Abstract
Cardiac-specific overexpression of a constitutively active form of calcineurin A (CNA) leads directly to cardiac hypertrophy in the CNA mouse model. Because cardiac hypertrophy is a prominent characteristic of many cardiomyopathies, we deduced that delineating the proteomic profile of ventricular tissue from this model might identify novel, widely applicable therapeutic targets. Proteomic analysis was carried out by subjecting fractionated cardiac samples from CNA mice and their WT littermates to gel-free liquid chromatography linked to shotgun tandem mass spectrometry. We identified 1,918 proteins with high confidence, of which 290 were differentially expressed. Microarray analysis of the same tissue provided us with alterations in the ventricular transcriptome. Because bioinformatic analyses of both the proteome and transcriptome demonstrated the up-regulation of endoplasmic reticulum stress, we validated its occurrence in adult CNA hearts through a series of immunoblots and RT-PCR analyses. Endoplasmic reticulum stress often leads to increased apoptosis, but apoptosis was minimal in CNA hearts, suggesting that activated calcineurin might protect against apoptosis. Indeed, the viability of cultured neonatal mouse cardiomyocytes (NCMs) from CNA mice was higher than WT after serum starvation, an apoptotic trigger. Proteomic data identified α-crystallin B (Cryab) as a potential mediator of this protective effect and we showed that silencing of Cryab via lentivector-mediated transduction of shRNAs in NCMs led to a significant reduction in NCM viability and loss of protection against apoptosis. The identification of Cryab as a downstream effector of calcineurin-induced protection against apoptosis will permit elucidation of its role in cardiac apoptosis and its potential as a therapeutic target.
Despite major advancements in the field of cardiovascular medicine, heart disease remains the leading cause of mortality in the developed world (1, 2). To meet this challenge, we require further understanding of the molecular mechanisms that trigger progression to cardiac disease. Knowledge of global changes in protein composition during disease progression will be critical to the elucidation of these molecular mechanisms. Previously, we described a proteomic analysis of cardiac tissue from a mouse model of dilated cardiomyopathy, the PLNR9C mouse, with an Arg9 to Cys mutation in phospholamban, a key regulator of cardiac contractility (3). PLNR9C mice progress directly to dilated cardiomyopathy, with decreased cardiac function, ventricular wall thinning, and early mortality (4). Endoplasmic reticulum (ER) stress and apoptosis are prominent features identified by bioinformatic analysis of the changes in protein composition observed during progression of the disease.
Here our aim was to carry out a similar analysis on a well-established model of hypertrophic cardiomyopathy that arises from the transgenic, cardiac-specific overexpression of a constitutively active form of calcineurin (calcineurin A, CNA) (5). Indeed, patients with cardiac hypertrophy exhibit increased calcineurin expression (6), thus our objective was to identify alterations in protein expression that accompany the pathophysiological mechanisms associated with this form of cardiac disease.
CNA mice manifest with a severe hypertrophic phenotype as early as 2 wk after birth; they demonstrate significantly elevated left ventricular mass at 2, 4, and ∼10 wk of age (7). CNA mice also demonstrate a progressive decrease in both systolic and diastolic cardiac function (8), and exhibit histo-pathological signs, with an increase in cardiomyocyte disarray and interstitial fibrosis. Dong et al. (9), demonstrated that the majority of CNA transgenic mice die by 6 mo, with death attributed to atrioventricular heart block, potentially caused by a decrease of transient outward K+ currents. This finding is in accord with the observation that the primary cause of death in patients with hypertrophic cardiomyopathy is sudden cardiac death.
Results
Proteomic Analysis of Cardiac Tissue from CNA Transgenic Mice.
Transgenic mice were examined for the presence of hypertrophic cardiomyopathy. Indeed, we found that transgenic mice demonstrated significantly increased heart mass index, increased cardiomyocyte cross sectional area, and interstitial fibrosis (Fig. S1 and Table S1). Through echocardiographic analysis, we demonstrated significant increases in ventricular wall thicknesses and decreased fractional shortening (Fig. S1 and Table S1).
Proteomic analysis was carried out on cardiac ventricular tissue from 14-wk-old and 24-wk-old CNA transgenic mice and their WT littermates. We subfractionated cardiac tissue lysates into cytosolic, microsomal, mitochondrial matrix, and mitochondrial membrane fractions by differential centrifugation, as described previously (4). Because the contractile proteins are several orders of magnitude more abundant than most other proteins, we did not include the sarcomeric fraction in our proteomic analysis. Nevertheless, large amounts of the contractile proteins were still well represented in the soluble cytosolic fraction. Samples were subjected to gel-free shotgun liquid chromatography-tandem mass spectrometry, as described previously (10). All proteomic sample runs from all fractions were combined to produce 4,893,830 spectra, which were mapped to a nonredundant mouse protein sequence database using the SEQUEST search algorithm (Fig. S2). We selected only spectra matching to peptides with ≥99% confidence. To further refine this dataset and thus reduce false positives, we selected only those proteins identified by four or more peptides. This latter criterion resulted in a proteome with 1,918 high-confidence proteins (Fig. 1 A and B and Table S2) with false-discovery rates of 0.0007 and 0.012 at the peptide and protein levels, respectively.
