Abstract
The p53 tumor suppressor is mutated in a high percentage of human tumors. However, many other tumors retain wild-type (wt) p53 expression, raising the intriguing possibility that they actually benefit from it. Recent studies imply a role for p53 in regulation of autophagy, a catabolic pathway by which eukaryotic cells degrade and recycle macromolecules and organelles, particularly under conditions of nutrient deprivation. Here, we show that, in many cell types, p53 confers increased survival in the face of chronic starvation. We implicate regulation of autophagy in this effect. In HCT116 human colorectal cancer cells exposed to prolonged nutrient deprivation, the endogenous wt p53 posttranscriptionally down-regulates LC3, a pivotal component of the autophagic machinery. This enables reduced, yet sustainable autophagic flux. Loss of p53 impairs autophagic flux and causes excessive LC3 accumulation upon starvation, culminating in apoptosis. Thus, p53 increases cell fitness by maintaining better autophagic homeostasis, adjusting the rate of autophagy to changing circumstances. We propose that some cancer cells retain wt p53 to benefit from the resultant increased fitness under limited nutrient supply.
Keywords: apoptosis, posttranscriptional regulation
The p53 tumor suppressor is a major barrier against cancer (1, 2). Accordingly, about one-half of all human tumors harbor p53 gene mutations. However, many other tumors retain expression of wild-type (wt) p53. This is commonly thought to reflect the presence of other genetic or epigenetic alterations that render p53 nonfunctional. However, in some cases, wt p53 may be retained because it provides a benefit to the cancer cells; such cells may misuse wt p53 to reap a selective advantage (3, 4).
Autophagy is a major catabolic pathway for delivery of proteins and organelles to lysosomes or the vacuole, where they are degraded and recycled. Autophagy is activated during nutrient deprivation and is often deregulated under pathological conditions (reviewed in refs. 5 and 6). Autophagy is initiated by the surrounding of cytoplasmic constituents with a crescent-shaped isolation membrane (phagophore), which forms a closed double-membrane autophagosome. The autophagosome fuses with a lysosome to become an autolysosome, where its content is degraded. Elongation of the autophagosomal membrane entails conjugation of MAP1-light chain 3 (LC3) proteins to phosphatidylethanolamine (7). Lipidated LC3 remains associated with autophagosomes until fusion with lysosomes, at which point intra-autophagosomal LC3 is degraded (8, 9).
Many links exist between autophagy and cancer (10–14). An anticancer role for autophagy is implied from its activation by tumor suppressor proteins, including Beclin 1, tuberous sclerosis (TSC)1, TSC2, death associated protein-kinase 1 (DAPK1), and alternate reading frame (ARF). Autophagy was also shown to mediate oncogene-induced senescence (15), a key anticancer mechanism. However, cancer cells encountering limited nutrient supplies in the growing tumor may exploit autophagy for survival; such improved autophagic capabilities will actually benefit the cancer cell (9–13). The complex relationship between autophagy and cancer is also underscored by studies on p53.
p53 modulates the expression of target genes, leading to diverse cellular responses including apoptosis, cell cycle arrest, and senescence (1, 2). p53 promotes autophagy after genotoxic stress through AMP-kinase (AMPK) activation and mammalian target of rapamycine (mTOR) inhibition (16). Transcriptional targets of p53 include sestrins, which activate AMPK and inhibit mTOR (17–19), and damage regulated autophagy modulator (DRAM), a lysosomal protein mediating autophagic cell death (20). However, p53 can also suppress autophagy (21), an effect attributable to cytoplasmic rather than nuclear p53. This is yet another example where, depending on the nature and extent of the inducing stress, p53 may modulate the same process differently to achieve different biological outcomes (12, 13, 22).
We investigated the impact of autophagy regulation by p53 on cancer cells chronically exposed to an unfavorable environment. We report that wt p53 promotes cell survival by enabling a better autophagic response to chronic nutrient deprivation, and we implicate regulation of LC3 in this process.
Results
p53 Loss Causes Aberrant Autophagosome Accumulation in Chronically Starved HCT116 Cancer Cells.
