Abstract
Computational studies of biological networks can help to identify components and wirings responsible for observed phenotypes. However, studying stochastic networks controlling many biological processes is challenging. Similar to Schrödinger’s equation in quantum mechanics, the chemical master equation (CME) provides a basic framework for understanding stochastic networks. However, except for simple problems, the CME cannot be solved analytically. Here we use a method called discrete chemical master equation (dCME) to compute directly the full steady-state probability landscape of the lysogeny maintenance network in phage lambda from its CME. Results show that wild-type phage lambda can maintain a constant level of repressor over a wide range of repressor degradation rate and is stable against UV irradiation, ensuring heritability of the lysogenic state. Furthermore, it can switch efficiently to the lytic state once repressor degradation increases past a high threshold by a small amount. We find that beyond bistability and nonlinear dimerization, cooperativity between repressors bound to OR1 and OR2 is required for stable and heritable epigenetic state of lysogeny that can switch efficiently. Mutants of phage lambda lack stability and do not possess a high threshold. Instead, they are leaky and respond to gradual changes in degradation rate. Our computation faithfully reproduces the hair triggers for UV-induced lysis observed in mutants and the limitation in robustness against mutations. The landscape approach computed from dCME is general and can be applied to study broad issues in systems biology.
Keywords: bistable switch, discrete chemical master equation, epigenetic control, stochasticity, stochastic switch
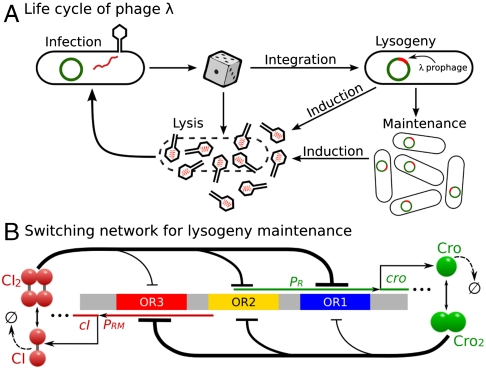
Bacteriophage lambda is a virus that infects Escherichia coli cells. It has served as a model system for studying regulatory networks and for engineering gene circuits (1–5). Of central importance is the molecular circuitry that controls phage lambda to choose between two productive modes of development, namely, the lysogenic state and the lytic state (Fig 1A). In the lysogenic state, phage lambda represses its developmental function, integrates its DNA into the chromosome of the host E. coli bacterium, and is replicated in cell cycles for potentially many generations. When threatening DNA damage occurs, phage lambda switches from the epigenetic state of lysogeny to the lytic state and undergoes massive replications in a single cell cycle, releases 50–100 progeny phages upon lysis of the E. coli cell. This switching process is called prophage induction (5).
Fig. 1.
Different selection of cell fate of E. coli infected by phage lambda and a model of the epigenetic circuit for lysogeny maintenance. (A) The lysogenic and lytic states of phage lambda. (B) A simplified model of the epigenetic switch for lysogeny maintenance.
The molecular network that controls the choice between these two different physiological states has been studied extensively during the past 40 y (5–9). All of the major molecular components of the network have been identified, binding constants and reaction rates characterized, and there is a good experimental understanding of the general mechanism of the molecular switch (5). Theoretical studies have also contributed to the illumination of the central role of stochasticity (3) and the stability of lysogen against spontaneous switching (4, 10). With the advent of systems biology, studying the switching network of phage lambda and lysogeny maintenance has gained added importance, because it provides an ideal ground for developing models and algorithms to study regulatory networks.
However, a general bottleneck problem for computational studies of regulatory networks is the limitation of existing techniques for studying stochastic networks. Because reactions often involve only low copy numbers of molecules and have large separations in timescale, stochasticity has a strong influence on the behavior of molecular networks (3, 4, 10). Deterministic models based on the principles of mass action are often incapable of capturing the multistable nature of the network when copy numbers are small (11). Although the theory of the chemical master equation (CME) provides a general framework for studying stochastic networks (12, 13), there are no analytical solutions to the CME except for simple toy problems (14).
One approach is to approximate the CME through various formulations of stochastic differential equations, including the Langevin and the Fokker–Planck formulation (12). However, the consequences of such approximations for realistic problems involving many molecular species and complex reactions are unknown. Another approach is to carry out extensive stochastic simulations. Powerful simulation tools, including the Gillespie algorithm, have been developed (13, 15). With this approach, many trajectories of simulated reaction events are followed, which are analyzed to reconstruct a probabilistic picture of the stochastic network.
