Abstract
Rare circulating tumor cells (CTCs) present in the bloodstream of patients with cancer provide a potentially accessible source for detection, characterization, and monitoring of nonhematological cancers. We previously demonstrated the effectiveness of a microfluidic device, the CTC-Chip, in capturing these epithelial cell adhesion molecule (EpCAM)-expressing cells using antibody-coated microposts. Here, we describe a high-throughput microfluidic mixing device, the herringbone-chip, or “HB-Chip,” which provides an enhanced platform for CTC isolation. The HB-Chip design applies passive mixing of blood cells through the generation of microvortices to significantly increase the number of interactions between target CTCs and the antibody-coated chip surface. Efficient cell capture was validated using defined numbers of cancer cells spiked into control blood, and clinical utility was demonstrated in specimens from patients with prostate cancer. CTCs were detected in 14 of 15 (93%) patients with metastatic disease (median = 63 CTCs/mL, mean = 386 ± 238 CTCs/mL), and the tumor-specific TMPRSS2-ERG translocation was readily identified following RNA isolation and RT-PCR analysis. The use of transparent materials allowed for imaging of the captured CTCs using standard clinical histopathological stains, in addition to immunofluorescence-conjugated antibodies. In a subset of patient samples, the low shear design of the HB-Chip revealed microclusters of CTCs, previously unappreciated tumor cell aggregates that may contribute to the hematogenous dissemination of cancer.
Keywords: microfluidics, ctcs, prostate cancer, clusters, point-of-care
Circulating tumor cells (CTCs) provide a potential source of cells derived from primary or metastatic sites that may be amenable for analysis in patients with epithelial cancers. However, isolating these cells is technically challenging due to their rare numbers (1 in 109 blood cells) and their low recovery following traditional batch purification techniques. Multiple batch approaches have been employed to detect CTCs, ranging from cell size based separation (1, 2) and fast scanning cytometry (3) to commercially available technology based on the use of immunomagnetic beads (4–10). To date, the low number of CTCs isolated from most patients (median ≤ 1 CTC/mL in ∼half of patients with known metastatic cancer) has allowed demonstration of their correlation with poor overall survival in metastatic prostate, breast, and colon cancers (6, 8, 11–13). However, higher CTC recovery rates are required to enable detailed molecular analyses and to provide reliable information for clinical monitoring of individual patients undergoing therapy.
Microfluidic devices provide important advantages over batch purification techniques in that they enable highly efficient processing of complex cellular fluids, with minimal damage to sensitive cell populations due to shear forces or need for cell fixation (14–16). However, under the laminar, uniaxial flow conditions present in microfluidic devices, cells follow streamlines and display minimal molecular diffusion across flow channels. This lack of mixing results in a limited number of interactions with the antibody-coated surface of the device, which is critical for target cell capture. In our first generation device called “the CTC-Chip,” 78,000 antibody-functionalized microposts were used to break up the streamlines and enhance cell-surface interactions. The CTC-Chip provided improved yield and purity of captured CTCs (17), enabling noninvasive serial genotyping of lung cancers carrying epidermal growth factor receptor (EGFR) mutations during the course of therapy with targeted kinase inhibitors (18) and monitoring of patients with either localized or metastatic prostate cancer (19). While effective at isolating CTCs from a cohort of patients with metastatic epithelial cancers (17), the CTC-Chip relied on laminar flow limiting the interactions of target cells with surfaces, and the complex micropost structure proved challenging to scale up for high-throughput production and larger-scale clinical applications. The alternative strategy developed here involves the use of surface ridges or herringbones in the wall of the device to disrupt streamlines, maximizing collisions between target cells and the antibody-coated walls themselves, thus not requiring the construction and functionalization of a complex micropost geometry.
A strategy for mixing of distinct solutions using such transverse flows within microchannels to induce fluidic microvortices was originally presented by Stroock et al. (20). In extending this principle to the capture of CTCs from whole blood, we developed a high-throughput microvortex mixing device that ensures effective contacts of cells with antibody-coated surfaces, while designed with simple geometry that is amenable for large-scale manufacturing. The Herringbone (HB)-Chip effectively captured CTCs from patients with metastatic prostate cancer, and its low shear flow properties revealed the presence of previously unappreciated microclusters of CTCs.
