 1A). We grew HeLa cells in DMEM containing either heavy [13C6, 15N4]-arginine and [13C6, 15N2]-lysine or light arginine and lysine to label the proteome of each population differentially. After labeling, HeLa cells grown in light medium were treated with 50 J/m2 UV light to induce DNA damage and were incubated for 90 min at 37 °C to allow time for recruitment to chromatin. This high dose of UV induces a variety of DNA lesions, including DSBs, and activates multiple DNA damage response pathways as evidenced by the monoubiquitination of FANCD2 and phosphorylation of CHK1 and histone H2AX at Ser-317 and Ser-139, respectively (Fig. 1B). Both mock- and UV-damaged cells were harvested, mixed at a 1:1 ratio, and fractionated to enrich for chromatin-bound proteins. Soluble nuclear and cytoplasmic proteins were released in fraction S2 and washed away; chromatin-bound proteins such as monoubiquitinated FANCD2 and histones were extracted and collected in fraction S3 + S4, leaving behind proteins associated with the nuclear matrix in the P4 fraction (Fig. 1C). The chromatin-enriched fraction (S3 + S4) was separated by SDS/PAGE, digested with trypsin, and analyzed by LC-MS/MS. Using a high-accuracy mass spectrometer, we were able to determine the relative abundance of light and heavy peptides present in the chromatin-enriched fraction from damaged (light) and undamaged (heavy) cells. A total of 10,529 unique peptide pairs were sequenced that correspond to 1,129 proteins. We determined the relative abundance of each protein from either damaged or undamaged cells by calculating a median ratio from the corresponding peptide pair(s) and normalizing to account for total protein levels. Proteins with a calculated median light:heavy (damaged:undamaged) ratio between 1 and 50 are plotted in Fig. 1D. In this graph, a median ratio >1 suggests a preferential enrichment of the protein in the chromatin fraction after DNA damage. A list of the chromatin-associated proteins shown in Fig. 1D and their median vista ratios can be found in Table S1. Importantly, we recovered multiple proteins known to be recruited to sites of DNA damage, including MDC1, 53BP1, cohesins, PARP2, LIG3, mutS homolog 2 (MSH2), mutS homolog 6 (MSH6), excision repair cross-complementing rodent repair deficiency, complementation group 3-6 (ERCC3-6), FANCD2, and FANCI. We also observed enrichment, although to a lesser extent, of nucleolar proteins in the chromatin fraction following DNA damage. Interestingly, three members of the polycomb complex [enhancer of zeste homolog 2 (EZH2), chromobox homolog 8 (CBX8), and suppressor of zeste 12 homolog (SUZ12)] were found to be enriched on damaged chromatin. This observation was of particular interest, because EZH2 was identified independently in an shRNA screen for genes involved in IR resistance (Fig. S1A) (19).
Fig. 1.
A proteomics screen to identify proteins that are recruited to sites of DNA damage. (A) Overview of the proteomics strategy used to identify proteins that are preferentially recruited to sites of DNA damage. Arg, arginine; Lys, lysine. (B) UV induced monoubiquitination of FANCD2 as well as phosphorylation of Chk1 and H2AX. (C) Untreated HeLa cells were mixed with cells exposed to UV light and fractionated. The effectiveness of separation of soluble, chromatin-bound, and nuclear matrix-associated proteins was determined by Western blotting. (D) Proteins in the chromatin fraction with a median vista ratio >1 are plotted in the order of increasing ratio. A ratio >1 suggests enrichment in the chromatin fraction after DNA damage.