Because the CME plays roles in systems biology equivalent to the roles the Schrödinger equation played in quantum mechanics, the development of computational solutions to the CME has important implications, just as the development of techniques for solving the Schrödinger equation for systems with many atoms does (16, 17). However, currently no numeric algorithms can solve the CME directly. Stochastic simulations can follow many cellular events, but have difficulty characterizing rare events that may be biologically critical. As the switching network in phage lambda is stable against random fluctuation, the transition from lysogeny to lysis occurs rarely under normal conditions. In this case, it is difficult to determine whether adequate sampling has been achieved, and for each trajectory whether the simulation time is sufficient.
In this paper, we describe a general approach to study molecular stochastic networks, called discrete CME (dCME), by directly obtaining accurate steady-state solutions to the CME that underlies a molecular network. Using a model of the network for the maintenance of lysogeny in phage lambda, we calculate its steady-state probability landscape, including those associated with the transition phase from lysogeny to lysis. Such a full stochastic characterization was previously computationally inaccessible. To understand the basic properties required for the maintenance of lysogeny, we characterize the probability landscape of the networks of wild-type and mutant phage lambda at different physiological conditions and identify the molecular and architectural determinants of the observed biological behavior. We aim to understand the origin of the network’s stability against environmental fluctuations in UV irradiation, the basis of its efficiency in switching, its robustness against changes in network components, as well as the mechanism of the heritability of the epigenetic state of lysogeny.
Results
Model of Epigenetic Switch of Phage Lambda.
To study how lysogeny is maintained and how it transitions to the lytic state, we assume that lysogeny has already been established. We use a simplified stochastic model for the molecular regulatory network that controls the epigenetic switch in phage lambda (Fig 1B). Using a total of 54 biochemical reactions involving 13 molecular species, our model explicitly includes key components, essential reactions, and cooperativities of the phage lambda decision circuitry. Details can be found in Methods and SI Appendix.
A stochastic biochemical network is characterized at any instant by the probability associated with each microstate of the network, namely, the probability of a specific combination of copy numbers of the molecular species. If the probabilities for all possible microstates at an instant are known, we have the probability landscape of the biochemical network for that instant. For a given initial condition, this landscape usually evolves with time. Our interest here is the steady-state probabilistic landscape, which describes the overall behavior of phage lambda in the steady state. This landscape also governs the transient chemical reaction dynamics of the system. As the microstates of a biochemical network in general are too numerous, the probabilistic landscape usually cannot be directly computed. Here we show the probability landscape of the decision circuit of phage lambda can be studied using the newly developed method for optimal enumeration of microstates and for exact calculation of the steady-state probability landscape (18). Briefly, this method slices through the high-dimensional space of microstates following the submanifold of accessible microstates for a given initial condition, without visiting the vast space outside this submanifold. Technical details of this algorithm can be found in ref. 18.
Probability Landscape of Phage Lambda in Lysogenic and Lytic States.
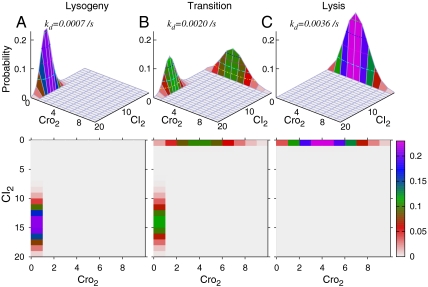
The epigenetic network model shown in Fig 1B can reach around 1.7 million microstates. We have calculated through dCME the steady-state probability associated with each of these microstates. Because the space of the microstate is 13D for 13 molecular species, we project the landscape to the 2D subspace and record only the total copy numbers of CI dimer (CI2) and Cro dimer (Cro2) molecules. With a high copy number of CI2, also known as repressor, the lysogenic state of the phage lambda is maintained, whereas a high copy number of Cro2 protein signifies the lytic state (6). The CI2 copy number therefore can be regarded as an indicator of the physiologic state of the phage. Fig 2A shows a probability landscape of the lysogenic state, which has one pronounced peak centered at around the location of 14 copies of CI2 and 0 copies of Cro2. As a range of values of CI2 copy numbers all have high probability in the lysogenic state, our results suggest that minor changes in CI2 concentration during cell growth and cell division will not affect this epigenetic state. This insensitivity to fluctuation in CI degradation is important to ensure the heritability of the lysogenic state. Such a conclusion cannot be drawn from studies obtained using the ordinary differential equation (ODE) model, which would give only a fixed point of CI2 concentration for the lysogenic state, rather than a probability distribution.
Fig. 2.