Results
Design of the Herringbone-Chip and Flow Visualization Studies.
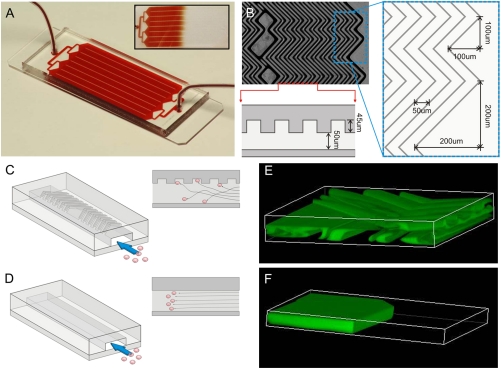
The HB-Chip consists of a 1” × 3” glass slide bonded to a polydimethylsiloxane (PDMS) structure, containing eight microchannels with patterned chevrons or herringbones on their upper surface (Fig. 1A). The internal walls of the device are made chemically active by in-line functionalization and coating with antibodies against EpCAM (see Materials and Methods). The geometric design of the device is based upon the general strategy for inducing chaotic mixing at low Re numbers (20), adapted for isolation of rare cells from whole blood by varying the ratio of height of the grooves to that of the channel, chevron dimensions, and periodicity. The herringbone grooves are staggered periodically, with each mixing cycle defined by two sequential regions of ten chevrons shifted asymmetrically (Fig. 1B). The final dimensions of the device were selected based on the optimization studies: overall height of channel (h) 50 μm, with the ratio of the height of the grooves to that of the channel (α) set to 0.8; angle between the herringbones and the axis of the channel (θ) 45°, and principal wave vector, q = 2π/100 μm (20, 21). A branching inlet header was designed to feed into eight individual channels to promote mechanical integrity and uniform flow distribution across the device (Fig. 1 A).
Fig. 1.
(A) The HB-Chip consists of a microfluidic array of channels with a single inlet and exit. Inset illustrates the uniform blood flow through the device. (B) A micrograph of the grooved surface illustrates the asymmetry and periodicity of the herringbone grooves. Cartoon illustrating the cell-surface interactions in (C) the HB-Chip, and (D) a traditional flat-walled microfluidic device. Flow visualization studies using two paired streams of the same viscosity (one stream is green, the other clear) demonstrate (E) the chaotic microvortices generated by the herringbone grooves, and the lack of mixing in (F) traditional flat-walled devices.
In comparison to a traditional flat-walled microfluidic device (Fig. 1D), the herringbone-induced microvortices disrupt the laminar flow streamlines that cells travel, causing them to “shift” (Fig. 1C), increasing the number of cell-surface interactions in the antibody-coated device. Flow visualization studies demonstrate this concept, showing in small scale versions of the flat and herringbone devices that two separate streams of equal viscosity (one fluorescently labeled green, one clear) fail to mix in the former but mix rapidly in the latter (Fig. S1 A, B). Three-dimensional projections of confocal images illustrate the high degree of mixing within one cycle of herringbone grooves, with the two streams of fluid folding over one another (Fig. 1 E, F) (see Movie S1, Movie S2, Movie S3, and Movie S4 for complete visualization of the flow fields; Fig. S2A–D). As predicted (20), flow rate does not impact the degree of mixing for the range of Re that is relevant in this system (Fig. S1 A, B). Thus, any interpatient variances in the rheological properties of the blood will not impact the extent of mixing in the herringbone CTC-Chip.
HB-Chip Optimization and Validation by Capture of Cancer Cell Lines.