The polycomb group (PcG) proteins belong to a family of transcriptional repressors known for their role in the silencing of homeobox (Hox) genes during development (20, 21). PcG proteins form two distinct multiprotein complexes. In flies, polycomb repressive complex 1 (PRC1) is composed of stoichiometric amounts of four proteins, polycomb (Pc), polyhomeotic (Ph), posterior sex combs (Psc), and sex combs extra (Sce) (22). Polycomb repressive complex 2 (PRC2) contains extra sex combs (Esc), enhancer of zeste E(z), and SUZ12 (23). In the current model, transcriptional repression is initiated by PRC2 (initiation complex), which possesses histone methyltransferase activity from the Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domain on E(z). PRC1 is recruited either by the binding of the H3K27me3 mark via the chromodomain on Pc or through a PRC2-independent mechanism. There are multiple orthologs of each PRC1 component in mammals, with five orthologs of Pc (CBX2, -4, -6, -7, -8), three orthologs of Ph (PHC1–3), two orthologs of Psc [really interesting new gene 1 (RING1) and ring finger 2 (RNF2)], and six orthologs of Sce [polycomb group ring finger 1–6 (PCGF1–6)]. In addition, there are two mammalian orthologs of E(z), EZH1 and EZH2, with EZH2 responsible for the bulk of trimethylated lysine 27 of histone H3 (H3K27me3) detected in cells (24, 25).
The recruitment of PcG proteins to damaged chromatin suggests that they may play a direct role in the cellular response to DNA damage. We therefore tested whether depletion of PRC2 proteins will cause DNA damage sensitivity using a multicolor competition assay with short hairpins that deplete either EZH2 or firefly luciferase (FF) as a control. Cells expressing EZH2 shRNAs and Discosoma sp. red fluorescent protein (dsRed) were mixed with uncolored cells, left untreated or exposed to IR, and analyzed by FACS after 10 d. In this assay, sensitivity to IR treatment is determined by monitoring the number of dsRed-stained cells expressing EZH2 shRNA relative to the uncolored control cells, after normalization to the relative cell numbers in the untreated control mixture. Depletion of EZH2 by four independent and unrelated shRNAs renders U2OS cells sensitive to ionizing radiation (Fig. 2A and Fig. S1B). Similar sensitivities were detected for cells depleted of the additional PRC2 components embryonic ectoderm development (EED), SUZ12, and EZH1 (Fig. S1 C and D). Together, these results indicate that 20–40% of the cell's IR resistance is controlled by the function of PRC2.
Fig. 2.
PcG proteins confer protection against ionizing radiation and accumulate at DNA lesions in a PARP1/2-dependent manner. (A) U2OS cells expressing dsRed and the indicated shRNA were mixed with U2OS cells expressing control FF shRNA, were mock-treated or exposed to IR, and were analyzed by FACS after 10 d in a competition assay. Relative fitness reflects the percentage of IR-treated red cells remaining in the mixture relative to mock-treated cells. (B) mes-2 mutant animals show increased sensitivity to ionizing radiation. Embryonic viability of the offspring of untreated and irradiated animals is quantified 24 h postirradiation by assessing the number of live offspring relative to the number of eggs laid by each animal. Error bars indicate SEM for 15–30 animals in three independent experiments. (C) MEFs were microirradiated with a UV laser, fixed after 10 min, and immunostained with anti-EZH2 and anti-γH2AX antibodies. (D) MEFs were microirradiated with a UV laser, fixed after 5 min, and immunostained with anti-H3K27me3 and anti-RPA32 antibodies. (E) Endogenous MEL-18 and BMI1 were recruited within 10–12 min to UV laser-induced sites of damage in HeLa cells. (F) HeLa cells treated with either DMSO or 1 μM KU-0058948 (PARP1/2 inhibitor) were microirradiated with a UV laser and immunostained with anti-MEL-18 and anti-γH2AX antibodies.
To determine if a role for the PRC in DNA damage resistance was evolutionarily conserved, we took advantage of a mutation in the EZH2 ortholog mes-2 in Caenorhabditis elegans (26) and tested it for sensitivity to ionizing radiation. Young adult wild-type, mes-2 heterozygous, and mes-2 homozygous mutant animals were treated with IR, then singled and scored for embryonic survival. Both the heterozygous and homozygous mes-2 mutants exhibited mild but statistically significant IR sensitivity compared with wild type, with P values of 0.0433 and 0.0002, respectively (Fig. 2B). Furthermore, the homozygous mes-2 mutant worms exhibited a more severe phenotype than their heterozygous counterpart. Together, our data show that PcG proteins play an evolutionarily conserved role in preventing sensitivity to DNA damage.