The probability landscape of the epigenetic circuits of lysogeny maintenance in phage lambda. (A) At the CI degradation rate of kd = 7.0 × 10-4/s, probability landscape centers at locations with high copy numbers of CI and close to 0 copy of Cro. This landscape corresponds to the lysogenic state of phage lambda. The landscape is shown both in three- and two-dimensional projections. (B) The probability landscape in transition with simultaneously two peaks when phage lambda is being induced to the lytic state. When kd increases from kd = 1.8 × 10-3/s to 2.2 × 10-3/s, the peak located at lysogenic states gradually diminishes, whereas the peak located at lytic states gradually increases. At about kd = 2.0 × 10-3/s, phage lambda has about equal probability to be in either lysogenic or lytic state (see SI Appendix for more information). (C) When CI is degraded at a faster rate of kd = 3.6 × 10-3/s, the probability landscape centers at locations where there are higher copy numbers of Cro dimer and close to 0 copy of CI. This landscape corresponds to the lytic state of phage lambda.
We then examine the effects of accelerated CI degradation due to DNA damage from UV irradiation that leads to the activation of RecA-mediated CI cleavage (5, 19). Fig 2C shows that when the CI degradation rate is raised from kd = 7.0 × 10-4/s to kd = 3.6 × 10-3/s, the probability landscape peaks at a different location, with about four copies of Cro2 and 0 copies of CI2.
Our results show that the steady-state probability landscapes change adaptively. In the lysogenic state, the probability landscape has one peak, which accounts for the vast majority of locales where there is a significant amount of CI2 molecules and few Cro2 molecules. The probability for the system to have high Cro2 copy numbers spontaneously is very small. In conditions that eventually lead to the lytic state, the probabilistic landscape adapts and changes, becoming dominated by a peak at different locations where there is a large amount of Cro and few CI molecules. At the same time, the peak for the lysogenic state disappears (Fig 2C).
High Threshold and Efficient Switching of Phage Lambda.
To model the effects of UV irradiation on RecA-mediated acceleration in CI degradation (5, 19), we have systematically examined the behavior of the network by computing its probability landscape at different CI degradation rates. To summarize our results, we calculated the mean copy numbers of CI2 and Cro2 of the network from the probability landscape. For example, for the probability landscape shown in Fig 2A, we integrate with properly weighted probabilities of the copy numbers of CI2 and Cro2 at each microstate, and obtain the mean copy numbers of 14.3 and 0.0. This calculation is repeated for 36 different values of CI degradation rate representing different dosage levels of UV irradiation. The results are summarized in computationally generated titration curves shown in Fig 3A (solid lines).
Fig. 3.
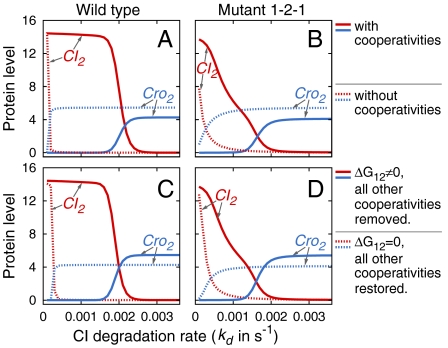
Stability of the epigenetic network for lysogeny maintenance against fluctuation in UV irradiation and its switching efficiency in wild-type phage lambda and key role of cooperativity between CI2 bound on neighboring OR1 and OR2 sites in maintaining lysogeny. The mean copy number of CI2 and Cro2 are plotted against the CI degradation rate kd (in per second unit). (A, solid lines) The lysogenic state in wild-type phage lambda is stable against fluctuation of UV-induced CI degradation rate. The level of CI2 is constant for a wide range of CI degradation rate. The threshold for switching to the lytic state is deep. Switching occurs efficiently once the threshold (kd = 1.8 × 10-3/s) is reached: A further small increase of 0.4 × 10-4/s completely throws the phage lambda to the lytic state. (Dashed lines) When all cooperativities are removed, phage lambda cannot generate sufficient amount of CI2 and therefore cannot maintain lysogenic state. (B) The stability and the deep threshold are missing if all other cooperativities except that between neighbor CI2 dimers on OR1 and OR2 are restored (ΔG12 = 0, dashed line). In contrast, if all other cooperativities are missing, but that between CI2 dimers on OR1 and OR2 is restored (ΔG12 ≠ 0, solid line), phage lambda recovers to a large extent the stability and gained significant depth in its switching threshold. (C and D) The same phenomenon is observed in mutant 1-2-1 studied by Little et al. (8). With cooperativity between OR1 and OR2, this mutant can still enter the lysogenic state, albeit without much stability and the threshold is shallow. Without this cooperativity (ΔG12 = 0), mutant 1-2-1 cannot maintain lysogeny (see SI Appendix for more details).
Our results show that the epigenetic network of phage lambda is very stable against changes in CI degradation rate due to environmental fluctuation of UV irradiation (Fig 3A, solid lines). Phage lambda stays firmly in the lysogenic state at normal condition. Even when UV irradiation is at a dosage that leads to doubling of the degradation rate of CI, the expected copy number of CI2 in the system changes very little. Over a wide range of CI degradation rates (1.0 × 10-4–1.7 × 10-3/s), the lysogenic state is maintained. The switching threshold for the lytic state is high.