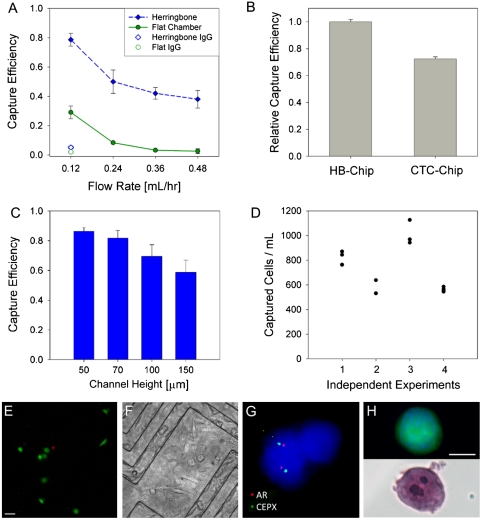
To demonstrate the impact of herringbone-mediated mixing, the HB-Chip was first compared with a traditional smooth wall microfluidic channel of similar dimensions (Fig. 2A, and Fig. S3 A, B). We used a single channel of either the HB-Chip or the smooth chamber device, connected to a microfluidic waste collection chamber (Fig. S4). This small footprint serial chamber setup allowed for accurate counting of target PC3 prostate cancer cells captured in the devices, as well as enumeration of noncaptured cells flowing into a microfluidic waste chamber, thus enabling a true mass-balance for cell capture efficiency (see Materials and Methods) (Fig. 2A). Compared with other epithelial cancer cell lines, PC3 cells express relatively low levels of cell-surface EpCAM (51,667 EpCAM molecules per cell), making them an appropriate choice for optimization of cell capture platforms (22). As predicted, the HB-Chip proved highly efficient at capturing PC3 cells (Fig. 2A), whereas significantly less capture was measured with the flat chamber device (p < 0.05 for all flow rates tested). At very low flow rates (0.12 mL/hr, translating to 1.2 mL/hr in the large-scale HB-Chip), the overall capture efficiencies in the flat chamber (29% ± 4.3% (n = 4)) and HB-Chip (79% ± 4.5% (n = 8)) reflect the settling of the PC3 cells towards the bottom surface of the device. However, as the flow rate is increased, the average capture efficiency for the flat chamber device drops to < 8%, whereas the HB-Chip maintains a target cell capture efficiency > 40% for all flow rates up to 0.48 mL/hr (or 4.8 mL/hr in the large HB-Chip). The capability to run at higher flow rates is an additional benefit in comparison to the CTC-Chip, previously shown to have a significant drop in capture efficiency at flow rates above 2–3 mL/hr (17).
Fig. 2.
(A) Device proof-of-principle studies were conducted using PC3 cells spiked into whole blood at 1,000 cells/mL and processed with a small version of the HB-Chip (Fig. S4, channel height 70 μm, α = 0.43) and a flat-walled device (channel height 100 μm). Capture efficiency is shown for both devices in addition to IgG controls. (B) Relative capture efficiency for head to head comparisons run between the full-sized HB-Chip (channel height 50 μm, α = 0.8) and the silicon CTC-Chip (17). For the data in (B), a single preparation of blood was spiked with PC3 cells at a concentration of 500 cells/mL, divided into equal parts and processed on both platforms. Due to slight errors in spiking concentration, the HB-Chip capture efficiency was determined to be greater than 100% (112.4% ± 1.8%, n = 4). Consequently, the comparison data is normalized against the HB-Chip. (C) Variations in device chamber height showed a minimal impact on capture efficiency, until the chamber height was increased threefold. (D) Variability in device performance is shown for four independent experiments, each with different spiking concentrations of PC3 cells in blood. (E) Micrograph of spiked cancer cells captured on the HB-Chip, representative of capture cell viability (LIVE/DEAD). (Scale bar: 40 μm). (F) Micrograph of spiked cells captured on the herringbone chip and subsequently cultured on the device for 21 d. (G) On-chip FISH of captured LNCaP cells with nuclei stained with DAPI, CEPX (green) and AR gene locus (red). (H) Micrograph of a fluorescently labeled PC3 cell captured on the HB-Chip (top) and subsequent micrograph taken of the same cell stained with H and E. (Scale bar: 10 μm).