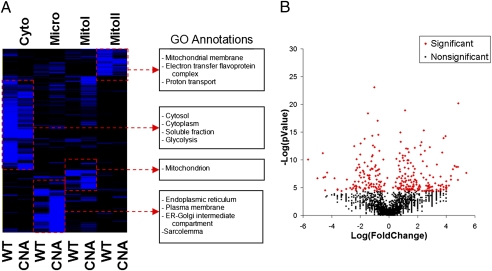
Fig. 1.
Hierarchical clustering and statistical analysis of cardiac proteins in WT and CNA hearts. (A) A heat map demonstrating the clustering of proteins based on their expression intensities in the cytosolic (Cyto), microsomal (Micro), mitochondrial matrix (mitoI), and mitochondrial membrane (MitoII) fractions for WT and CNA mice. To the right of the heat map are selected annotations found to be enriched in the protein clusters shown. (B) A volcano plot showing the CNA/WT protein expression ratio (Log2 scale) of proteins against their P value (−Log2). Red diamonds indicate the 290 significantly altered proteins.
Hierarchical clustering analysis demonstrates that proteins enriched in particular fractions cluster together. In accord with our previous findings using the same protocol (11), proteins that were most abundant in the cytosolic fraction were enriched in Gene Ontology (GO) annotations related to that fraction including “cytosol,” “cytoplasm,” and “soluble fraction” (Fig. 1A); proteins most abundant in the microsomal fraction, which consists of membraneous structures including the sarco(endo)plasmic reticulum, the golgi apparatus, and the plasma membrane, were enriched with annotations for these constituents; proteins most abundant in the mitochondrial matrix and membrane fractions were enriched in GO annotations for mitochondria.
Validation and Bioinformatic Characterization of Significantly Altered Proteins.
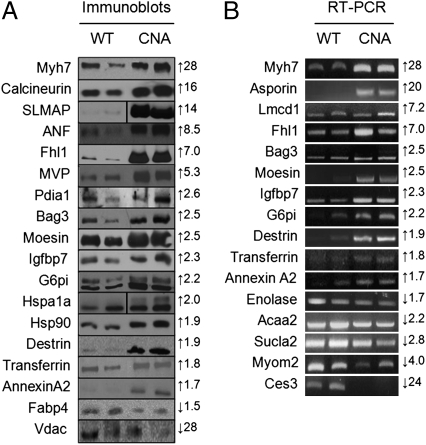
Using a linear model-based regression analysis that we published previously (4), we identified 290 significantly altered proteins (116 down-regulated and 174 up-regulated) (Fig. 1B and Table S2). We confirmed the altered expression of several proteins by both immunoblot (where antibodies were commercially available) and RT-PCR analyses (Fig. 2 A and B). All 25 of 25 immunoblots and RT-PCR analyses demonstrated expression changes in CNA mice that were concordant with our proteomic data.
Fig. 2.
Immunoblot and RT-PCR expression analysis of significantly altered genes. Protein expression levels were analyzed by immunoblot (A) or RT-PCR (B). Numbers on the right of the panels indicate the fold-change of the respective proteins in the proteomic analysis and arrows indicate up- or down-regulation.
Next, we characterized this set of 290 altered proteins using GO term-enrichment analysis. A wide variety of GO terms were enriched in the pool of significantly altered proteins (Tables S3 and S4). Significantly down-regulated proteins were highly enriched in a multitude of GO terms associated with metabolism, including “fatty acid metabolism,” “tricarboxylic acid cycle,” and “acetyl-CoA catabolism.” Significantly up-regulated proteins were enriched in GO terms related to ER stress and the unfolded protein response, including “response to stress,” “response to unfolded protein,” and “response to heat,” suggesting that the CNA hearts were subject to ER stress. Several GO terms were also related to programmed cell death, including “apoptosis,” “regulation of apoptosis,” and “negative regulation of programmed cell death.”
Microarray Analysis of Cardiac Tissue from CNA Transgenic Mice.
In parallel, we performed microarray analysis on total cellular mRNA from hearts of 14-wk-old CNA transgenic mice and their WT littermates to complement the proteomic profile for the stage at which hypertrophy is fully expressed. We found that the expression of 722 genes was significantly altered (with P < 0.05 and a fold-change >2) in the disease model (168 down-regulated and 554 up-regulated) (Table S5). To broadly characterize this differential gene set, we searched for enriched GO terms associated with the latter genes (Tables S6 and S7). Down-regulated genes were again associated with GO terms related to energy metabolism, including “fatty acid metabolism,” but up-regulated genes were again associated with GO terms related to ER stress and apoptosis, including “response to stress,” “response to heat,” and “negative regulation of programmed cell death.” In addition, up-regulated genes were also associated with GO terms related to the extracellular matrix, including “extracellular matrix organization and biogenesis,” as well as several GO terms related to ion channels, including “regulation of action potential” and “ion transport,” Which is congruent with the observed interstitial fibrosis and previous reports indicating electrophysiological abnormalities in these hearts (9).