To gather additional support for the recruitment of PRC complexes, we induced site-specific DNA damage by UV laser microirradiation and examined colocalization with γH2AX or replication protein A2 (RPA32) by immunofluorescence. In this process, cells are presensitized by the incorporation of BrdU, which upon exposure to an UV laser results in photolysis and generates DNA lesions along the path of the laser. The cells subsequently are fixed with formaldehyde and immunostained to allow the detection of specific proteins to defined areas of DNA damage within the nucleus. Although EZH2 generally is associated with chromatin, it also was recruited to sites of DNA damage, colocalizing with γH2AX (Fig. 2C). Because EZH2 is a methyltransferase that trimethylates lysine 27 of histone H3 (H3K27), we examined H3 K27 methylation and found H3K27me3 that colocalized with RPA32 at damage sites (Fig. 2D). The association of EZH2 with DSB-flanking chromatin is rapid, and the H3K27me3 mark is detected almost immediately after laser microirradiation but dissipates with time. PRC2 is most likely responsible for the observed H3K27me3 mark at DNA breaks, although we have not ruled out a possible contribution from another unidentified H3K27 methyltransferase.
Mammalian Pc contains a chromodomain that binds H3K27me3 and potentially can recruit the PRC1 complex. We therefore hypothesized that components of PRC1 also may be recruited. Indeed, we observed rapid recruitment of endogenous MEL-18/PCGF2 and BMI1/PCGF4 to sites of DNA damage (Fig. 2E). The specificity of MEL-18 and BMI1 antibodies was demonstrated in Fig. S2. Furthermore, using GFP fusions, we showed that EZH1, RING1, RNF2, PHC1, PHC2, PCGF3, CBX4, CBX6, CBX7, and CBX8 are recruited to sites of DNA damage (Fig. S3A). Interestingly, we noticed that some PRC1 components (e.g., MEL-18) are retained only transiently at DNA lesions, whereas others (e.g., BMI1) are readily detected more than 2 h after damage (Fig. S3 B and C). Finally, we were unable to observe recruitment of GFP-tagged PCGF1, PCGF5, PCGF6, PHC3, and CBX2, perhaps because of signal-to-noise problems or possibly because the GFP tag interfered with the localization of the fusion protein.
To determine whether PRC recruitment depends on the PIKKs ATM and ATR, we treated HeLa cells with an ATM kinase inhibitor (KU-55933), siRNAs that target ATR, or a combination of both. As shown in Fig. S4A, recruitment of MEL-18 to DNA breaks was not noticeably affected by inhibition of ATM, depletion of ATR, or both. To eliminate an effect of residual H2AX phosphorylation, we examined localization in immortalized H2AX+/+ and H2AX−/− mouse embryonic fibroblasts (MEFs) and readily observed H3K27me3 at lesions in H2AX−/− MEFs (Fig. S4B, Upper). Similar results were observed for MEL-18 recruitment (Fig. S4B, Lower), suggesting that both PRC1 and PRC2 localize to sites of damage independently of H2AX.
Because H2AX had no role in PRC recruitment, we initiated a search for other pathways responsible for PRC recruitment. The PARP family of proteins has been implicated previously in recruitment of proteins to sites of damage and is known to localize rapidly to sites of damage (17). To explore a role for PARP, we asked whether an inhibitor of PARP1 and PARP2 (KU-0058948) affects the recruitment of MEL-18 to sites of DNA damage. Cells were treated 1 h before UV laser microirradiation with either DMSO or the PARP inhibitor, fixed with formaldehyde, and stained for endogenous MEL-18. MEL-18 is recruited efficiently to DNA lesions in control-treated cells, but recruitment is largely abrogated in cells treated with the PARP inhibitor or transfected with siRNAs targeting PARP1 and PARP2 (Fig. 2F and Fig. S4C). Thus, PARP activity is required for the recruitment of PcG proteins to sites of DNA damage.