Once the switching threshold of degradation rate kd = 1.8 × 10-3/s is reached (Fig 3A, solid lines), a further small increase of 0.4 × 10-3/s turns the expected copy number of CI2 from 88% of its maximum value at kd = 1.8 × 10-3/s to 16%. The system therefore can be fully thrown efficiently to the lytic state with a small additional increase in CI degradation at the threshold.
Cooperativity between OR1 and OR2 Enables the Lysogenic State.
Next we studied the effects of cooperativity. It is well known that repressor binds cooperatively to neighboring operator sites (20). Similarly, Cro dimers also bind cooperatively, although to a lesser extent (21). These cooperativities are fully incorporated in our model (see SI Appendix for details). In addition, we assume that the looping cooperativity always exists in the form of enhanced CI synthesis when OR2 is occupied by a CI2.
We first studied the effects of removal of all cooperativities between neighboring repressors (CI dimers) and between neighboring Cro dimers. The results show that without these cooperativities, phage lambda cannot maintain the lysogenic state (Fig 3A, dashed lines). As soon as CI degradation becomes slightly above zero, repressors are depleted in the steady state, and Cro dimers accumulate.
The strength of the cooperativity between CI2 dimers and between Cro2 dimers at different operator sites is uneven. An interesting question is whether the coordination of multiple cooperativities is required, or whether one key cooperativity is sufficient to lead phage lambda to the lysogenic state. We tested different alternative possibilities. When all of the five cooperativities are removed (CI2 binding to OR1-OR2, to OR2-OR3, Cro2 to OR1-OR2, to OR2-OR3, and to OR1-OR2-OR3, respectively), phage lambda cannot lysogenize (Fig 3A, dashed lines, and details shown in the SI Appendix). Restoring the cooperativity between CI2 dimers binding to OR1 and OR2 alone can recover the lysogenic state of phage lambda (Fig 3B, solid lines, and Fig S4 in SI). In contrast, the stability and the high threshold are still missing when all cooperativities other than that between CI2s binding to OR1-OR2 are restored (Fig 3b, dashed lines, and the SI Appendix). This finding shows the cooperativity of CI dimers binding to OR1 and OR2 is a key enabling factor for phage lambda to maintain the lysogenic state.
Our results are consistent with the experimental observation that the cooperativity between CI2 dimer binding to OR1 and OR2 plays the dominant role (20, 22, 23). The stronger affinity and higher occupancy of OR1 bound by CI2 dimer leads to cooperative binding of CI2 to the neighboring operator site OR2. This CI2 dimer precludes the alternative cooperative binding of CI2 dimers between OR2 and OR3 (5). Our findings support the model that stability against prophage induction largely results from cooperative DNA binding by CI2 to the OR1 and OR2 sites (22, 23). Zhu et al. also showed the importance of cooperativity between CI dimers in an earlier computational study (10). The finding emerging from this study is that the cooperativity of CI2 dimers between OR1 and OR2 is the dominant enabling factor, and it alone is sufficient to endow phage lambda with the ability to adopt the lysogenic life style.
Effects of Altered Operators: Mechanism of Hair-Triggers of the Little Mutants.
To assess the robustness of phage lambda against changes in the molecular components of the epigenetic network, Little et al. replaced the ordered operator sites of OR321 in the wild-type phage lambda with the symmetric variants of OR323, OR121, and OR3′23′, respectively. In OR3′23′, the OR3′ site has one of the OR3 nucleotides replaced by that of OR1 (8). In this seminal work, all of the mutants are found to have functional epigenetic circuits, all have the ability to form lysogens, to grow lytically, and to undergo prophage induction upon UV irradiation. However, these mutants have markedly different tolerance to UV irradiation. The wild-type lysogen has a high threshold and requires the highest level of UV irradiation for prophage induction to occur. In contrast, all mutant variants exhibit the behavior of a hair-trigger, and require much less UV irradiation for the onset of prophage induction.
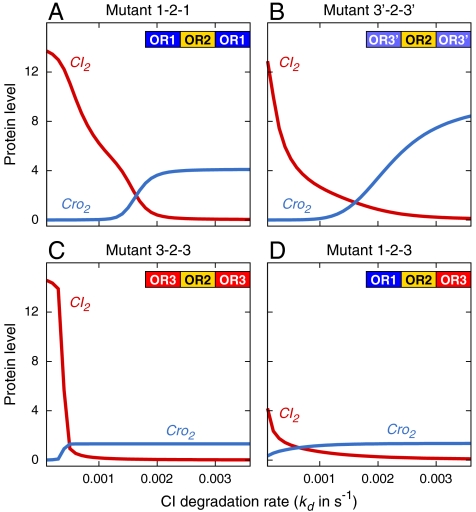
To study the effects of different dosage of UV irradiation on the Little et al. mutants (8), we calculated their probability landscapes at different CI degradation rates (Fig 4, and details shown in SI Appendix). Overall, we find that the Little et al. mutants all exhibit threshold behavior in our model, as was found experimentally (8). However, these mutants are generally defective, with reduced thresholds for prophage induction, and are hair-triggered. As seen in Fig 3A (solid lines), wild-type phage lambda has a deep threshold for prophage induction (kd = 2.0 × 10-3/s at about 50% induction). In contrast, although all mutants have thresholds for the lytic response, these thresholds are much shallower (≤ 1.0 × 10-3/s) (Fig 4). These results are consistent with the experimental finding that they are hair-triggered (8).