Comparison of the proteome to the microarray dataset resulted in an overlap of 1,649 proteins between the two datasets. Although the overall correlation of expression ratios was rather low for this global perspective (r = 0.44) (Fig. S3A and Table S8), we did find that 70% of the proteins demonstrated concordant expression ratios (i.e., were increased in both proteome and microarray or decreased in both) (Fig. S3D). Next, we assessed whether there was increased correlation of relative expression ratios found in the subset of 290 significantly altered proteins. Indeed, this latter protein subset demonstrated an increased correlation with the total microarray data (r = 0.65) (Fig. S3 B and E and Table S9). Finally, we compared only the significantly altered proteins to the subset of microarray containing only significantly altered genes. This process resulted in a very high correlation, with r = 0.88; however, only 63 proteins were represented in the subset of significantly altered genes (Figure S3 C and F and Table S10).
Assessment of ER Stress and Apoptosis in CNA Hearts.
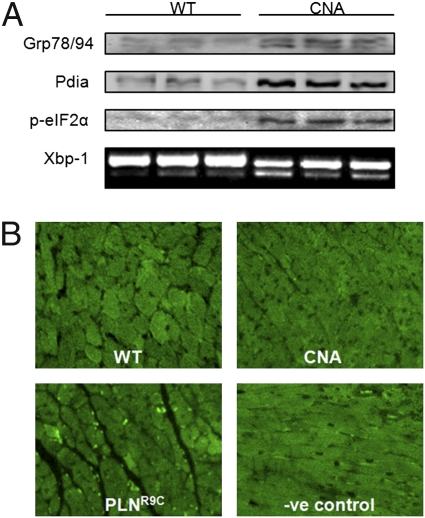
As several GO terms related to ER stress and the unfolded protein response were enriched in both our sets of significantly up-regulated genes and proteins, we investigated the degree of ER stress in CNA mice. We evaluated the expression of four markers of ER stress, including Grp78, Pdia1, phosphorylated eIF2α, and spliced Xbp-1 (Fig. 3A). Indeed, these markers were all up-regulated in adult ventricular tissue from CNA mice, supporting the presence of ER stress in CNA transgenic animals.
Fig. 3.
ER stress and cardiac apoptosis. (A) Immunoblots demonstrating increased Grp78, Pdia1, and phosphorylated eIF2α in adult CNA hearts, compared with their WT littermates and a PCR gel demonstrating increased ratio of spliced (active) Xbp1 (sXbp1) to unspliced Xbp1 (uXbp1) in CNA hearts, compared with WT hearts. (B) Photomicrographs demonstrating negligible TUNEL labeling of a WT heart section; a CNA heart section demonstrating negligible TUNEL labeling, a heart section from the PLNR9C mouse model of dilated cardiomyocyte demonstrating a high degree of TUNEL labeling (bright spots) and, the absence of TUNEL labeling in a negative control section in which the labeling enzyme was omitted.
Because ER stress can lead to apoptosis, we evaluated the basal level of apoptosis in CNA hearts. Paradoxically, we found that apoptosis, as evidenced by the presence of TUNEL reactive nuclei, was not elevated in CNA hearts compared with WT hearts (Fig. 3B). In contrast, hearts from the PLNR9C mouse model of dilated cardiomyopathy, previously demonstrated to have an elevated rate of apoptosis (4), exhibited prominent TUNEL labeling.
This unexpected finding suggested that activated calcineurin might mediate protection from cell death following an apoptotic stimulus. To evaluate this question, we subjected neonatal cardiomyocytes (NCMs) from CNA mice and their WT littermates to serum starvation for 72 h, a protocol that normally triggers apoptosis. NCMs from CNA transgenic animals exhibited significantly higher retention of viability following 72 h serum starvation, compared with WT (P = 0.006) (Fig. S4), indicating that the expression of constitutively active calcineurin in cardiomyocytes might play a role in protection against apoptosis.
Identification and Knockdown of Protective Mediator.