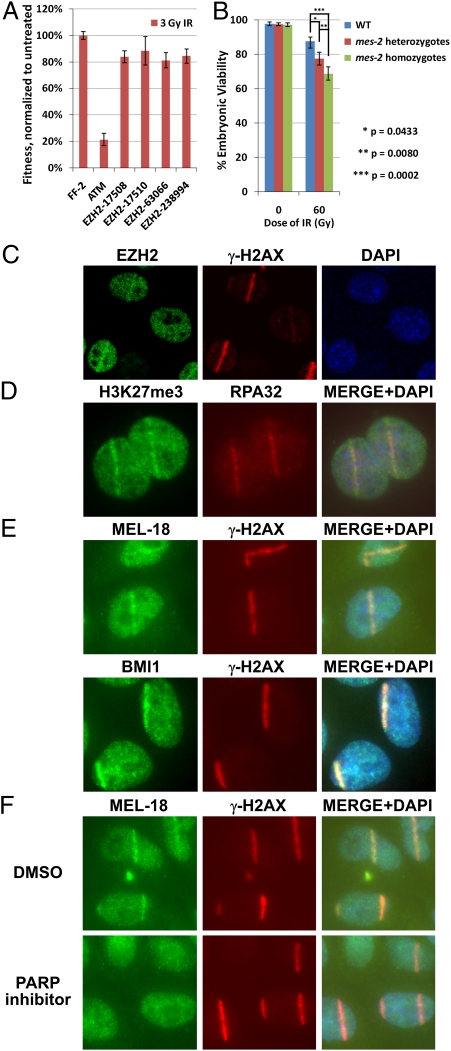
In a separate ongoing screen for proteins that show increased ubiquitination in response to DNA damage, two proteins, metastasis associated 1 (MTA1) and chromodomain helicase DNA binding protein 4 (CHD4), were implicated in the DNA damage response and were found to localize to sites of DNA damage (Fig. 3 A and B). These proteins are components of the nucleosome remodeling and deacetylase (NuRD) complex, which has been implicated in transcriptional repression (27). Because they were recruited very rapidly and transiently to sites of damage, we asked if they also might be controlled by PARP. Inhibition of PARP clearly suppressed the localization of these two NuRD components to sites of microirradiation (Fig. 3 A–C), indicating that PARP recruits at least two distinct transcriptional repression complexes to sites of DNA damage. Interestingly, we also identified MTA1 in an shRNA screen for genes required for IR resistance (Fig. S5A), prompting us to test whether the two NuRD components would protect cells against IR sensitivity. Indeed, depletion of MTA1 by three independent siRNAs rendered U2OS cells sensitive to IR (Fig. 3D and Fig. S5B). However, it was not possible to ascertain whether CHD4 depletion leads to DNA damage sensitivity because of the toxicity associated with CHD4 depletion.
Fig. 3.
NuRD components CHD4 and MTA1 accumulate at DNA lesions following UV laser microirradiation. (A) U2OS cells expressing MTA1-GFP pretreated for 1 h with either DMSO (control) or 1 μM PARP inhibitor KU-0058948 were microirradiated with a UV laser, fixed after 10 min, and immunostained with anti-GFP and anti-γH2AX antibodies. (B) U2OS cells pretreated for 1 h with DMSO or 2 μM PARP inhibitor KU-0058948 were microirradiated with a UV laser, fixed after 10 min, and immunostained with anti-CHD4 and anti-γH2AX antibodies. (C) Quantification of the data shown in A. (D) U2OS cells expressing GFP were transfected with the indicated siRNAs, mixed with cells expressing dsRed, mock-treated or exposed to various doses of IR, and analyzed by FACS after 9 d in a competition assay.