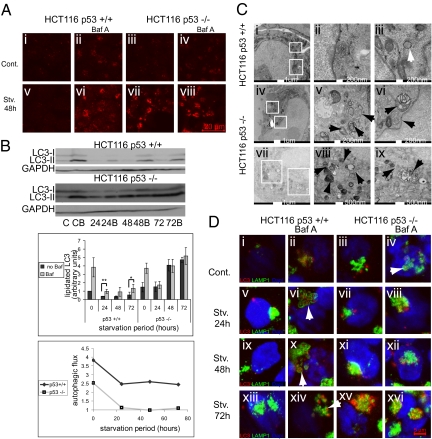
To explore the impact of p53 on autophagy in cancer cells facing chronic nutrient deprivation, we compared human colorectal carcinoma-derived HCT116 cells that retain wt p53 (HCT116 p53+/+) with their isogenic derivatives, in which the p53 gene had been somatically knocked out (HCT116 p53−/−). For nutrient deprivation, cells were placed in Earle's balanced salts solution (EBSS); except for glucose (1 g/L), EBSS contains no nutrients or growth factors. Fig. 1A shows immunostaining of LC3, a canonical marker for autophagosomes. Under standard conditions (Cont), autophagosomes were present in similar small numbers in both p53+/+ and p53−/− cells (Fig. 1A, i and iii). Moreover, both cell types displayed efficient autophagosome recycling (autophagic flux), evident as accumulation of autophagosomes in the presence of the lysosomal inhibitor Bafilomycin A (BafA) (23) (Fig. 1A, ii and iv). Short nutrient deprivation, up to 8–12 h, mildly increased autophagy in both cell types (Fig. S1A). However, on prolonged starvation (24 h or longer), autophagosomes accumulated massively in p53−/− but not in p53+/+ cells (Fig. 1A, v and vii). This could imply increased functional autophagy in the p53−/− cells. However, assessment of autophagic flux refuted this notion. Whereas BafA increased LC3 staining in p53+/+ cells, indicative of productive autophagic flux (Fig. 1A, vi), it failed to do so in p53−/− cells (Fig. 1A, viii), implying that, in the absence of p53, autophagosome recycling became ineffective under prolonged starvation and that the high LC3 staining was actually because of the stalling of autophagy, causing aberrant accumulation of static autophagosomes. This was confirmed by a functional bulk protein degradation assay monitoring autophagic activity. Although protein degradation increased markedly in HCT116 p53+/+ cells after 24 h of starvation (Fig. S1C), the effect was attenuated in p53−/− cells.
Fig. 1.
p53 reduces autophagy initiation during prolonged starvation of HCT116 cells, and lack of p53 causes aberrant accumulation of LC3 and autophagosomes. p53+/+ or p53−/− cells were incubated in control medium (McCoy's medium supplemented with 10% FCS and 2 mM l-glutamine) or Earle's balanced salts solution (EBSS) for 48 h; 1 mM Bafilomycin A (BafA) was added, where indicated, for the last 3 h of incubation, and cells were subsequently (A) fixed for immunostaining with anti-LC3 antibodies (the experiment was repeated three times, and representative images are shown) or (B) lysed as explained in Materials and Methods, resolved on 15% SDS/PAGE, Western blotted with the indicated antibodies, and quantified using Image-J software. For each sample, lipidation was calculated as the ratio between LC3II and GAPDH. Lipidation of nonstarved p53+/+ cells was set to 1, and the rest of the samples were normalized accordingly. Average results of three separate experiments are presented. Autophagic flux was calculated by dividing the value of lipidated LC3 in the presence of BafA by that without BafA. *P value < 0.05. **P < 0.01. (C) Cells starved for 48 h in EBSS were fixed in Epon and imaged by transmission EM (TEM). Representative images of a p53+/+ cell (Top) and two p53−/− cells (Middle and Bottom) are shown. White rectangles in Left indicate regions enlarged in Center and Right. White arrow indicates an intact autophagosome; black arrows indicate aberrant accumulation of autophagosomes. (D) HCT116 p53+/+ or p53−/− cells kept in control medium or starved in EBSS as indicated, in the presence or absence of 1 mM BafA for the last 3 h of incubation where indicated, were fixed for immunostaining with anti-LC3 antibodies (red), anti-LAMP1 antibodies (green), and DAPI (blue). Arrows indicate autolysosomes.