For clinical samples, a larger version of the HB-Chip was created (Fig. 1A). Repeating the spiked cell experiments at 1.2 mL/hr (Fig. 2A), the large HB-Chip yielded an average capture efficiency of 91.8% ± 5.2% (n = 6) for PC3 cells spiked into whole blood. In comparing the HB-Chip and the micropost-based CTC-Chip, operating at a flow rate that favors high performance in both devices (1 mL/hr), the HB-Chip yielded a significantly improved capture efficiency in comparison with the CTC-Chip (26.3% improvement, p = 0.0001, Fig. 2B). The purity of spiked cancer cells captured among contaminating leukoctyes was 14.0% ± 0.1% for the HB-Chip, compared with 9.2% ± 0.1% for the CTC-Chip (p = 0.033). The kinetics of flow through the HB-Chip allowed an increased inner chamber height to 100 μm (corresponding to a twofold increase in blood volume throughput) without a significant drop in capture efficiency (Fig. 2C, p > 0.05). High data reproducibility (average variance = 5% ± 1.5%, n = 12)) was also evident with the HB-Chip when analyzing duplicate samples at different cell spiking densities (Fig. 2D).
The viability of cancer cells spiked into whole blood and captured on the HB-Chip was high (95% ± 0.6%, n = 4, Fig. 2E), and these cells could be cultured in vitro following separation of the top and bottom surfaces of the device (Fig. 2F). The relative simplicity of the HB-Chip design allows for manufacture using transparent, chemically stable materials, enabling high magnification imaging and assays such as FISH. Androgen receptor (AR) gene copy number was demonstrated using on-chip FISH in the LNCaP prostate cancer cell line, following spiking into blood and capture on the device (Fig. 2G). Two copies of the AR gene were detected in LNCaP cells, consistent with the known X chromosome copy number in this cell line (23). Imaging CTCs captured on a transparent device also allows for the use of standard cytology stains, which require immunohistochemical analysis using transmitted light. Hematoxylin and eosin staining was adapted for flow-through microfluidic conditions (see Materials and Methods), producing characteristic images of captured cancer cells (Fig. 2H).
Capture of CTCs from Patients with Metastatic Prostate Cancer.
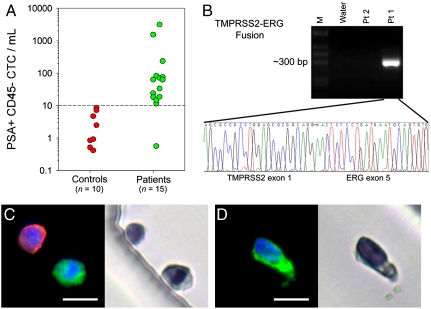
Blood samples from 15 patients at various stages of treatment for metastatic prostate cancer were processed with the HB-Chip. We also processed blood from healthy individuals as controls (n = 10). All cells captured on the HB-Chip were stained for DNA content (4,6-diamidino-2-phenylindole or DAPI), prostate-specific antigen (PSA, a highly specific marker for prostate epithelial cells), and a leukocyte marker (CD45). All devices were imaged in a blinded fashion, using our automated imaging algorithm (19). Rare “false positive” events were observed in healthy donors (median = 1 CTC/mL, mean = 3 ± 1 CTCs/mL, range 0 to 8 CTCs/mL), allowing us to set a detection threshold for significance in patients at ≥10 CTCs/mL. Among men with prostate cancer, CTCs were captured in 14 of 15 cases (93%), with counts ranging from 12 to 3,167 CTCs/mL (median = 63 CTCs/mL, mean = 386 ± 238 CTCs/mL) (Fig. 3A, see Table S1 for clinical characteristics). We have recently reported capture of CTCs in metastatic prostate cancer in 23 of 36 (64%) patients using the micropost CTC-Chip, under similar capture and enumeration conditions (19). Thus, in this patient population the HB-Chip demonstrates equal or improved capture efficiency. To validate the HB-Chip for molecular characterization of CTCs, we undertook on-chip lysis of captured cells and isolated RNA of sufficient quality for RT-PCR assays, identifying the known TMPRSS2-ERG chimeric transcript, confirmed by nucleotide sequencing, in a patient whose primary tumor harbored this translocation, but not in a patient whose tumor did not have the chromosome fusion (Fig. 3B).
Fig. 3.