Fig. 4.
Instability, shallow threshold, and switching inefficiency of the network against fluctuation in UV irradiation in mutant phage lambda (8). (A–B). In contrast to wild type (Fig 3A, solid lines), mutant 1-2-1 and 3′-2-3′ do not maintain a stable level of CI2. They are leaky and responds gradually to graded changes in kd. Their thresholds and that of 3-2-3 (C) for lytic transition are much shallower. (D) Mutant 1-2-3 does not maintain a sufficient amount of CI2, and therefore cannot maintain lysogeny.
Our model can also reproduce detailed differences in UV responses among the mutants. Both OR121 (Fig 4A) and OR3′23′ (Fig 4B) mutants are experimentally found to have a higher threshold of UV irradiation than mutant OR323 (Fig 4C) for prophage induction (8), which we reproduced in our model. We find that the degradation rate at which the amount of Cro2 surpasses CI2 for both mutants OR121 and OR3′23′ is kd = 1.6 - 1.7 × 10-3/s, whereas the OR323 mutant has a lower value of kd = 4.0 - 5.0 × 10-4/s. There is also another subtle difference in the behavior among the Little et al. mutants (8). Mutant OR121 is found experimentally to be slightly more stable than OR3′23′, according to figure 3 in ref. 8, which is again reflected in our results. When CI degradation is small, the mutant OR121 (Fig 4A) has a larger amount of CI repressor molecules than the mutant OR3′23′ (Fig 4B), hence it has a higher tolerance for UV irradiation.
We find that these mutants cannot maintain a constant level of CI2. Although it has been suggested that the Little et al. mutants (8) have lower levels of CI, which contributes to the reduced threshold, the full mechanism of how the switching network functions in these mutants is not well understood. From our model, we find that although prophage inductions can occur in both OR121 and OR3′23′ mutants, their lysogenic states are different from that of wild-type and can be characterized as “leaky.” There is a gradual reduction in the level of CI in response to a gradual increase in the CI degradation rate (Fig 4 A and B), corresponding to graded amounts of DNA damage. When the degradation rate of CI increases, the level of CI also decreases, and this effect is cumulative, even though phage lambda remains overall in the lysogenic state. In contrast, wild-type phage lambda exhibits a true high-threshold behavior that is very stable: There is little change in the amount of CI, even when there is a large increase in the CI degradation rate. This state is maintained until the set point of switching threshold is reached (Fig 3A, solid lines). Even the best mutants OR121 and OR3′23′ with relatively high thresholds respond to gradual changes in CI degradation rate and do not have a high threshold.
The epigenetic circuit of lambda phage is generally robust against system parameter perturbations (8). However, there are limits to this robustness. As shown in ref. 8, a nonsymmetric mutant OR123, with the positions of OR1 and OR3 swapped could not lysogenize, a finding our results also reproduce. In our model (Fig 4D), this mutant has a severely impaired ability to generate CI repressor, even when there is little or no CI degradation due to UV damage.
Comparison with Other Methods.
To examine whether the same results can be obtained using stochastic simulation algorithm, we carried out simulation using the Gillespie algorithm (15). For rare events such as transition from lysogenic to lytic state (kd = 0.0020/s), the algorithm failed to converge to the steady-state probability distribution after > 6 times of more computation time compared to the dCME method, and the results strongly depend on the initial conditions. There remain significant residual errors at each of the initial conditions tested. Conclusions drawn from nonconverged simulations can give incorrect prediction that the system is still in the lysogenic state. Such failure of convergence can be difficult to detect. Residual error remains significant when the system is in the region of starting to enter the lytic state (kd = 0.0022/s). Furthermore, the expected copy number of CI2 can be overestimated by 300% in this region, and the small amount of Cro2 calculated in the lysogenic region can be off by three orders of magnitude with comparable computational time, which would lead to unreliable estimation of the frequency of very rare events such as spontaneous lysis in lysogenic state. Details of error analysis can be found in the SI Appendix.