LC3 is converted to lipidated LC3 (LC3-II) on induction of autophagy, slowing down its migration relative to nonlipidated LC3-I in SDS/PAGE. We monitored LC3-II levels under different conditions. Because the anti-LC3 antibodies react preferentially with LC3-II and less so with LC3-I, LC3-II levels were normalized to GAPDH rather than to total LC3 (24). Prolonged starvation reduced LC3-II levels in p53+/+ cells (Fig. 1B). Autophagic flux, deduced from the ratio between the amount of LC3-II in the presence of BafA and the amount in its absence, was modestly reduced but persisted overall (Fig. 1B). Hence, although the rate of initiation of autophagic events slowed down, initiated autophagosomes concluded the process successfully and were recycled. In stark contrast, LC3-II accumulated copiously in chronically starved p53−/− HCT116 cells but did not further increase with BafA (Fig. 1B), implying stalled autophagic flux. This trend began to emerge already at 8 h of starvation (Fig. S1B), becoming more prominent by 24 h (Fig. 1B).
When p53+/+ cells starved for 48 h were examined by transmission EM (TEM), very few autophagosomes were seen in most cells (Fig. 1C, i–iii). In contrast, the majority of p53−/− cells displayed extensive accumulation of double and multimembraned structures with a broad range of morphologies, presumably corresponding to stalled autophagosomes or autolysosomes (Fig. 1C, iv–ix). Similar structures were occasionally seen also in p53+/+ cells but at much lower abundance.
We next performed immunofluorescence analysis for LC3 and lysosomal-associated membrane protein (LAMP)-1 as markers for autophagosomes and lysosomes, respectively (Fig. 1D and Fig. S1D). In p53+/+ cells, extensive colocalization of autophagosomes and lysosomes was evident with but not without BafA, confirming functional autophagic flux (Fig. 1D, ii, vi, x, and xiv). A ring of LAMP-1 staining often encircled the LC3 staining (arrows), suggesting that BafA caused accumulation of autophagosomal material within autolysosomes. However, in p53−/− cells, whereas engulfment of autophagosomes by LAMP-1–decorated rings was observed under basal conditions in a BafA-dependent manner (Fig. 1D, iv), extended starvation led to BafA-independent LC3 accumulation (Fig. 1D, vii, xi, and xv). Remarkably, LC3 accumulated preferentially in large clusters localized next to but often not within lysosomes. This suggests that, in chronically starved HCT116 cells, p53 tunes down autophagy to an affordable rate. In the absence of p53, cells continue to form abundant autophagosomes that cannot be efficiently recycled and accumulate aberrantly without concluding their catabolic mission. Of note, lysosomal acidity did not decrease in p53−/− cells compared with the p53+/+ cells but rather, increased (Fig. S1E), suggesting that lysosomal quantity and integrity were maintained.
p53 Is Required for LC3 mRNA Down-Regulation in Chronically Starved HCT116 Cells.
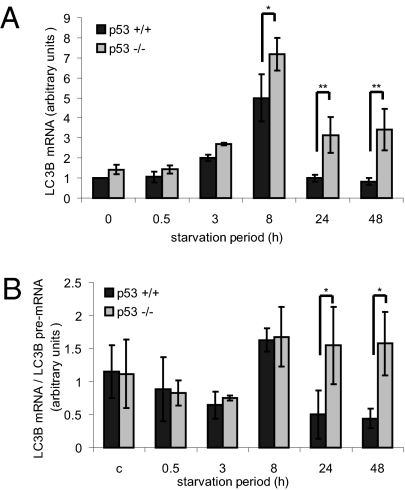
As noted in Fig. 1B, LC3 protein levels decreased in chronically starved p53+/+ but not p53−/− HCT116 cells. To further investigate this difference, we monitored LC3 mRNA. As seen in Fig. 2A, levels of mRNA encoding LC3B, one of the three human LC3 isoforms, did not change on very short starvation, as reported earlier (25). However, by 8 h, LC3B mRNA increased about fivefold; this occurred in both cell types, suggesting that p53 is not involved. On further starvation, LC3B mRNA began to decrease, reaching basal levels within 24 h in p53+/+ cells. In contrast, in p53−/− cells, LC3B mRNA remained approximately twofold higher than basal even after 48 h. Another LC3 isoform, LC3A, displayed a similar pattern (Fig. S2A). Hence, p53 is required for efficient down-regulation of LC3 mRNA in chronically starved HCT116 cells.