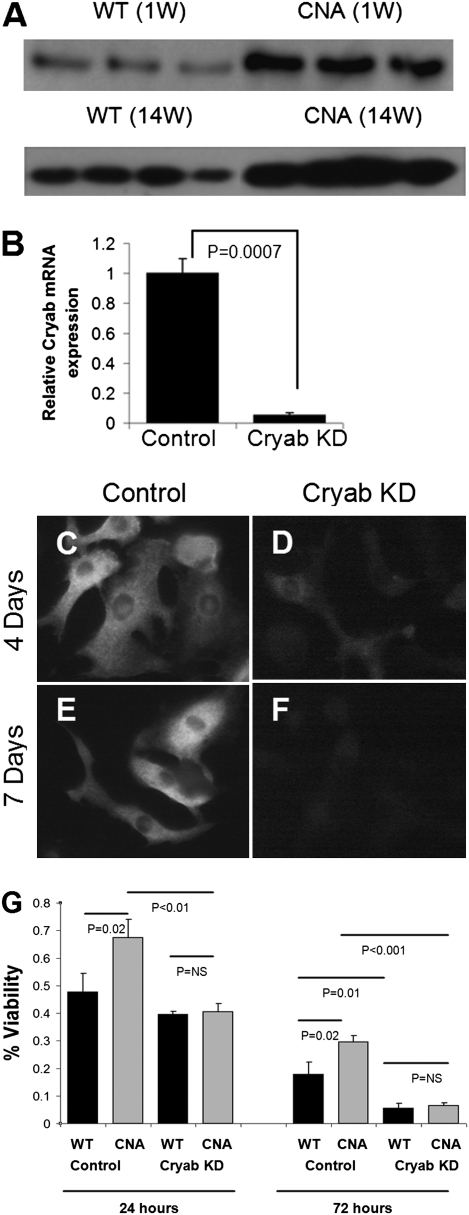
To identify which proteins contributed to the protection against cell death afforded by activated CNA overexpression, we data-mined the proteome for apoptosis regulatory proteins with differential expression. This process resulted in the identification of eight protein candidates previously shown to act as apoptotic regulators, including Reticulon-3, Reticulon-4, Catenin-α1, Ubiquilin-1, Bcl-associated athanogene-3, α-Crystallin B (Cryab), Cytochrome C (CyC), and Asc (Fig. S5). To identify those factors that might be responsible for the protective effects, we compared the expression values for these factors in the CNA proteome to their expression in the PLNR9C proteome, our aim being to filter out those factors that might be responsible for the protective effects from those factors that were simply up-regulated because of ER stress, as both the CNA and the PLNR9C models have elevated ER stress, yet the PLNR9C model shows increased apoptosis. The comparison identified four candidates with differential expression between the two proteomes. One candidate, Cryab, which is abundantly up-regulated in CNA mice but not significantly altered in the PLNR9C mouse model, was selected for further study.
Immunobloting confirmed that Cryab was overexpressed approximately twofold in adult CNA hearts and CNA transgenic NCMs (Fig. 4A). To test its protective effects against apoptosis, we silenced Cryab in NCMs using lentivectors expressing specific shRNAs targeting Cryab. Transduction of NCMs with the shRNA-expressing lentivectors resulted in an ∼20-fold reduction in Cryab mRNA expression relative to NCMs transduced with the control lentivectors expressing scrambled shRNAs (Fig. 4B). Furthermore, we validated the knockdown of Cryab at the protein level by immunofluorescence analysis (Fig. 4 C–F and Fig. S6). Next, we validated the functionality of the Cryab knockdown by demonstrating its effect on NCM viability. In this case, exposure to H2O2 was used as the trigger for induction of apoptosis (12). Mouse NCMs, transduced with either the Cryab knockdown lentivectors or the scrambled shRNA control lentivectors, were subjected to a viability assay following administration of 200 μmol/L H2O2 for 48 h. NCMs transduced with the Cryab-targeting shRNAs demonstrated a significant (P < 0.0001) reduction in viability compared with the NCMs transduced with the negative control construct (Figs. S7 and S8).
Fig. 4.
Cytoprotective candidate selection, knockdown and Cryab mediated cyto-protection. (A) Immunoblot validating the significantly increased Cryab protein levels in NCMs (1 wk old) and in CNA mouse hearts (14 wk old) compared with their WT littermates. (B) Bar graph demonstrating significantly decreased Cryab mRNA expression in WT NCMs following transduction with lentivectors expressing shRNAs targeting Cryab. (C and E) Photomicrograph (630× magnification) of WT NCMs immunofluorescently probed with anti-Cryab antibody 4 and 7 d after transduction with the control lentivectors expressing the scrambled shRNA. (D and F) Photomicrograph of WT NCMs immunofluorescently probed with anti-Cryab antibody 4 and 7 d after transduction with lentivectors expressing the Cryab targeting shRNAs. (G) Bar graph demonstrating the significant increase in viability in CNA NCMs, compared with WT NCMs (both transduced with the control lentivectors expressing the scrambled shRNAs). Both cultures were incubated with serum-free media and 200 μM H2O2 for 24 and 72 h. Abrogation of the protective effect in CNA NCMs incubated with serum-free media and 200 μmol/L H2O2 for 24 and 72 h, compared with WT NCMs, was observed when Cryab was silenced (Cryab KD).
Finally, we assessed the effect of Cryab knockdown in CNA NCMs compared with WT littermate NCMs. As expected, CNA NCMs demonstrated significantly higher retention of viability following 200 μmol/L H2O2 exposure for 24 and 72 h compared with the WT NCMs (Fig. 4G). This protective effect was abolished in CNA NCMs in which Cryab was silenced, thus indicating that Cryab is at least partially responsible for the protection afforded by calcineurin overexpression.
Discussion
Our aim in the present study was to investigate alterations in the proteome and transcriptome of ventricular tissue in a well-established mouse model of hypertrophic cardiomyopathy induced by cardiac-specific overexpression of constitutively active calcineurin (5). The proteome, which provides expression ratios for 1,918 high-confidence proteins, and the transcriptome, which provides expression ratios for 21,403 genes, are now publicly available resources. Stringent statistical analyses of these expression profiles resulted in the identification of 290 significantly altered proteins and 722 significantly altered genes. When these were compared, only 63 proteins overlapped the significantly altered genes subset, yet the correlation of their expression ratios was very high. In light of the relatively recent understanding of the role of microRNAs (miRs) in posttranscriptional regulation (13), a perfect concordance between mRNA and protein levels was not expected. It may be of particular relevance that mir-133 and calcineurin have been demonstrated to exhibit reciprocal repression of each other. Indeed, Dong et al. (14) also showed that calcineurin activity was increased in a model of cardiac hypertrophy induced by aortic banding, whereas mir-133 levels were decreased. Consistent with these findings, van Rooij et al. (15) demonstrated that mir-133 was one of the down-regulated microRNAs identified by microRNA microarray analysis in CNA transgenic mice. Therefore, a proportion of the discordance between the transcriptome and the proteome may represent genuine biological divergence between net mRNA and protein levels.