For this comparison, we assume that the steady state can be established during the life span of a cell. As the models of the reactions are Markovian, the probability landscape of the steady state computed by all stochastic methods should be independent of the initial condition. The issue of assessing differences in computed time-evolving probability landscape before reaching the steady state using different methods requires further investigations.
We also carried out calculation using both stochastic differential equation (SDE) and ODE models based on the studies of Santillán and Mackey (24) and Gillespie (25), with modifications so it is directly comparable to our model. There are significant qualitative differences in both cases. The SDE model failed to reach the correct steady state, with the landscape much further away than that from the stochastic simulation algorithm. In the ODE model, which can be regarded as the skeleton of the stochastic models, there is no stabilization of the CI2 concentration against fluctuations in the CI degradation rate, as the amount of CI2 rapidly decreases when the CI degradation rate increases. Wild-type phage lambda would be hair-triggered by this model, which disagrees with experimental data. In addition, the transition from lysogeny to lysis in the 1-2-1 and 3′-2-3′ mutants occurs at higher CI degradation rate than the wild type, which would lead to the erroneous conclusion that these mutants are more resistant to UV irradiation. Details of these comparison can be found in the SI Appendix.
Discussion
In this study, we characterized the properties of the epigenetic decision network of phage lambda through direct computation of its steady-state probability landscape using the method of dCME, an approach previously unfeasible. We find that the switching network of phage lambda is very stable and is strongly buffered with a high threshold against fluctuations in CI degradation rate due to environmental changes in UV irradiation, a behavior only observed in the wild-type phage lambda. This high threshold against environmental fluctuations is important for the self-perpetuating nature of the epigenetic state of E. coli cells, allowing lysogeny to be passed on to its offspring. We also find that once the degradation rate of CI reaches a threshold, phage lambda switches very efficiently to the lytic state, and this efficiency is not built at the expense of stability against random fluctuation. Phage lambda can integrate signaling in the form of different CI degradation rates and can distinguish a true signal above the high threshold from random noise fluctuating below this threshold.
Our results further indicate that the cooperativity of CI2 binding between OR1 and OR2 plays the key role in enabling the wild-type behavior of phage lambda. Nevertheless, the phenotype of a high threshold and robustness against mutations are best viewed overall properties of the network. In addition to this cooperativity, disruptions of other structural and architectural features may also result in the loss of these phenotypes. The distributive nature of such network properties have been discussed in ref. 26.
Our results are consistent with many experimental findings (8). Our results point out that the Little et al. (8) mutants have a leaky switch, compromising stability against fluctuation in UV irradiation and reducing switching efficiency. Our finding suggests that the leaky response to UV damage and the lack of a high threshold in mutants OR121 and OR3′23′ are responsible for the hair-trigger of prophage induction upon UV damage.
There exist a number of theoretical studies of phage lambda and its mutants (4, 10, 24). Here we discuss the main differences in biological findings. In a pioneering study, Aurell et al. (4) investigated the effects of two free parameters on the stability of lysogenic state in an SDE model of phage lambda based on measured lysis frequencies, with the goal of exploring unrecognized control mechanism for the stability of lysogeny. A major conclusion is that the total affinity of Cro for OR3 is a key factor in determining the stability of lysogenic state, which affects the transcription rate of derepressed PRM, and therefore the synthesis rate of CI. In addition to the differences in underlying model and computational techniques, a focus of our study is to explore the effects of cooperativity between CI dimers, with otherwise experimentally derived model parameters. We are able to pinpoint the cooperativity between CI dimers on OR1 and OR2 as the key factor for the stability of phage lambda. An effect of this cooperativity is the increased repression of PR, which promotes the production of CI. Santillán and Mackey developed an ODE model (24). Without stochasticity, this model shows neither stability against small increases of CI degradation rate, nor switching efficiency when CI degradation rate reaches the set-point threshold. The study of Zhu et al. was based on a potential surface reconstructed from an SDE model (10), with three free parameters adjusted for best fit of experimental data. All parameters in our model are derived from the literature. Although both studies show the importance of cooperativity between CI dimers, our results pinpoint to the important role of the cooperativity between CI dimers binding to OR1 and OR2 (see SI Appendix).
This work is also a significant improvement over a preliminary version of our model (27), which does not consider cooperativities between CI dimers or Cro dimers, and does not account for self-promoted synthesis of CI. Without such considerations, lysogeny can only be reached by increasing CI synthesis rate, which is unrealistic and cannot be used to model the effects of UV irradiation.
Overall, our study indicates that an important mechanistic understanding of the system behavior of a stochastic network can be gained through direct computing of the network probability landscape. The study of the effects of altered molecular components and altered wiring of the network further suggests that we could predict the outcome of manipulated phage lambda through computational studies, as elegantly laid out experimentally by Little et al. (8, 28). Our focus in this work is exploring the overall global behavior of the phage lambda switching network. There are many aspects of the model that can be further improved, for example, the nonspecific binding of CI molecules to DNA and the effects of cooperative looping between OR and OL are currently only modeled implicitly. In addition, if discrepancies in reported and in vivo parameters can be reconciled and obtained, we expect that better quantitative predictions can be made.