Fig. 2.
p53 down-regulates LC3 mRNA posttranscriptionally during chronic starvation of HCT116 cells. p53+/+ or p53−/− cells kept in control medium or starved in EBSS as indicated were harvested, and RNA was extracted and subjected to quantitative RT-PCR analysis. (A) qRT-PCR analysis of LC3B mRNA. Levels measured for nonstarved p53+/+ cells were set to 1, and the rest of the samples were normalized accordingly. Average results of three experiments are presented. *P < 0.05. **P < 0.01. (B) Time-dependent changes in the ratio between LC3B mature mRNA and premRNA. Results of three experiments performed as in Fig. S2B were analyzed so that the values for LC3B mRNA were divided by the corresponding values for LC3B premRNA and averaged. *P < 0.05.
p53 is a transcriptional regulator. Hence, it was plausible that it represses LC3 gene transcription. However, changes in mRNA abundance can also reflect posttranscriptional processes. We, therefore, assessed the impact of starvation on the relative amounts of primary LC3B transcript (LC3B pre-mRNA). quantitative RT-PCR was performed as in Fig. 2A but with primers derived from intronic LC3B sequences, which allowed quantification of immature pre-mRNA to better approximate relative transcription rates (26–28). Surprisingly, except for an early increase in LC3B pre-mRNA observed already after 3 h, the starvation-induced changes affected mainly the mature rather than the primary transcripts (Fig. S2B). The ratio between LC3B mRNA and pre-mRNA increased at 8 h in both p53+/+ and p53−/− cells (Fig. 2B), suggesting p53-independent stabilization of mature LC3B mRNA. Remarkably, longer starvation greatly decreased the LC3 mRNA/pre-mRNA ratio in p53+/+ cells but hardly in p53−/− cells. This strongly suggests that, in HCT116 cells, p53 promotes the posttranscriptional down-regulation of LC3 mRNA on prolonged nutrient deprivation.
LC3 has several functional homologs, including Golgi-associated ATPase enhancer (GATE)-16 and GABA-receptor associated protein (GABARAP); neither of the corresponding mRNAs changed during starvation (Fig. S2C). We also monitored the mRNA of p62, an important LC3 interactor in autophagy (29, 30). Like LC3 mRNA, p62 mRNA was up-regulated at 8 h, albeit more mildly, and down-regulated later (Fig. S2D); however, this was p53-independent.
p53 Enhances HCT116 Cell Survival Under Prolonged Starvation.
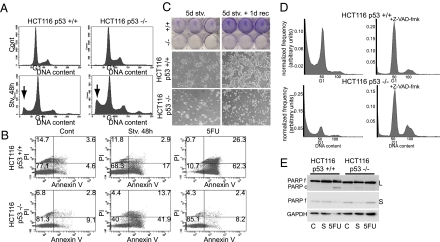
p53 can induce apoptosis in response to severe stress. We, therefore, tested the effect of p53 on starvation-induced cell death by performing propidium iodide (PI) staining followed by flow cytometry; apoptotic cells were identified by their sub-G1 DNA content. Surprisingly, prolonged starvation promoted more apoptosis in p53−/− HCT116 cells than in p53+/+ cells (Fig. 3A), implying that p53 protects these cancer cells from chronic starvation-induced death. This was further confirmed by Annexin V staining (Fig. 3B): over 50% of the p53−/− cells were either in early apoptosis (Annexin V+, PI−) or dead (Annexin V+, PI+) at 48 h compared with only 20% in p53+/+ cells. In contrast, p53+/+ cells died much more readily on treatment with 5-fluorouracil (5-FU), a known inducer of p53-dependent apoptosis. siRNA-mediated p53 down-regulation in the p53+/+ cells before starvation resulted in an increased sub-G1 population (Fig. S3A), further arguing that p53 inhibits apoptosis under these conditions. Notably, p21 knockdown did not affect cell death (Fig. S3A). Levels of phosphorylated acetyl-CoA-carboxylase (ACC), an AMPK substrate, did not increase, indicating that AMPK was not activated under these conditions (Fig. S3B). A protective effect of p53 during prolonged starvation was seen also in HepG2 hepatocellular carcinoma cells harboring wt p53 (Fig. S3C). However, p53 knockdown did not increase starvation-induced death of wt p53-expressing U2OS human osteosarcoma cells (Fig. S3D). Thus, in some but not all wt p53-positive cancer cell lines, p53 provides better protection against starvation-induced death. Interestingly, starved p53−/− HCT116 cells exhibited increased polyploidy (Fig. 3A), which became more prominent when death was blocked (Fig. 3D and Fig. S3F). This suggests that functional autophagic flux reduces the incidence of genomic instability, supporting earlier findings (31).