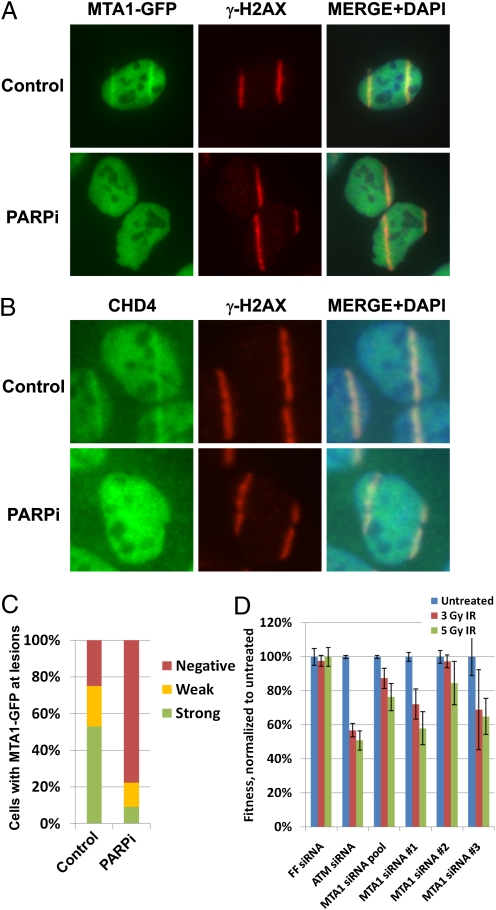
Because PcG and NuRD complexes promote transcriptional repression, we asked whether PARP regulates transcription at sites adjacent to DNA breaks. To visualize transcription, we used antibodies that recognize the 7-methylguanosine (m7G) cap structure present on nascent transcripts (28). Under these conditions we observed a clear absence of staining, essentially an antistripe of m7G staining, that coincided with γH2AX in both human mammary epithelial cells and U2OS cells (Fig. 4A and Fig. S6). Notably, antistripes are not a result of a general loss of DNA from the area exposed to the UV laser, because we can readily detect γH2AX as well as HA-histone H3 after UV microirradiation (Fig. S7A). Furthermore, another relatively abundant chromatin-associated protein, RE1-silencing transcription factor (REST), maintains its localization to chromatin along the laser stripe (Fig. S7B), and the DAPI signal is not decreased at the sites of DNA damage (Figs. S7B and S8), indicating that the antistripes are not an artifact of a general disruption of chromatin structure at the sites of damage.
Fig. 4.
PARP activity is required for inhibition of transcription at sites of DNA damage. (A) Human mammary epithelial cells were microirradiated with a UV laser, fixed after 10–15 min, and immunostained with an antibody that detects the mRNA m7G cap and an anti-γH2AX antibody. (B) HeLa cells were transfected with the indicated siRNAs and were mock-treated or incubated with the PARP inhibitor (PARPi) 1 h before microirradiation, fixed 20 min after irradiation, and immunostained with the anti-m7G cap and anti-γH2AX antibodies. (C) HeLa cells transfected with PARG siRNAs were either mock treated or incubated with the PARP inhibitor (PARPi) 1 hour prior to microirradiation, fixed after 20 min, and immunostained with anti-RNA polymerase II (RNAP2)-ser-2 and anti-γH2AX antibodies. (D) Quantification of the data shown in B and C. (E) HeLa cells transfected with PARG siRNAs were microirradiated, fixed after 20 min, and immunostained with anti-RNAP2 and anti-γH2AX antibodies.