Blood from metastatic prostate cancer patients and healthy donors was processed using the HB-Chip. (A) Healthy donors (n = 10, red circles) were used to select the signal intensity threshold in our automated imaging system (19). The metastatic prostate cancer patient data (n = 15, green circles) had significantly higher results, with 14∶15 patient samples presenting CTCs above-threshold levels. Additionally, (B) the TMPRSS2-ERG fusion transcript was detected from RNA isolated from the HB-Chip, using RT-PCR. Sequencing of the gel band identified the rare gene fusion of TMPRSS2 exon 1 and ERG exon 5. (C and D) Micrographs of CTCs isolated from patients with metastatic prostate cancer costained using antibodies against PSA (green), a leukocyte marker, CD45 (red), and a nuclear stain (DAPI). Corresponding micrographs of the same cells are shown to the right of the fluorescent images, demonstrating their appearance after subsequent staining with H and E. (Scale bars: 10 μm).
The transparent materials used in the manufacture of the HB-Chip made it possible to complement immunofluorescence staining of CTCs with stains used in standard pathology laboratories to identify tumor cells using light microscopy. Thus, a CTC identified using a specific fluorescence-conjugated antibody can be reevaluated under high magnification using transmitted light to gain additional insights into cytomorphologic and nuclear features (Fig. 3 C, D and Fig. S5C–F). Characteristics of tumor cells, such as large nuclei or visible nucleoli, are evident in some circulating cells identified by expression of PSA (Fig. 3D), prostate-specific membrane antigen (PSMA) (Fig. 4C), or cytokeratin (Fig. 4D and Fig. S5E–F).
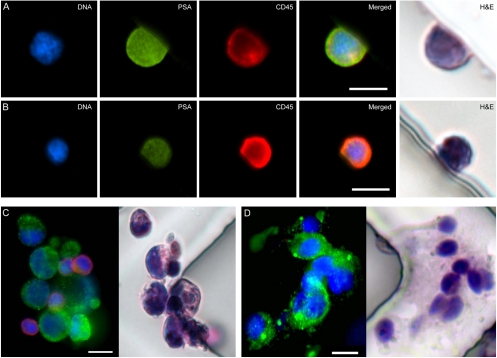
Fig. 4.
(A and B) Micrographs of cells isolated on the HB-Chip from a metastatic prostate cancer patient that were not classified as CTCs due to their dual-positivity for both PSA (green) and CD45 (red). Corresponding H and E images are shown. (C) Micrographs of a CTC cluster isolated from a metastatic prostate cancer patient on the HB-Chip; immunofluoresence staining (DNA (blue), prostate-specific membrane antigen (green), and CD45 (red)) and subsequent immunohistochemical staining (H and E) are shown. A three-dimensional projection of this cluster can be viewed in SI Text. (D) A CTC cluster isolated from a metastatic lung cancer patient on the HB-Chip; immunofluoresence staining (DNA (blue), cytokeratins 7/8 (green), and CD45 (red)) and subsequent immunohistochemical staining (H and E) are shown. All scale bars represent 10 μm.
The ability to combine immunofluorescence with light microscopic analysis provides insight into a common challenge in CTC isolation, namely the identification of cells that stain both for the epithelial and the leukocyte marker. While these “double positive” cells are typically excluded from CTC enumeration, they are more prevalent than CTCs, and their identity has been the subject of much debate: coating of CTCs with leukocyte-derived proteins may occur during their circulation in the bloodstream, while some monocytes may ingest tumor-derived markers. To begin to address this challenge, which has been described with all CTC isolation platforms, we identified double positive cells (PSA+, CD45+) captured by the HB-Chip from four patients with metastatic prostate cancer. In the majority of cases, hematoxylin and eosin staining revealed the characteristic morphology of leukocytes (Fig. 4B), suggesting that these cells are appropriately excluded from CTC enumeration. However, there are also a few cells with more ambiguous staining patterns in this population (Fig. 4A), whose size and staining patterns are more consistent with CTCs. To maintain a conservative approach in our CTC enumeration schema, these cells are still excluded from overall counts. It is of interest that double positive cells are observed at a significantly lower frequency in healthy blood donors, suggesting that these are not simply artifacts of postcapture staining. Further studies will be required to identify specific properties of these cells to explain their costaining for epithelial markers in patients with metastatic cancers.