Fig. 3.
p53 enhances HCT116 cell survival during chronic starvation. (A) p53+/+ or p53−/− cells kept in control medium or starved in EBSS for 48 h were trypsinized, fixed in methanol, stained with propidium iodide (PI), and analyzed in a FACSsort flow cytometer. A representative result of three experiments is shown. The position of G1 is indicated, and arrows indicate the subG1 subpopulation in starved cultures. (B) p53+/+ or p53−/− cells were kept in control medium, starved in EBSS for 48 h or treated with 5-FU for 16 h, trypsinized, and stained with Annexin V and PI. A representative result of two experiments is shown. The percentage of cells in each quadrant is indicated. (C) p53+/+ or p53−/− cells were starved in EBSS for 5 d, after which live cell images were taken (Middle Left and Bottom Left), and cells were fixed and stained with crystal violet (Top Left) or EBSS was replaced with control medium for 24 h before imaging and staining (Right). A representative result of three experiments is shown. (D) p53+/+ or p53−/− cells were starved in EBSS with or without Z-VAD-fmk for 48 h before fixation and analysis in an LSR-II flow cytometer. The experiment was repeated two times; a representative result is shown. Data analysis was performed by Matlab using FCS data reader script. (E) p53+/+ or p53−/− cells were kept in control medium, starved in EBSS for 48 h or treated with 5-FU for 16 h before lysis, and analysis on 10% SDS/PAGE. (Top) Anti-PARP, long exposure (L). (Middle) Anti-PARP, short exposure (S). (Bottom) Anti-GAPDH.
Extension of the starvation to 5 d further exacerbated the deleterious effect of p53 depletion: substantial numbers of cells were observable in the p53+/+ cultures, both macroscopically and microscopically, but not in the p53−/− ones (Fig. 3C). Importantly, replenishment with full medium led to a rapid increase in the number of p53+/+ cells within 1 d (Fig. 3C), showing that not only are the cells viable but they are also capable of resuming vigorous proliferation as soon as nutrient supply is resumed. Furthermore, when maintained for several weeks under continuous milder nutrient deprivation, many more cells remained in the p53+/+ than in the p53−/− cultures (Fig. S3E).
The general caspase inhibitor Z-VAD-FMK (fluoro methyl ketone) effectively rescued HCT116 cells of both genotypes from starvation-induced death, further attesting to the apoptotic nature of this death (Fig. 3D and Fig. S3F). Of note, prolonged starvation triggered poly(ADP ribose) polymerase (PARP) cleavage, a hallmark of apoptosis, in the p53−/− HCT116 cells; this was detectable as reduced amounts of the noncleaved form (Fig. 3E f) and appearance of the cleaved form (Fig. 3E c). As expected, an opposite impact of p53 status was observed on 5-FU treatment.
Together, these data imply that p53 can delay apoptosis and promote long-term survival in HCT116 cancer cells encountering prolonged nutrient deprivation.
LC3 Knockdown Rescues p53-Deficient HCT116 Cells from Apoptosis.
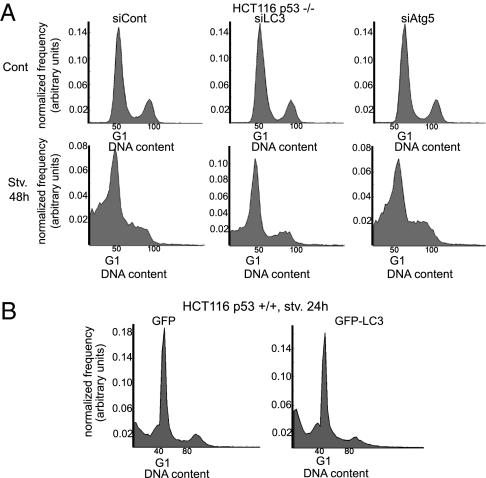
To assess whether the inability of p53-deficient HCT116 cells to down-regulate LC3 on prolonged starvation contributes to their increased death, apoptosis was monitored after siRNA-mediated knockdown of endogenous LC3. Combined knockdown of all three LC3 isoforms markedly reduced the sub-G1 population (Fig. 4A and Fig. S4A), strongly arguing that the excessive LC3 levels contribute to the enhanced apoptosis. Conversely, overexpression of GFP-LC3 modestly increased apoptosis in p53+/+ cells (Fig. 4B). Notably, knockdown of either Atg5 or Beclin 1, required for LC3 lipidation (6), also reduced apoptosis to some extent (Fig. 4A and Fig. S4D). However, although the knockdown was efficient (Fig. S4B) and autophagy was attenuated, as evident by reduced LC3-II flux and accumulation of LC3-I in siAtg5 cells (Fig. S4C), the protective effect was substantially milder. Hence, excessive LC3 may also promote cell death through additional mechanisms that do not depend on its lipidation and are not necessarily related to autophagy.