Some cell-to-cell variability in the m7G cap antistripes was apparent. We were able to enhance the antistripe phenotype and make it clearly apparent in all irradiated cells by interfering with the PARP antagonist poly(ADP ribose) glycohydrolase (PARG), an enzyme that counteracts the activity of PARP by disassembling PAR. siRNAs to PARG strongly enhanced the antistripe phenotype (Fig. 4B). To demonstrate that this enhancement resulted from the role of PARG in opposing PARP rather than from a second function of PARG or an off-target effect, we showed that the addition of a PARP inhibitor completely reversed the effects of the PARG siRNAs, indicating that poly-ADP ribosylation was responsible for the antistripes (Fig. 4 B and D).
Loss of nascent RNA could occur in several ways, one of which would be inhibiting transcription through the regulation of RNA polymerase II. RNA polymerase II assembles at start sites in an unphosphorylated form. The initiation of transcription is correlated with phosphorylation of serine 5 of the heptapeptide repeats at the carboxyl-terminal domain of RNA polymerase II. RNA polymerase II is phosphorylated subsequently at serine 2 of the heptapeptide repeats during transcription elongation. Using antibodies to phospho-Ser-2 of the heptad repeats, we readily detected transient loss of RNA polymerase II serine-2 phosphorylation at sites of breaks (Fig. S9A). This transient loss of the elongating RNA polymerase II occurs independently of ATM, ATR, or protein kinase, DNA-activated (DNA-PK) activity (Fig. S9B). As with the m7G antistripes, the phospho-Ser-2 antistripes were enhanced and stabilized for an extended period by siRNAs to PARG in a PARP-dependent fashion (Fig. 4 C and D and Fig. S9A). Although enhancing PARP activity by depletion of PARG strongly enhanced the antistripes observed, the weaker stripes observed in the presence of normal PARG levels were not blocked by PARP inhibition, ATM inhibition, or both, suggesting that the initial antistripes are PARP-independent (Fig. S9C).
Loss of RNA polymerase II serine-2 phosphorylation could occur by loss of the phospho-epitope by dephosphorylation or loss of the polymerase itself, or both. Therefore we investigated the presence of total RNA polymerase II at damage sites. Changes in RNA polymerase II at laser stripes were difficult to detect in normal cells. However, we detected loss of RNA polymerase II in PARG-depleted cells (Fig. 4E). The difference in sensitivity of the phospho-Ser-2 and m7G antibodies compared with RNA polymerase II antibodies for detecting antistripes could be caused by signal-to-noise issues. However, it is also possible that both removal of RNA polymerase II and serine-2 dephosphorylation occur to accomplish maximal repression of transcription.
Discussion
The sequential localization of proteins to sites of DNA damage provides cells with the ability to promote distinct repair activities in a temporally optimized fashion. In this study, we identified proteins that became enriched on chromatin in response to UV-induced DNA damage. Among these proteins were three core subunits of polycomb complexes. PcGs have been studied extensively for their role as transcriptional inhibitors in development, stem cell pluripotency, and cellular senescence. We discovered recruitment of multiple members of PcG complexes to sites of DNA damage induced by UV laser microirradiation.
During the course of our study, two studies reported a link between polycomb proteins and DNA damage that support our findings. The first reported a transient association of PHD finger protein 1 (PHF1), an accessory factor that associates with a minor fraction of PRC2, with DNA lesions (29). These data are consistent with our findings. However, this recruitment is not limited to PHF1, as the authors suggested in their study, but instead involves core members of PRC2 as well as a majority of the PRC1 components. In the second report, EZH2 was detected at exogenous E-cadherin promoter CpG islands by ChIP at the site of a DSB (30). This report is consistent with our findings, although given our observation of the recruitment of EZH2 to UV laser-induced DNA lesions, we believe the association is more likely a general phenomenon that is not limited to promoter CpG islands.