Overall, we have high confidence in this proteome through validation of a large series of these proteins through standard techniques. In addition, atrial natriuretic factor, a protein with a well-established association with cardiac hypertrophy (16), as well as angiotensin converting enzyme and β-myosin heavy chain, are all proteins reported to be up-regulated in cardiac disease (17, 18) and they were all up-regulated in the proteome.
The CNA model of hypertrophic cardiomyopathy exhibits severe cardiac pathology in addition to hypertrophy, which includes systolic dysfunction, interstitial fibrosis, cardiac arrhythmia, and sudden cardiac death. It is important to understand how changes in protein expression cause the observed phenotypes in this mouse model. For example, several genes directly involved in cardiac cyto-structure were substantially up-regulated, including actin, fibrillin, tubulin, filamin, spectrin, and annexin. Similarly, Fhl1, reported to be involved in the formation of sarcomeres by binding myosin-binding protein C (19), was found to be up-regulated sevenfold in CNA hearts and to be up-regulated in other studies of human hypertrophic cardiomyopathy (20) and in other models of cardiomyopathy (4, 21).
Conduction system defects in cardiac hypertrophy may result from redistributed gap junctional proteins, leading to a reduction in cell-to-cell coupling (8). Our results showing marked down-regulation of a gap junction protein, Gja1, are consistent with this pathology and may help explain the re-entrant circuits that lead to ventricular tachycardia (22), a common phenomenon in CNA mice (23). In addition, a transcriptional regulator, Lmcd1 (Lim and cysteine-rich domains protein 1) was increased sevenfold in CNA hearts. Lmcd1 restricts the transcription factor Gata6, an atrioventricular conduction system-specific regulatory factor (24) function by inhibiting DNA binding (25). Therefore, the resultant modulation of transcriptional regulation in these cells may contribute to the observed electrophysiological abnormalities.
The majority of down-regulated proteins found in our study can be classified as metabolic enzymes. Indicating that there is a significant generalized reduction in β-oxidation and a decrease in the shuttling of metabolites into the citric acid cycle in CNA hearts. This classification is in accordance with the documented shift from fatty acid oxidation to anaerobic glycolysis in hypertrophic hearts (26, 27).
GO enrichment analysis indicated a significant overrepresentation of proteins related to the unfolded protein response and response to stress. ER stress can lead to the unfolded protein response in which ER resident chaperones are elevated to restore homeostasis (28). An important ER stress response is the activation of protein kinase RNA-like ER kinase (PERK) by its release from Grp78 (28). PERK activation results in the phosphorylation of eIF2α, which, in turn, inhibits protein translation (28). Our proteomic data demonstrated that prominent ER chaperones involved in this response to stress, including Grp78, Grp94, and calreticulin, were all significantly up-regulated in the transgenic mice, as were additional markers/mediators of ER stress, including Pdia1, phosphorylation of eIF2α, and splicing of Xbp1, confirming ER stress-response pathway activation in this mouse model.
Prolonged ER stress can lead to apoptosis, which led us to evaluate the basal degree of apoptosis in unstimulated hearts. However, the negligible evidence of apoptosis in the CNA hearts, despite elevated ER stress, suggested that calcineurin overexpression may protect against apoptosis. This finding would be in accordance with reports demonstrating decreased apoptosis in CNA mice following ischemia/reperfusion injury (29), or that ablation of calcineurin predisposes the heart to apoptosis (30).
Accordingly, we evaluated whether CNA transgenic cardiomyocytes were resistant to cell death following apoptosis stimulation by either serum starvation or H2O2. Indeed we found a significant protective effect afforded by CNA overexpression, which supports demonstrations that either plasmid transfection or adenovirus-mediated overexpression of calcineurin significantly protected rat neonatal cardiomyocytes against either simulated ischemia/reperfusion, 2-deoxyglucose, or staurosporine induced apoptosis (29, 31).