Fig. 4.
Knockdown of LC3 rescues p53-deficient HCT116 cells from apoptosis. (A) p53−/− cells were transfected with 150 nM control (100 nM siLacZ plus 50 nM nontargeting siControl 2), LC3-specific (50 nM siLC3A, 50 nM siLC3B, 50 nM siLC3C), or Atg5-specific (50 nM siAtg5 plus 100 nM siLacZ) siRNA oligos; 48 h later, the cells were starved in EBSS or kept in control medium for 48 h before staining with PI and FACS analysis as in Fig. 3D. A representative result of three experiments is shown. (B) p53+/+ cells were transfected with enhanced-GFP (eGFP) vector or with eGFP-LC3; 24 h later, the cells were starved in EBSS for 24 h before fixation and FACS analysis as in Fig. 3D. A representative result of three experiments is shown.
p53 Enhances the Survival of Chronically Starved Nontransformed Cells.
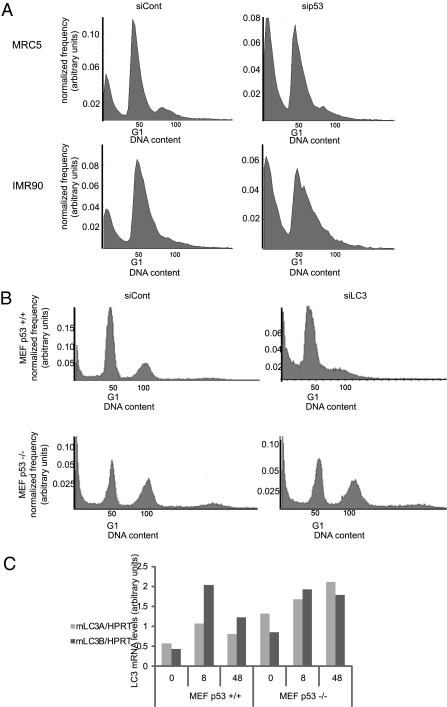
We next sought to determine whether p53 also protects noncancerous cells against starvation-induced apoptosis. As seen in Fig. 5A and Fig. S5A, p53 knockdown rendered both MRC5 and IMR90 human embryonic diploid lung fibroblasts more sensitive to death on extended nutrient deprivation. Similarly, starved p53-null mouse embryonic fibroblasts (MEFs) exhibited higher apoptosis rates than wt MEFs (Fig. 5B, siCont panels). Hence, this protective function of p53 is an inherent feature of normal cells.
Fig. 5.
p53 promotes survival of nontransformed cells during prolonged starvation. (A) MRC5 or IMR90 cells were transfected with 20 nM control (siLacZ) or p53-specific (sip53) siRNA oligonucleotides; 48 h later, cells were placed in EBSS for 48 h before staining with PI and FACS analysis as in Fig. 3D. A representative result of three experiments is shown. (B) Wt or p53 −/− MEFs were transfected with 50 nM control (siLacZ) or mouse LC3-specific (20 nM mLC3A plus 30 nM mLC3B) siRNA oligonucleotides; 48 h later, cells were placed in EBSS and analyzed as in A. A representative result of three experiments is shown. (C) Wt or p53 −/− MEFs kept in control medium or starved in EBSS as indicated were harvested, and RNA was extracted and subjected to qRT-PCR analysis with the indicated primers. Results were normalized for HPRT mRNA in the same samples. A representative result of three experiments is shown.
As in HCT116 cells, p53 was also required in MEFs for effective down-regulation of LC3 mRNA on extended starvation (Fig. 5C, 48 h). However, unlike in HCT116, LC3 knockdown did not affect the death of starved p53−/− MEFs; rather, it augmented the death of p53-proficient cells (Fig. 5B and Fig. S5B). Although this may seem at odds with the HCT116 data, there is actually a common denominator: in both cases, p53 seems to increase fitness by optimizing the autophagic response. In MEFs, basal autophagic flux is low (Fig. S5C), and the main challenge under starvation is to induce and maintain higher autophagy rates. This is at least partially p53-dependent (Fig. S5C Lower), as reported earlier (16). In contrast, basal autophagic flux in HCT116 is already quite high (Fig. 1B), presumably owing to the deranged metabolism of cancer cells and the subsequent constitutive damage (3); hence, preventing excessive autophagy is more critical for the chronically starved cancer cells. In either case, p53 loss compromises the cell's ability to adjust autophagy to changing circumstances over time, rendering it more vulnerable to the detrimental effects of prolonged starvation.