In addition to patients with metastatic prostate cancer, we isolated CTCs from patients with lung cancer (n = 4), using antiEpCAM capture followed by staining for DAPI, CK7/8, and CD45 (Fig. S5). As for prostate cancer patients, the variable size (8–16 μm in diameter) and the morphological heterogeneity of CTCs is well illustrated (Fig. S5 A–D). Of note, the lower size limit of CTCs is comparable to the median size of leukocytes, suggesting that CTC isolation strategies relying solely on size-based filtration may exclude a significant portion of the CTC population.
Identification of CTC Microclusters in the Bloodstream of Patients with Cancer.
While most of the CTCs captured on the HB-Chip were single cells, we observed rare clusters of 4–12 cells (Fig. 4 A, D). Such clumps of tumor cells were never identified using the CTC-Chip, where interpost distances are designed to allow flow of single cells (17), nor have they been reported after the batch immunomagnetic bead purification process (4–9). Microclusters were identified in specimens from 3 out of 19 patients with either metastatic prostate or lung cancer. These were analyzed using immunofluorescence (PSMA or keratin), as well as hematoxylin and eosin stains (Figs. 4 C–D). Microclusters have never been identified following spiking of cancer cell lines into control blood specimens in our experiments, suggesting that they are not simply cell aggregations resulting from physical properties of the HB-Chip. Three-dimensional imaging of the microclusters also demonstrates that they have preserved shape and orientation, and are not groupings of cells flattened onto the walls or grooves of the device (rotating three-dimensional projection Movie S5).
Discussion
The application of microfluidic technology to the isolation of CTCs provides an opportunity for improved capture yield for these rare cells and for their preserved viability, allowing for detailed molecular and eventually functional characterization. However, the constraints inherent in large-scale production and chemical modification of a complex microfluidic device, combined with the need to image rare cells captured in a nontransparent three-dimensional array of microposts provide a challenge for “point-of-care” application of our previously described CTC-Chip (24). Here we demonstrate the application of a microfluidic vortex generator, the HB-Chip, which uses herringbone grooves within the transparent wall of the device to create microvortices that direct target cells toward capture on the antibody-coated walls. This “second generation” device allows for higher blood volume throughput, and increased CTC capture efficiency and purity. In addition to the imaging benefits provided by its transparent walls, the less complex three-dimensional structures of the HB-Chip greatly facilitates scaled up device production, and will thus enable the initiation of larger-scale clinical studies. In this initial proof of principle, the HB-Chip successfully isolated CTCs from 14 of 15 (93%) patients with metastatic prostate cancer (median 63 CTCs/mL) and allowed RNA-based detection of the TMPRSS2-ERG fusion transcript, showing a further improvement over our previous generation CTC-Chip (19).
The application of standard histopathology imaging platforms and metrics to the study of CTCs is essential to their broad application in cancer diagnostics and to realize the potential of a “blood biopsy” for epithelial cancers. To date, the very low numbers of CTCs isolated by commercial platforms, together with the reliance on relatively low resolution immunofluorescence-based imaging has limited such applications. As such, the ability to collect significant numbers of CTCs on a transparent slide is likely to facilitate the use of standardized criteria for analysis of tumor cells, as commonly used in the cytologic examination of ascites or pleural effusions. Previous studies using straight deposition of blood onto glass slides postlysis of red blood cells have shown that some Wright-Giemsa-stained CTCs may share morphological appearance with cells within the primary tumor biopsy (25, 26). As shown here, significant heterogeneity in hematoxylin and eosin staining intensity is evident among CTCs, despite uniform staining of contaminating leukocytes. Consistent with standard cytology observations, cells with reduced staining intensity are likely to be at early stages of autolysis, a fate that would be expected for most CTCs in the circulation. In addition to the detailed morphological analysis now possible for some captured CTCs, the combined use of immunofluorescence and light microscopy-based stains also provides insight into the identity of the so-called “double positive” cells, positive for both epithelial and hematopoietic markers. While these cells may greatly outnumber the “single positive” epithelial cells that are typically scored as CTCs, their identity and significance has been the subject of debate. The demonstration that most of these cells have the morphology of mononuclear leukocytes confirms their appropriate exclusion from CTC enumeration.
