There is a broad consensus that genetic alterations of normal body cells are the basis of cancer progression. Throughout the lifetime of an individual, her or his cells have to divide often, which is associated with occasional genetic changes. Some of the changes lead to uncontrolled cell proliferation and, at later stages of cancer progression, to blood vessel formation in the tumor tissue and distribution of tumor tissue across the body. The molecular genetics of cancer is a very advanced field: Many genetic alterations that predispose an individual to a certain cancer have been identified, and specific genetic pathways of cancer development have been elucidated (1). More recently, studies have been conducted on the scale of the entire genome to identify cancer-associated mutations (2–5). However, our understanding of the population genetic aspects of cancer development, that is, those aspects that relate to the dynamics of cancer cell replication, survival, and evolution, have not yet caught up with the advances in molecular genetics. A study by Bozic et al. (6) in PNAS brings the understanding of the population genetics of cancer cells closer to the edge defined by recent studies of the molecular genetics and genomics of various cancers.
Mathematical studies of cancer development date back to the 1950s (7). The studies in this tradition (8–12) focus on the age-specific incidence of cancers. It is remarkable how much can be learned about molecular genetics from the analysis of incidence patterns. For example, in a seminal paper, Knudson (8) anticipated the discovery of tumor suppressor genes by comparing the incidence of retinoblastoma in groups with and without a family history of this cancer. Despite these successes, the analysis of incidence patterns has clear limitations. The mathematical models used to explain the relationship between age and cancer incidence conceptualize cancer progression as a series of component failures of a complex system [e.g., monograph by Frank (11)] and remain quite vague on the dynamics and genetics of tumor cells. With more and more data becoming available on the accumulation of mutations in tumor cell lineages (2–5), mathematical approaches need to be developed to infer the parameters characterizing the population genetics of cancer development.
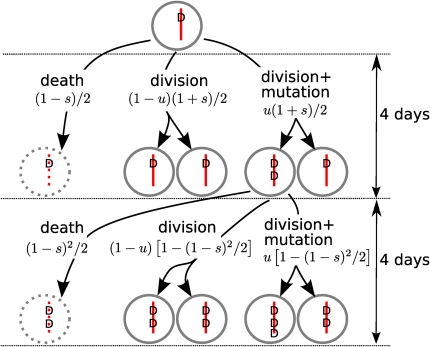
Bozic et al. (6) develop a mathematical model for tumorigenesis based on the multitype branching process—a stochastic process often used in population genetics. The model assumes that cells divide and accumulate mutations (Fig. 1). Some of these mutations, referred to as drivers, increase the fitness of the tumor cell, whereas others, called passengers, are neutral. The model can be used to predict the expected number of passenger mutations in a tumor cell with a certain number of driver mutations. This relationship between passenger and driver mutations depends on the selective advantage that a single driver mutation confers to the tumor cell lineage on average. Bozic et al. (6) then extract the number of driver and passenger mutations carried by tumor cells from previously published genomic data obtained from glioblastoma multiforme (the most common type of brain tumor) (4) and pancreatic adenocarcinoma (the most common type of pancreatic cancer) (5). To this end, they use the computational method called “cancer-specific high-throughput annotation of somatic mutations” (13). By fitting their model to the extracted numbers of driver and passenger mutations, Bozic et al. (6) estimate the average selective advantage of a driver mutation as 0.4% for both glioblastoma multiforme and pancreatic adenocarcinoma. Interestingly, the estimated selective advantage of drivers is almost the same for both cancer types and may therefore constitute a universal quantity not specific to the particular cancer type. Bozic et al. (6) further find that, with this estimate of the selective advantage, predictions of their mathematical model agree with the mean number of tumors and the tumor size observed in the clinical studies on familial adenomatous polyposis (14, 15).
Fig. 1.
Sketch of the multitype branching process developed by Bozic et al. (6). A tumor start with one cell carrying a single driver mutation (D). This cell may either die or divide, and it can further mutate. The probabilities of these events are indicated. They depend on the number of driver mutations the cell carries and the selection coefficient s. Driver mutations arise in one of the daughter cells at division with a probability of u = 3.4 × 105. The model assumes discrete generations with a length of 4 d. Passenger mutations (not shown in this sketch) are accumulated at a rate of v = 0.016 from generation to generation.
Why should we care about the fitness changes of tumor cell lineages during the progression to cancer? Fitness is at the heart of population genetics. To go beyond theoretical considerations and to derive specific predictions from population genetical models, fitness estimates are needed. The more quantitative picture of the population biology of tumor cells that Bozic et al. (6) develop will lead to a better understanding of cancer progression. Particularly important is the insight that the average selective advantage of driver mutations is very low at 0.4%, which translates into a high extinction rate of driver mutations: More than 99% of driver mutations that emerge during tumor cell replication will become extinct. This provides an explanation for the large variance in cancer progression rates. Furthermore, by parametrizing their population genetic model for tumor growth, Bozic et al. (6) provide a baseline model that is useful for the assessment of treatment strategies, especially with respect to minimizing the emergence of resistance to anticancer drugs.