To identify potential mediators of this protective effect, we surveyed the subset of significantly altered proteins in the CNA proteome. We identified potential candidates related to apoptosis regulation and compared their expression in the CNA proteome to our previously published proteome of the PLNR9C model of dilated cardiomyopathy. This R9C model was demonstrated to have markers of elevated ER stress but also of increased apoptosis. Therefore, by comparison of the CNA proteome to the PLNR9C proteome, we eliminated proteins that were commonly up-regulated because of ER stress and identified differentially expressed proteins potentially responsible for the CNA mediated cyto-protection. Four proteins were differentially expressed between the two proteomes, including Reticulon-3, Ubiquilin-1, Cryab, and CyC. Reticulon-3 has been demonstrated to be both a proapoptotic factor (32), and an antiapoptotic factor (33, 34). CyC, on the other hand, is a well-established mediator of apoptosis through its role in the formation of the apoptosome; however, it was only down-regulated at 24 wk, when it was unlikely to be the cause of the cyto-protection we observed in NCMs. The remaining proteins were Ubquilin and Cryab. Cryab is a heat-shock protein that is essential for maintenance of lens cells in the eye. Several studies have demonstrated an antiapoptotic role for this factor (35–38). Of note, Manukyan et al. (38) demonstrated that the calcineurin/NFAT pathway directly activates the Cryab promoter as evidenced by direct binding of Nfatc3 to the cryab promoter, which was increased in CNA transgenic hearts, and abrogated by the expression of Cain, a calcineurin inhibitor. Accordingly, we demonstrated by proteomic analysis and then by immunoblotting that Cryab is overexpressed in CNA hearts. We also showed that silencing of Cryab via lentivector-mediated transduction of NCMs with Cryab targeting shRNAs led to a significant reduction in cardiomyocyte viability. These observations correlate with a report that a significant increase in apoptosis occurs in Cryab-deficient hearts following ischemia/reperfusion injury in the mouse (39). The protection afforded by calcineurin overexpression in NCMs was shown here to be largely ablated following silencing of Cryab.
The exact mechanism of Cryab cyto-protection has yet to be elucidated; however, there is evidence indicating that cellular stresses promote the translocation of this chaperone to the mitochondria. Indeed, following ischemia reperfusion in cardiomyocytes, Cryab was shown to translocate from a predominantly cytosolic location to the mitochondria (40, 41). Furthermore, adenoviral-mediated overexpression of Cryab led to a reduction in H2O2-mediated CyC release (41). This protective effect may be enhanced by p38 MAPK induced phosphorylation of the serine 59 residue (41). Another study has demonstrated that Cryab binds to, and sequesters, Bax and Bcl-X(s), thereby preventing their proapoptotic effects on the mitochondria (42). Two different mutants of Cryab exhibit decreased binding of these latter proapoptotic factors, which, in turn, decreased Cryab-mediated cyto-protection. Alternatively, Cryab induced cyto-protection may be mediated by the maintenance of the intracellular integrity. It is possible that cellular stresses in cardiac myocytes may activate Cryab expression and mobilize cytosolic Cryab to the cytoskeleton and contractile filaments, where its chaperone functions stabilize the cytoskeleton and myofilaments. Indeed, Verschuure et al. demonstrated that when ER stress was induced by proteosomal inhibition, crystallin was mobilized to the actin cytoskeleton in H9c2 cells, a rat cardiac myoblast cell line (43). In addition, a human disease-causing mutation in Cryab (R120G) causes desmin-related myopathy (44–46) that presents with amyloid-positive aggregrates, contractile dysfunction, and progresses to heart failure (47).
In conclusion, using our proteomic-derived list of altered proteins we have demonstrated compensated ER stress in CNA mice and identified Cryab as a specific mediator of the cyto-protective effects of calcineurin.
Methods
Animals.
CNA mice expressing the constitutively active CNA catalytic subunit driven by the α- myosin heavy chain promoter were generously provided by Jeff Molkentin (Cincinnati Children's Hospital Medical Center, Cincinnati, OH) and have been described previously (5). Cardiac hypertrophy was evaluated in CNA mice by echocardiography and histology. All animal care protocols were approved by and conformed with the guidelines of the institutional animal care and use committees at all universities involved in this study.
Proteomic Analysis and Microarray Analysis.
Hearts were pooled and then fractionated as described in SI Methods and elsewhere (10). Protein were then digested and treated for liquid chromatography-tandem mass spectrometry as described in SI Methods and elsewhere (4, 10).
Hierarchical clustering analysis was carried out using Cluster 3.0 and Treview Software. Statistical analysis was carried out as described in SI Methods and elsewhere (4, 10). Microarray analysis was carried out as described in SI Methods and elsewhere (4, 10).
Immunoblotting and RT-PCR.
Immunoblotting and RT-PCR was carried out using standard techniques as described in SI Methods and elsewhere (4, 10).
Apoptosis and Viability Assays and Gene Knockdown.
Mouse hearts were analyzed by TUNEL as per the manufacturer's instructions. Lentivector-mediated transduction of neonatal cardiomyocytes was carried out as previously described (48). Neonatal cardiomyocytes were subjected to either serum starvation for 72 h or 200 μM H2O2 for 24 or 72 h. Viability assays were carried out using WST-8 (Dojindo). See SI Methods for more discussion.
Supplementary Material
Acknowledgments
We thank Dr. Jeff Molkentin (Cincinnati Children's Hospital Medical Center, Cincinnati, OH) for the gift of the calcineurin A mouse model, Dr. Jason Moffat for assistance and helpful discussions regarding the lentiviral vectors, and Wen-Ping Li and Vijay Khanna for expert technical assistance. This work was supported by Genome Canada through the Ontario Genomics Institute, the Heart and Stroke Foundation of Ontario (T-6281 and T-5042), the Canadian Institutes of Health Research (MT 125450), the Canadian Foundation for Innovation, and the Heart and Stroke/Richard Lewar Centre of Excellence. N.B. is a fellow of the Heart and Stroke Foundation of Canada. A.O.G. and T.K. are Canada Research Chairs, and A.O.G. is a New Investigator of the Heart and Stroke Foundation of Canada. A.E. is the Ontario Chair in Biomarkers.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013555107/-/DCSupplemental.