The most unexpected finding of this study was the isolation using the HB-Chip of clusters of CTCs from the blood of patients with metastatic cancer. In some cases, the size of these clusters was quite large, consisting of up to 14 CTCs, with cells within the cluster having highly variable cytomorphologic features. These clusters have not been reported using the commercial immunomagnetic bead platform, presumably because they do not withstand the multiple steps associated with batch purification, and they were also not evident using the CTC-Chip, since the tightly packed spacing of microposts may have prevented their passage. While we cannot formally exclude the possibility that in vitro clustering of CTCs might result from HB-Chip-induced flow patterns, this is unlikely, since we have never identified such clusters in experiments with cancer cells spiked into control blood specimens, nor is the presence of CTC clusters correlated with the absolute number of CTCs in clinical specimens. The existence of clusters of CTCs in the blood of patients with cancer may provide insight into the process of metastasis in human cancer (27). Given technological limitations to date, the mechanisms underlying cancer metastasis have been studied exclusively in mouse models. These have emphasized the altered motility of single cells intravasating from the primary tumor into the circulation, followed by their active extravasation at distant sites of metastasis. Circulating CTC clusters could either constitute microemboli breaking off from a tumor mass or, alternatively, they could result from intravascular proliferation of tumor cells (28). By whatever mechanism they arise, CTC clusters have the potential to lodge in distal capillary beds, thereby initiating metastatic lesions. Further characterization of these clusters in additional cancer types and at various stages of cancer progression will be required to ascertain their potential contribution to the metastatic spread of cancer.
Materials and Methods
Device Fabrication and Surface Modification.
Microfluidic devices were made of PDMS bonded to glass substrates using soft lithography techniques (29). The microfluidic devices were functionalized with epithelial cell adhesion molecule (EpCAM, R and D Systems) antibody using avidin-biotin chemistry. The immobilization of antibody on the glass and PDMS surfaces was achieved using a previously described method (30). See SI Text for additional details.
Study Subjects and Blood Processing.
Patients with advanced prostate and lung cancer were recruited according to a protocol approved by the institutional review board (IRB). Blood specimens from healthy volunteers were collected under a separate IRB-approved protocol. A total of 19 cancer patients (15 prostate, 4 lung), who were treated at the Massachusetts General Hospital Cancer Center, donated 10–20 mL of blood on one or more occasions for analysis on the HB-Chip. All specimens were collected into vacutainer (Becton-Dickinson) tubes containing the anticoagulant EDTA and were processed through the HB-Chip within 6 h of blood draw. Samples were run on the previously described microfluidic processing machine (17). Briefly, a 5 mL aliquot of blood was placed in an air-tight conical tube on a rocker assembly, and ∼4 mL of blood were pneumatically driven through the chip at a flow rate of 1.5–2.5 mL/hr. Following, the HB-Chip was flushed with 2.5 mL of PBS at 2.5 mL/hr to remove any nonspecifically bound cells.
CTC Staining and Enumeration.
After processing of blood on the HB-Chip, cells were fixed with 4% paraformaldehyde, and subsequently permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) in PBS. Cells were immunostained with primary antibodies, either a rabbit polyclonal antibody to PSA (DAKO), monoclonal antibody against cytokeratin 7/8 (CAM 5.2 clone, BD Biosciences), or a monoclonal mouse IgG2b antibody to PSMA (Santa Cruz Biotechnology). Following, the appropriately matched secondary AlexaFluor 488-conjugated antibody (Invitrogen) was used to identify epithelial cells. Nuclei were stained with DAPI. All samples were counterstained with mouse IgG1 antihuman anti-CD45 (Clone H130, BD Biosciences), followed by Alexa Fluor 594 fluorophore (Invitrogen) to identify any bound leukocytes. After completion of staining, all devices were washed PBS and stored at 4 °C. All devices were imaged using our previously described imaging system (19). See SI Text for additional details.
Statistical Analysis.
Data are reported as mean ± standard error of the mean as noted. If groups had a normal distribution and homogenous variances, the group means were compared by an independent t-test. Differences were considered significant at the 95% confidence level (p < 0.05).
See SI Text for details on the hematoxylin and eosin staining, on-chip FISH, RT-PCR analysis and sequencing, cell line preparation, flow visualization, and cancer cell line experiments.