The integration of recent genomic data on cancer into the framework of theoretical population genetics of carcinogenesis requires strong assumptions and simplifications. The mathematical implementation of too much detail would introduce too many unknown parameters, and thus would make a statistical analysis impossible. One simplification that Bozic et al. (6) make concerns the life span of tumor cells, which is set to be exactly 4 d in their main model. After 4 d, a cell in their model either dies or divides, and, as a consequence, the cell generations are synchronous. A fixed life span of tumor cells is an obvious oversimplification. However, in the supplementary information for their paper, Bozic et al. (6) show that assuming exponentially distributed life spans of tumor cells leaves the relation between driver and passenger mutations unchanged. (Exponentially distributed life spans represent the extreme opposite of fixed life spans.) Thus, the estimate of an average selective advantage of 0.4% of driver mutations is robust with respect to the assumptions of the life span of tumor cells.
More than 99% of driver mutations that emerge during tumor cell replication will become extinct.
Another simplification concerns the growth patterns of tumors. According to the mathematical model of Bozic et al. (6), tumors grow exponentially. However, because many types of tumors grow spherically, which leads to the suffocation of cells in the core, they do not display the exponential growth that a branching process predicts [reviewed by Byrne (16)]. This so-called “Gompertzian” growth of tumors has been tied into the population genetic models (17), and it may provide a better model for the analysis of genomic data in the future, especially if the later stages of tumor growth are considered.
Bozic et al. (6) also make strong assumptions about the effect of driver mutations. Each driver mutation is assumed to affect the fitness of the tumor cell lineage equally. Bozic et al. (6) partially assess how sensitive their findings are to this assumption by comparing the fits of their model with one in which the selective advantages of driver mutations are sampled from a normal distribution. However, potential differences between the effects of driver mutations on cell fitness are a very central issue, and should be addressed in greater detail in the future. It is certainly futile to attempt to determine the selection coefficient of each individual driver mutation from the data that Bozic et al. (6) analyzed. Nevertheless, it may be possible to go beyond the analysis of Bozic et al. (6). One could try to determine the variance of selection coefficients across all driver mutations, in addition to their mean, using standard frailty modeling approaches. One could also roughly classify the driver mutations according to their function (e.g., oncogene, tumor suppressor, stability gene) or with respect to the pathways they affect, and estimate function- or pathway-specific effects. Bozic et al. (6) also assume that the effect of one driver mutation is independent of the other driver mutations carried by the cell lineage. In the terminology of population genetics, they neglect “epistatic interactions.” However, it is very likely that such interactions exist. For example, the initial mutation hitting an apoptosis pathway should, on average, have a larger effect simply because an intact pathway is easier to damage. Thus, the fitness landscape, in which tumor cells evolve, may be much rougher than conceived by Bozic et al. (6).
Despite the bold assumptions typical of studies breaking previously unexplored ground, the value of the approach by Bozic et al. (6) lies in paving the way to integrate genomic data on cancer with the theoretical framework for population genetics of carcinogenesis. In 1986, Moolgavkar (18) wrote about the state of mathematical modeling of cancer development: “We are still struggling with the melody. Harmony and counterpoint must come later.” After reading Bozic et al. (6), I hear a nice melody and a driving beat.
Footnotes
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 3.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moolgavkar SH. Carcinogenesis models: An overview. Basic Life Sci. 1991;58:387–396. doi: 10.1007/978-1-4684-7627-9_14. discussion 396–399. [DOI] [PubMed] [Google Scholar]
- 10.Michor F, Iwasa Y, Nowak MA. The age incidence of chronic myeloid leukemia can be explained by a one-mutation model. Proc Natl Acad Sci USA. 2006;103:14931–14934. doi: 10.1073/pnas.0607006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank SA. Dynamics of Cancer: Incidence, Inheritance, and Evolution. Princeton: Princeton Univ Press; 2007. [PubMed] [Google Scholar]
- 12.Meza R, Jeon J, Moolgavkar SH, Luebeck EG. Age-specific incidence of cancer: Phases, transitions, and biological implications. Proc Natl Acad Sci USA. 2008;105:16284–16289. doi: 10.1073/pnas.0801151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: Computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giardiello FM, et al. Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med. 1993;328:1313–1316. doi: 10.1056/NEJM199305063281805. [DOI] [PubMed] [Google Scholar]
- 15.Giardiello FM, et al. Primary chemoprevention of familial adenomatous polyposis with sulindac. N Engl J Med. 2002;346:1054–1059. doi: 10.1056/NEJMoa012015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne HM. Dissecting cancer through mathematics: From the cell to the animal model. Nat Rev Cancer. 2010;10:221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 17.Dewanji A, Luebeck EG, Moolgavkar SH. A generalized Luria-Delbrück model. Math Biosci. 2005;197:140–152. doi: 10.1016/j.mbs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Moolgavkar SH. Carcinogenesis modeling: From molecular biology to epidemiology. Annu Rev Public Health. 1986;7:151–169. doi: 10.1146/annurev.pu.07.050186.001055. [DOI] [PubMed] [Google Scholar]