Computational study of biological systems allows exploration of many alternative hypotheses and facilitates the identification of key elements responsible for phenotypic observations (3, 4, 10, 29, 30). We show that the approach of dCME can offer fresh insight and generate testable predictions. An advantage of this approach is that rare events can be characterized, without difficulties associated with stochastic simulation in determining whether millions or billions of samples are required.
The approach of obtaining exact solutions of simplified models with enumerated microstates advocated here has an analogy in previous studies of protein folding. Models such as lattice self-avoiding walks with only hydrophobic and polar interactions allow the complete enumeration of all feasible conformations and the calculation of exact thermodynamics as well as folding dynamics from the master equation for model molecules. Such studies have played important roles in elucidating the principles of protein folding (31–34).
It is likely that direct solutions to simplified but realistic stochastic networks can lead to the elucidation of the mechanisms of many biological processes. Because the molecular mechanism underlying the control of phage lambda applies to many other biological regulatory processes as well, and because similar processes likely underlie many developmental and epigenetic processes, including cooperative control of histone coding (26), the decision control of phage lambda has offered us a paradigm for studying broad issues in cell development and cell fate (4, 5, 10, 35, 36). The approach described in this study is generally applicable and can be extended to these other systems as well.
Materials and Methods
Our model includes the repressor protein CI, its dimer CI2, the Cro protein, and its dimer Cro2. The synthesis and degradation of CI monomer and Cro monomer, as well as their dimerizations are modeled explicitly. In addition, the three operator sites, OR1, OR2, and OR3, are modeled to bind to either CI2 dimer or Cro2 dimer with different affinities. These sites can also be unoccupied. The cooperativity between CI2 dimers and between Cro2 dimers on neighboring sites is included. In addition, the enhanced CI synthesis when OR2 is occupied by CI dimer (currently understood to be due to the looping effect between OL and OR) is also included (37, 38). The self repression of CI2 at high concentration is also included. The values of protein-operator binding affinities, cooperativity, protein synthesis rate, and protein degradation rates are based on experimental measurement and are described in detail in the SI Appendix. In our model, we assume that there are a total of about 100 copies of CI repressors (4, 39), of which about 50 copies are free in a cell volume available to bind to the operator sites in lysogen. The remaining 50 copies are assumed to bind to DNA nonspecifically, because it is expected that a significant amount of CI repressors are bound to DNA in regions other than the operators, and the copy number of free CI repressor may be as few as 10–20 (39). The rationale of this assumption is further described in the SI Appendix.
Supplementary Material
Acknowledgments.
We thank Drs. Ping Ao, William Hendrickson, Linda Kenney, Konstantin Mischaikow, Shoudan Liang, Qing Nie, Garyk Papoian, Hong Qian, Michael Samoilov, and Kim Sneppen for helpful discussions; Dr. Zhu Chen and Zhifeng Shao for encouragement; and the computing center at Shanghai Center for Systems Biomedicine for machine time. This work was supported by a phase II 985 Project, 973 Grant 2007CB914703, National Institutes of Health Grants GM079804-01A1 and GM081682, National Science Foundation Grant DMS-0800257, and a grant from the Chicago Biomedical Consortium.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001455107/-/DCSupplemental.