Discussion
p53 has been linked with regulation of autophagy; however, the exact nature of this link remains seemingly controversial. We now suggest that p53 can modulate autophagy in a context-dependent manner and propose that p53 helps maintain autophagic homeostasis. Thus, p53 is neither merely a positive nor negative regulator of autophagy but rather acts as a rheostat that continuously adjusts the rate of autophagy to changing circumstances. By so doing, p53 endows cells with increased fitness and better long-term survival. Its loss compromises the ability to adjust the autophagic response properly.
We show that extension of cell survival in the face of prolonged nutrient deficiency is an inherent feature of p53, observable in a variety of normal cells. Indeed, although p53 is better known for its proapoptotic activities, it can also possess prosurvival effects, particularly under mild stress conditions (32–34). Such cytoprotective function of p53 may be part of its increasingly appreciated role in maintaining intracellular metabolic homeostasis. In a healthy organism, this may reduce the incidence of cancer-promoting cellular derangements. However, some cancer cells may take advantage of this prosurvival feature of p53: if they successfully complete the transformation process without acquiring a p53 mutation, subsequent retention of wt p53 might render them more resilient under conditions of changing nutrient availability. Indeed, blockage of autophagic flux may promote tumor cell death in vivo under conditions of stress (35). Our findings join a growing number of examples where cancer cells misuse p53 functions meant to serve the wellbeing of the organism (3, 4).
The mechanisms whereby p53 maintains autophagic homeostasis likely differ among different cell types. In the case of HCT116 cells, p53 promotes the selective down-regulation of LC3 RNA and protein under conditions of prolonged nutrient deprivation. This presumably enables the tuning down of autophagy initiation to rates compatible with the gradually diminishing metabolic capabilities of these cells. Consequently, sustainable autophagic flux is maintained over a longer period. Conversely, p53 loss drives excessive LC3 production, forcing an attempt to maintain high autophagy rates. However, owing to continued depletion of essential resources, the process is eventually stalled, slowing down autophagic flux and causing accumulation of excessive aberrant intermediates and eventually, apoptotic cell death.
p53 is primarily a transcriptional regulator. Surprisingly, we find that the impact of p53 on LC3 mRNA down-regulation is chiefly posttranscriptional. This raises the intriguing possibility that, in line with the observation that cytoplasmic p53 modulates autophagy (21), p53 may promote the degradation of mature LC3 mRNA, perhaps through its RNA binding activity (36). Alternatively, p53 may regulate genes encoding other RNA binding proteins. Either way, this deserves further investigation.
The extent to which our in vitro observations pertain to a variety of actual in vivo situations remains to be rigorously determined in both normal and cancerous tissues. However, it is noteworthy that, whereas starvation leads to induction of autophagy in several tissues of wt mice, this does not happen in p53-null mice, where autophagosomes are present in higher numbers under optimal conditions but starvation does not induce any further change (21).
Altogether, our findings imply that p53 ensures autophagic homeostasis, tuning it up or down as dictated by changes in nutrient availability. On the one hand, this could serve as an anticancer mechanism (e.g., by maintaining genomic stability) (31), in which case tumorigenesis would select for loss of wt p53 as is indeed seen in many cancers. On the other hand, however, the prosurvival effects described above are likely to drive a selective pressure for retaining wt p53 in some cancer cells. Which alternative eventually wins is likely to depend largely on the particular combination of additional genetic and epigenetic events in a particular tumor as well as the environmental stresses that it has been exposed to during its progression.
Materials and Methods
Cell Culture, Reagents, and siRNA Transfection.
Cell culture growth and starvation conditions, reagents, and transfection details are listed in SI Materials and Methods.
EM.
Cells were grown on coverslips and fixed with 0.3% glutaraldehyde and 0.3% paraformaldehyde in cacodylate buffer (cacodylate 0.1 m, pH 7.4). Samples were dyed in 1% OsO4, 0.3% potassium dichromate, and 0.3% potassium ferrocyanide and embedded in Epon. Sections of 70–100 nm were viewed by TEM (Technai-12; Phillips).
Protein Analysis.