The association of polycomb proteins with DNA lesions occurs independently of ATM, ATR, or histone H2AX. Instead, we find that PcG components are recruited by a PARP1/2-specific pathway. PARP1 and 2 are known to recruit multiple repair factors associated with base excision repair, such as XRCC1, LIG3, and POLB to sites of DNA damage (31). In addition, DNA damage-induced PARP activation has been shown to recruit and activate chromodomain helicase DNA-binding protein 1-like (ALC1), a member of the SNF2 ATPase superfamily that promotes nucleosome sliding (32, 33). PARylation at sites of DNA damage is transient and is reversed by PARG. We found that PARG depletion can enhance the localization of BMI1 to DNA lesions (Fig. S9D), probably by allowing the persistence of PAR structures after PARP has self-inactivated. We also discovered that components of the NuRD complex, MTA1 and CHD4, are recruited to sites of DNA damage in a PARP-dependent manner. The NuRD complex consists of multiple proteins, including two histone deacetylases, HDAC1 and -2, which are known to down-regulate transcription (27). The discovery that PARP recruits both the PcG and NuRD complexes, both negative regulators, suggested that part of PARP’s regulatory program involves transcriptional repression. Indeed, we found that sites of laser microirradiation result in the rapid loss of nascent RNA and elongating RNA polymerase from these regions. This removal is enhanced by depletion of PARG, unless PARG depletion is coupled with PARP inhibition. How this repression is exerted is currently unclear. Members of the PcG proteins have been shown to possess ubiquitin ligase activity toward histone H2A (34–36), suggesting that the recruitment of polycomb proteins may be responsible for the ubiquitination of histone H2A at DNA lesions. However, loss of either polycomb components or MTA1 and CHD4 failed to block removal of the elongating RNA polymerase (Fig. S9 E and F), although the possibility of incomplete depletion via siRNA precludes a definitive interpretation. However, depletion of PcG proteins or MTA1 in mammals or mutation of Ezh2 in C. elegans conferred a DNA damage-sensitivity phenotype, highlighting their importance in promoting DNA repair. It is possible that PARP recruits additional factors that act alone or in concert with PcG and NuRD complexes to block transcription.
RNA polymerases stalled at bulky lesions are known to be removed in part by ubiquitinylation by the damaged DNA-binding protein (DDB1)-Cockayne syndrome A (CS-A)-cullin-4 (CUL4) E3 ubiquitin ligase and degraded by the proteosome in the process called “transcription-coupled repair” (37). However, neither the depletion of CS-A with siRNAs nor inhibition of the proteosome with MG132 affected the removal of RNA polymerase at microirradiation sites (Fig. S9 G and H). Thus, dissection of the mechanism by which the elongating RNA polymerase is removed will require a complete catalog of factors recruited by PAR structures and investigation of their roles alone and in combination. Nevertheless, it is tempting to speculate that the recruitment of PcG and NuRD complexes to DNA lesions may act to inhibit transcription in the vicinity of the DNA lesion and prevent active RNA polymerase II complexes from interfering with the recruitment of repair proteins. Alternatively, the local inhibition of transcriptional activity may be important to prevent the synthesis of truncated mRNA molecules. This notion is reminiscent of the finding that RNA polymerase I transcription can be transiently inhibited in response to DNA damage (38). Similar to the ATM-dependent displacement of RNA polymerase I from ribosomal genes in response to chromosome breaks, the PARP-dependent recruitment of transcription inhibitory factors may act in a parallel pathway to inhibit RNA polymerase II in response to damage outside the nucleolus. We also note that the presence of persistent DNA damage, such as that which results in induction of cellular senescence, might constitutively recruit polycomb complexes to sites of DNA damage, thereby depleting the complexes from other cellular locations, such as the INK4A/ARF locus, and thereby could aid in induction of p16 and other genes during senescence. Although much remains to be learned, the connection between PARP and transient transcriptional repression described here opens an area of investigation in DNA damage response that is likely to be important for the control of genomic stability.
Materials and Methods
Chromatin Fractionation.