References
- 1.Kung H-C, Hoyert DL, Xu J, Murphy SL. Deaths: Final data for 2005. Natl Vital Stat Rep. 2008;56(10):1–120. [PubMed] [Google Scholar]
- 2.Rosamond W, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics—2008 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–e146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan DH, Kranias EG. Phospholamban: A crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 4.Gramolini AO, et al. Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol Cell Proteomics. 2008;7:519–533. doi: 10.1074/mcp.M700245-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haq S, et al. Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation. 2001;103:670–677. doi: 10.1161/01.cir.103.5.670. [DOI] [PubMed] [Google Scholar]
- 7.Semeniuk LM, et al. Time-dependent systolic and diastolic function in mice overexpressing calcineurin. Am J Physiol Heart Circ Physiol. 2003;284:H425–H430. doi: 10.1152/ajpheart.00546.2002. [DOI] [PubMed] [Google Scholar]
- 8.Chu G, et al. Enhanced myocyte contractility and Ca2+ handling in a calcineurin transgenic model of heart failure. Cardiovasc Res. 2002;54:105–116. doi: 10.1016/s0008-6363(02)00230-4. [DOI] [PubMed] [Google Scholar]
- 9.Dong D, et al. Overexpression of calcineurin in mouse causes sudden cardiac death associated with decreased density of K+ channels. Cardiovasc Res. 2003;57:320–332. doi: 10.1016/s0008-6363(02)00661-2. [DOI] [PubMed] [Google Scholar]
- 10.Gramolini AO, Kislinger T, Liu P, MacLennan DH, Emili A. Analyzing the cardiac muscle proteome by liquid chromatography-mass spectrometry-based expression proteomics. Methods Mol Biol. 2007;357:15–31. doi: 10.1385/1-59745-214-9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousette N, et al. Large-scale characterization and analysis of the murine cardiac proteome. J Proteome Res. 2009;8:1887–1901. doi: 10.1021/pr800845a. [DOI] [PubMed] [Google Scholar]
- 12.Peng W, et al. Cardioprotection by CaMKII-deltaB is mediated by phosphorylation of heat shock factor 1 and subsequent expression of inducible heat shock protein 70. Circ Res. 2010;106:102–110. doi: 10.1161/CIRCRESAHA.109.210914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong DL, et al. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: A novel mechanism for progressive cardiac hypertrophy. Hypertension. 2010;55:946–952. doi: 10.1161/HYPERTENSIONAHA.109.139519. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takemura G, et al. Expression and distribution of atrial natriuretic peptide in human hypertrophic ventricle of hypertensive hearts and hearts with hypertrophic cardiomyopathy. Circulation. 1991;83:181–190. doi: 10.1161/01.cir.83.1.181. [DOI] [PubMed] [Google Scholar]
- 17.Schunkert H, et al. Increased rat cardiac angiotensin converting enzyme activity and mRNA expression in pressure overload left ventricular hypertrophy. Effects on coronary resistance, contractility, and relaxation. J Clin Invest. 1990;86:1913–1920. doi: 10.1172/JCI114924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiser PJ, Portman MA, Ning XH, Schomisch Moravec C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol. 2001;280:H1814–H1820. doi: 10.1152/ajpheart.2001.280.4.H1814. [DOI] [PubMed] [Google Scholar]
- 19.McGrath MJ, et al. Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. J Biol Chem. 2006;281:7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- 20.Lim DS, Roberts R, Marian AJ. Expression profiling of cardiac genes in human hypertrophic cardiomyopathy: Insight into the pathogenesis of phenotypes. J Am Coll Cardiol. 2001;38:1175–1180. doi: 10.1016/s0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaussin V, et al. Common genomic response in different mouse models of beta-adrenergic-induced cardiomyopathy. Circulation. 2003;108:2926–2933. doi: 10.1161/01.CIR.0000101922.18151.7B. [DOI] [PubMed] [Google Scholar]
- 22.Peters NS, Coromilas J, Severs NJ, Wit AL. Disturbed connexin43 gap junction distribution correlates with the location of reentrant circuits in the epicardial border zone of healing canine infarcts that cause ventricular tachycardia. Circulation. 1997;95:988–996. doi: 10.1161/01.cir.95.4.988. [DOI] [PubMed] [Google Scholar]
- 23.Gillis AM, et al. Heart block in mice overexpressing calcineurin but not NF-AT3. Cardiovasc Res. 2004;64:488–495. doi: 10.1016/j.cardiores.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Davis DL, et al. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–119. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- 25.Rath N, Wang Z, Lu MM, Morrisey EE. LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding. Mol Cell Biol. 2005;25:8864–8873. doi: 10.1128/MCB.25.20.8864-8873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allard MF, Schönekess BO, Henning SL, English DR, Lopaschuk GD. Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am J Physiol. 1994;267:H742–H750. doi: 10.1152/ajpheart.1994.267.2.H742. [DOI] [PubMed] [Google Scholar]