The following commercial antibodies and reagents were used: anti-LC3B (L7543; Sigma); anti-GAPDH (Chemicon); anti-PARP (SA-250; Biomol), anti-LAMP1 (h4a3; Developmental Studies Hybridoma Bank, University of Iowa), DAPI (Sigma), and LysoTracker Red (DND-99; Molecular Probes). For Western blot analysis, cells were lysed in radioimmunoprecipitation (RIPA) buffer (0.1 M NaCl, 5 mM EDTA, 0.1 M Na2HPO4/NaH2PO4, pH 7.5, 1% Triton, 0.5% deoxycholate (DOC), 0.1% SDS, and protease inhibitors), and 40 μg of lysate were loaded to SDS/PAGE. For immunofluoresence, cells were fixed in methanol, permeabilized with acetone, incubated with the indicated antibodies, and visualized by a Zeiss LSM-510 or Zeiss LSM-710 confocal microscope.
RNA Analysis.
Total RNA was isolated using the mirVana miRNA isolation kit (Ambion). cDNA and real-time PCR were performed as described (37). For LC3B premRNA analysis, RNA was incubated with (RT) or without (non-RT) reverse transcriptase in the presence of random hexamer primers before being subjected to real-time PCR. The non-RT real-time PCR values were subtracted from the RT values to obtain the data presented in Fig. 2B and Fig. S3D. Primer sequences are detailed in Table S1.
FACS Analysis.
FACS-assisted cell cycle analysis for DNA content was performed as described (37). Analysis of apoptotic cells was performed using a FITC-annexin-V apoptosis detection kit (Roche). Samples were analyzed using a FACS sorter (Becton Dickinson) or LSR-II (BD Biosciences) flow cytometer. Results from the LSR-II flow cytometer were analyzed by Matlab using an FCS data reader script.
Supplementary Material
Acknowledgments
We thank Adi Kimchi (Weizmann Institute, Rehovot, Israel) and her team for helpful discussions and reagents, Vera Shinder for helpful discussions and technical guidance, Dan Michael for sharing ideas, Gilad Fuchs, Gil Hornung, and Noa Levi for scientific and technical assistance, and Bert Vogelstein (Johns Hopkins University, Baltimore) and Varda Rotter (Weizmann Institute, Rehovot, Israel) for the gift of cell lines. R.S.-S. is recipient of the Sir Charles Clore postdoctoral fellowship. Z.E. is incumbent of the Harold Korda Chair of Biology; M.O. is incumbent of the Andere Lwoff chair in Molecular Biology. This work was supported in part by Grant R37 CA40099 from the National Cancer Institute, a grant from the Robert Bosch Foundation, the Israel Science Foundation and the Israeli Cancer Research Foundation (to Z.E.), and the M. D. Moross Institute for Cancer Research and the Yad Abraham Center for Cancer Diagnosis and Therapy (to M.O.). The electron microscopy studies were conducted at the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging at The Weizmann Institute of Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006124107/-/DCSupplemental.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim E, Giese A, Deppert W. Wild-type p53 in cancer cells: When a guardian turns into a blackguard. Biochem Pharmacol. 2009;77:11–20. doi: 10.1016/j.bcp.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. Physiological functions of autophagy. Curr Top Microbiol Immunol. 2009;335:71–84. doi: 10.1007/978-3-642-00302-8_3. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 10.Corcelle EA, Puustinen P, Jäättelä M. Apoptosis and autophagy: Targeting autophagy signalling in cancer cells—‘trick or treats’? FEBS J. 2009;276:6084–6096. doi: 10.1111/j.1742-4658.2009.07332.x. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 12.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiuri MC, et al. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 14.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiuri MC, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 20.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 21.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 24.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda M, et al. Effects of 3-methylcholanthrene on the transcriptional activity and mRNA accumulation of the oncogene hWAPL. Cancer Lett. 2005;221:21–28. doi: 10.1016/j.canlet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Phelps ED, Updike DL, Bullen EC, Grammas P, Howard EW. Transcriptional and posttranscriptional regulation of angiopoietin-2 expression mediated by IGF and PDGF in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C352–C361. doi: 10.1152/ajpcell.00050.2005. [DOI] [PubMed] [Google Scholar]
- 28.Shema E, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassus P, Ferlin M, Piette J, Hibner U. Anti-apoptotic activity of low levels of wild-type p53. EMBO J. 1996;15:4566–4573. [PMC free article] [PubMed] [Google Scholar]
- 34.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosner J, et al. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aylon Y, et al. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.