HeLa cells were either mock-treated or exposed to 50 J/m2 UV light, harvested by trypsinization, mixed at a 1:1 ratio in the SILAC experiment, and fractionated as described previously (39), with the exception that 1,000 U/mL DNase I was used, and the chromatin fractions (S3 and S4) were collected together for MS analysis.
Competition Assay.
Cell-competition assays were performed essentially as described previously (16). Cells expressing indicated shRNA or siRNA were mixed, either mock-treated or exposed to ionizing radiation, and analyzed on a BD LSRII flow cytometer (BD Biosciences) after 9 or 10 d. The ratio of colorless cells to cells expressing dsRed or humanized Renilla reniformis green fluorescent protein (hrGFP) in the treated population was normalized to the ratio in the untreated population to determine relative fitness after IR.
Immunofluorescence.
Cells were grown on LabTek II chamber-slides (Thermo Scientific) in the presence of 10 μM BrdU for 24 h before induction of DNA damage by a UV-A laser (λ = 355 nm, 40% energy) using a Zeiss Observer.Z1 inverted microscope with a Palm microbeam laser microdissection workstation. After irradiation, the cells were incubated at 37 °C for the indicated time, fixed with 3.7% formaldehyde in PBS, permeabilized with 0.5% Nonidet P-40, and incubated with primary antibodies, followed by secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen). Images were acquired with either an Olympus Fluoview confocal microscope or a Zeiss Axioplan 2 fluorescence microscope.
C. elegans DNA Damage Sensitivity Assay.
C. elegans strains were cultured under standard conditions (40). Bristol N2 worms were used as the wild-type background. Strain SS186 [mes-2(bn11) unc-4(e120)/mnC1 dpy-10(e128) unc-52(e444)II] was used to obtain mes-2 heterozygous and mes-2 homozygous animals. To assess IR sensitivity, young adult animals (24 h after larval stage 4) were irradiated, then singled and scored for embryonic survival (live offspring/eggs laid) over the next 24 h. We assessed 15–30 animals during three separate experiments. Statistical comparisons between genotypes were performed using the two-tailed Mann–Whitney test, 95% confidence interval (InStat3 software).
Supplementary Material
Acknowledgments
We thank J. Jin (University of Texas-Houston Medical School, Houston) for the MSCV-N-GFP gateway expression construct, M. Vidal (Harvard Medical School, Boston) for ORFeome clones, M. Kirschner (Harvard Medical School, Boston) for the HeLa line, A. Nussenzweig (National Cancer Institute, Bethesda, MD) for the H2AX+/+ and H2AX−/− MEFs, S. Boulton (London Research Institute - Clare Hall Laboratories, UK) for the PARP1/2 inhibitor (KU-0058948), and S. Mango (Harvard University, Cambridge, MA) for the mes-2 C. elegans strain. We also thank S. Matsuoka, A. Ciccia, and S. Buratowski for helpful discussions. D.M.C. was supported in part by a National Science Foundation Pre-doctoral Fellowship. X.T. is a Fellow of the Damon Runyon Cancer Research Foundation. K.E.H. is a Special Fellow of the Leukemia and Lymphoma Society. This work was supported by a National Institutes of Health grant (to S.J.E.) and by National Institutes of Health Grants R01GM072551 and R01GM058012 (to M.P.C.) and Grant HG3456 (to S.G.). S.J.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012946107/-/DCSupplemental.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 7.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 13.Dorsman JC, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 15.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: Functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 19.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24:117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- 21.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 23.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- 27.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 28.Reddy R, Singh R, Shimba S. Methylated cap structures in eukaryotic RNAs: Structure, synthesis and functions. Pharmacol Ther. 1992;54:249–267. doi: 10.1016/0163-7258(92)90002-h. [DOI] [PubMed] [Google Scholar]
- 29.Hong Z, et al. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 32.Gottschalk AJ, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Elderkin S, et al. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 37.O’Connell BC, Harper JW. Ubiquitin proteasome system (UPS): What can chromatin do for you? Curr Opin Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Kruhlak M, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.