- 27.Sambandam N, Lopaschuk GD, Brownsey RW, Allard MF. Energy metabolism in the hypertrophied heart. Heart Fail Rev. 2002;7:161–173. doi: 10.1023/a:1015380609464. [DOI] [PubMed] [Google Scholar]
- 28.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: Cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Windt LJ, et al. Calcineurin-mediated hypertrophy protects cardiomyocytes from apoptosis in vitro and in vivo: An apoptosis-independent model of dilated heart failure. Circ Res. 2000;86:255–263. doi: 10.1161/01.res.86.3.255. [DOI] [PubMed] [Google Scholar]
- 30.Bueno OF, et al. Calcineurin Abeta gene targeting predisposes the myocardium to acute ischemia-induced apoptosis and dysfunction. Circ Res. 2004;94:91–99. doi: 10.1161/01.RES.0000107197.99679.77. [DOI] [PubMed] [Google Scholar]
- 31.Obasanjo-Blackshire K, et al. Calcineurin regulates NFAT-dependent iNOS expression and protection of cardiomyocytes: Co-operation with Src tyrosine kinase. Cardiovasc Res. 2006;71:672–683. doi: 10.1016/j.cardiores.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Lee JT, Lee TJ, Kim CH, Kim NS, Kwon TK. Over-expression of Reticulon 3 (RTN3) enhances TRAIL-mediated apoptosis via up-regulation of death receptor 5 (DR5) and down-regulation of c-FLIP. Cancer Lett. 2009;279:185–192. doi: 10.1016/j.canlet.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 33.Zhu L, Xiang R, Dong W, Liu Y, Qi Y. Anti-apoptotic activity of Bcl-2 is enhanced by its interaction with RTN3. Cell Biol Int. 2007;31:825–830. doi: 10.1016/j.cellbi.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Wan Q, et al. Reticulon 3 mediates Bcl-2 accumulation in mitochondria in response to endoplasmic reticulum stress. Apoptosis. 2007;12:319–328. doi: 10.1007/s10495-006-0574-y. [DOI] [PubMed] [Google Scholar]
- 35.Mehlen P, Kretz-Remy C, Préville X, Arrigo AP. Human hsp27, Drosophila hsp27 and human alphaB-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNFalpha-induced cell death. EMBO J. 1996;15:2695–2706. [PMC free article] [PubMed] [Google Scholar]
- 36.Mehlen P, Schulze-Osthoff K, Arrigo AP. Small stress proteins as novel regulators of apoptosis. Heat shock protein 27 blocks Fas/APO-1- and staurosporine-induced cell death. J Biol Chem. 1996;271:16510–16514. doi: 10.1074/jbc.271.28.16510. [DOI] [PubMed] [Google Scholar]
- 37.Kamradt MC, Chen F, Cryns VL. The small heat shock protein alpha B-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J Biol Chem. 2001;276:16059–16063. doi: 10.1074/jbc.C100107200. [DOI] [PubMed] [Google Scholar]
- 38.Manukyan I, Galatioto J, Mascareno E, Bhaduri S, Siddiqui MA. Cross-talk between calcineurin/NFAT and Jak/STAT signalling induces cardioprotective alphaB-crystallin gene expression in response to hypertrophic stimuli. J Cell Mol Med. 2009;14:1707–1716. doi: 10.1111/j.1582-4934.2009.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison LE, Whittaker RJ, Klepper RE, Wawrousek EF, Glembotski CC. Roles for alphaB-crystallin and HSPB2 in protecting the myocardium from ischemia-reperfusion-induced damage in a KO mouse model. Am J Physiol Heart Circ Physiol. 2004;286:H847–H855. doi: 10.1152/ajpheart.00715.2003. [DOI] [PubMed] [Google Scholar]
- 40.Jin JK, et al. Localization of phosphorylated alphaB-crystallin to heart mitochondria during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2008;294:H337–H344. doi: 10.1152/ajpheart.00881.2007. [DOI] [PubMed] [Google Scholar]
- 41.Whittaker R, et al. Kinetics of the translocation and phosphorylation of alphaB-crystallin in mouse heart mitochondria during ex vivo ischemia. Am J Physiol Heart Circ Physiol. 2009;296:H1633–H1642. doi: 10.1152/ajpheart.01227.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao YW, Liu JP, Xiang H, Li DW. Human alphaA- and alphaB-crystallins bind to Bax and Bcl-X(S) to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. 2004;11:512–526. doi: 10.1038/sj.cdd.4401384. [DOI] [PubMed] [Google Scholar]
- 43.Verschuure P, et al. Translocation of small heat shock proteins to the actin cytoskeleton upon proteasomal inhibition. J Mol Cell Cardiol. 2002;34:117–128. doi: 10.1006/jmcc.2001.1493. [DOI] [PubMed] [Google Scholar]
- 44.Vicart P, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 45.Bova MP, et al. Mutation R120G in alphaB-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proc Natl Acad Sci USA. 1999;96:6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 47.Maloyan A, et al. Biochemical and mechanical dysfunction in a mouse model of desmin-related myopathy. Circ Res. 2009;104:1021–1028. doi: 10.1161/CIRCRESAHA.108.193516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.