References
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Shea MA, Ackers GK. The OR control system of bacteriophage lambda a physical-chemical model for gene regulation. J Mol Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 3.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurell E, Brown S, Johanson J, Sneppen K. Stability puzzles in phage λ. Phys Rev E. 2002;65:051914. doi: 10.1103/PhysRevE.65.051914. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M. A Genetic Switch: Phage Lambda Revisited. 3 edition. Plainview, NY: Cold Spring Harbor Laboratory Press; 2004. pp. 1–66. [Google Scholar]
- 6.Johnson AD, et al. Lambda repressor and cro-components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 7.Ptashne M, et al. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- 8.Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson LM, Yang H. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc Natl Acad Sci USA. 2008;105:5827–5832. doi: 10.1073/pnas.0705570105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu XM, Yin L, Hood L, Ao P. Robustness, stability and efficiency of phage lambda genetic switch: Dynamical structure analysis. J Bioinform Comput Biol. 2004;2:785–817. doi: 10.1142/s0219720004000946. [DOI] [PubMed] [Google Scholar]
- 11.Artyomov MN, Mathur M, Samoilov MS, Chakraborty AK. Stochastic bimodalities in deterministically monostable reversible chemical networks due to network topology reduction. J Chem Phys. 2009;131:195103. doi: 10.1063/1.3264948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Kampen NG. Stochastic Processes in Physics and Chemistry. 3rd Ed. Amsterdam: North–Holland; 2007. pp. 96–133.pp. 193–200. [Google Scholar]
- 13.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 14.Vellela M, Qian H. Stochastic dynamics and non-equilibrium thermodynamics of a bistable chemical system: The schlogl model revisited. J R Soc Interface. 2009;6:925–940. doi: 10.1098/rsif.2008.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Cao Y, Petzold LR, Gillespie DT. Algorithms and software for stochastic simulation of biochemical reacting systems. Biotechnol Progr. 2008;24:56–61. doi: 10.1021/bp070255h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohn W, et al. Self-consistent equations including exchange and correlation effects. Phys Rev. 1965;140:A1133–A1138. [Google Scholar]
- 17.Kohanoff J. Electronic Structure Calculations for Solids and Molecules: Theory and Computational Methods. Cambridge, UK: Cambridge Univ Press; 2006. pp. 51–69. [Google Scholar]
- 18.Cao Y, Liang J. Optimal enumeration of state space of finitely buffered stochastic molecular networks and exact computation of steady state landscape probability. BMC Syst Biol. 2008;2:30. doi: 10.1186/1752-0509-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer RT, Ross MJ, Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J Biol Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 20.Koblan KS, Ackers GK. Site-specific enthalpic regulation of DNA transcription at bacteriphage λ OR. Biochemistry. 1992;31:57–65. doi: 10.1021/bi00116a010. [DOI] [PubMed] [Google Scholar]
- 21.Darling PJ, Holt JM, Ackers GK. Coupled energetics of λ cro repressor self-assembly and site-specific DNA operator binding II: Cooperative interactions of cro dimers. J Mol Biol. 2000;302:625–638. doi: 10.1006/jmbi.2000.4050. [DOI] [PubMed] [Google Scholar]
- 22.Johnson AD, Meyer BJ, Ptashne M. Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc Natl Acad Sci USA. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci USA. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santillán M, Mackey MC. Why the lysogenic state of phage lambda is so stable: A mathematical modeling approach. Biophys J. 2004;86:75–84. doi: 10.1016/S0006-3495(04)74085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie DT. The chemical Langevin equation. J Chem Phys. 2000;113:297–306. [Google Scholar]
- 26.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 27.Cao Y, Lu HM, Liang J. Stochastic probability landscape model for switching efficiency, robustness, and differential threshold for induction of genetic circuit in phage λ. IEEE EMBS. 2008:611–614. doi: 10.1109/IEMBS.2008.4649227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atsumi S, Little JW. A synthetic phage λ regulatory circuit. Proc Natl Acad Sci USA. 2006;103:19045–19050. doi: 10.1073/pnas.0603052103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapidus S, Han B, Wang J. Intrinsic noise, dissipation cost, and robustness of cellular networks: The underlying energy landscape of MAPK signal transduction. Proc Natl Acad Sci USA. 2008;105:6039–6044. doi: 10.1073/pnas.0708708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Xu L, Wang E. Potential landscape and flux framework of nonequilibrium networks: Robustness, dissipation, and coherence of biochemical oscillations. Proc Natl Acad Sci USA. 2008;105:12271–12276. doi: 10.1073/pnas.0800579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dill KA, et al. Principles of protein-folding—a perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Socci ND, Onuchic JN. Folding kinetics of proteinlike heteropolymer. J Chem Phys. 1994;101:1519–1528. [Google Scholar]
- 33.Ozkan SB, Bahar I, Dill KA. Transition states and the meaning of ϕ-values in protein folding kinetics. Fold Des. 1998;3:R45–R58. [Google Scholar]
- 34.Kachalo S, Lu H, Liang J. Protein folding dynamics via quantification of kinematic energy landscape. Phys Rev Lett. 2006;96:058106. doi: 10.1103/PhysRevLett.96.058106. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne M. Binding reactions: Epigenetic switches, signal transduction and cancer. Curr Biol. 2009;19:R234–R241. doi: 10.1016/j.cub.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Liang J, Qian H. Computational cellular dynamics based on the chemical master equation: A challenge for understanding complexity. J Comput Sci Technol. 2010;25:154–168. doi: 10.1007/s11390-010-9312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer BJ, Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. III lambda Repressor directly activates gene transcription. J Mol Biol. 1980;139:195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- 38.Révet B, et al. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr Biol. 1999;9:151–154. doi: 10.1016/s0960-9822(99)80069-4. [DOI] [PubMed] [Google Scholar]
- 39.Bakk A, Metzler R. Nonspecific binding of the OR repressors CI and Cro of bacteriophage lambda. J Theor Biol. 2004;231:525–533. doi: 10.1016/j.jtbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.