Supplementary Material
Acknowledgments.
We express our gratitude to the all patients who participated in this study and the healthy volunteers who contributed blood samples. We are also grateful to Octavio Hurtado, Jochen Lennerz, Douglas Rubinson, Vijay Ambravaneswaran, Salil Desai, Ravi Kapur, John Walsh, Tom Barber, Justin Wong, Zev Nakamura, Matthew Ulman, Suchismita Paul, Brian Brannigan, Alessandra Moore, Maria Kempner, Brooke Nentwig, and Laura Libby for expert technical support. This work was supported by Dream Team Awards from the Prostate Cancer Foundation and Stand Up To Cancer (to D.A.H. and M.T.), a Quantum Grant from the National Institute for Biomedical Imaging and Bioengineering (to M.T.), National Institutes of Health/National Cancer Institute (NIH)/(NCI) Grant CA89138 (to S.M.), and generous donations from the Ellison Foundation, AstraZeneca, the Martell Foundation, Alex and Sonja Spier, the Monell Foundation, and institutional support from Massachusetts General Hospital. S.L.S. is supported through an American Cancer Society New England Division Research Grant, S.N. is supported through an NIH Director’s New Innovator Award, R.J.L. is supported by a Physician Research Training Award (Department of Defense, Prostate Cancer Research Program) and a Career Development Award (The American Society of Clinical Oncology (ASCO) Cancer Foundation), and D.A.H. is supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012539107/-/DCSupplemental.
References
- 1.Vona G, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Giorgi V, et al. Application of a filtration- and isolation-by-size technique for the detection of circulating tumor cells in cutaneous melanoma. J Invest Dermatol. 2010;130:2440–2447. doi: 10.1038/jid.2010.141. [DOI] [PubMed] [Google Scholar]
- 3.Krivacic RT, et al. A rare-cell detector for cancer. Proc Natl Acad Sci USA. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehm T, et al. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin Cancer Res. 2002;8:2073–2084. [PubMed] [Google Scholar]
- 5.Moreno JG, et al. Changes in circulating carcinoma cells in patients with metastatic prostate cancer correlate with disease status. Urology. 2001;58:386–392. doi: 10.1016/s0090-4295(01)01191-8. [DOI] [PubMed] [Google Scholar]
- 6.Moreno JG, et al. Circulating tumor cells predict survival in patients with metastatic prostate cancer. Urology. 2005;65:713–718. doi: 10.1016/j.urology.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Fizazi K, et al. High detection rate of circulating tumor cells in blood of patients with prostate cancer using telomerase activity. Ann Oncol. 2007;18:518–521. doi: 10.1093/annonc/mdl419. [DOI] [PubMed] [Google Scholar]
- 8.Danila DC, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZP, et al. Identification and characterization of circulating prostate carcinoma cells. Cancer. 2000;88:2787–2795. doi: 10.1002/1097-0142(20000615)88:12<2787::aid-cncr18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono JS, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 12.Olmos D, et al. Circulating tumor cell (CTC) counts as intermediate end points in castration-resistant prostate cancer (CRPC): a single-center experience. Ann Oncol. 2009;20:27–33. doi: 10.1093/annonc/mdn544. [DOI] [PubMed] [Google Scholar]
- 13.Scher HI, et al. Circulating tumor cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharmasiri U, et al. Highly efficient capture and enumeration of low abundance prostate cancer cells using prostate-specific membrane antigen aptamers immobilized to a polymeric microfluidic device. Electrophoresis. 2009;30:3289–3300. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams AA, et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagrath S, et al. Isolation of rare circulating tumor cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stott SL, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroock AD, et al. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CH, Di Carlo D, Chen C, Irimia D, Toner M. Microvortex for focusing, guiding, and sorting of particles. Lab Chip. 2008;8:2128–2134. doi: 10.1039/b813434k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao CG, et al. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005;27:49–57. [PubMed] [Google Scholar]
- 23.Pan Y, et al. Characterization of chromosomal abnormalities in prostate cancer cell lines by spectral karyotyping. Cytogenet Cell Genet. 1999;87:225–232. doi: 10.1159/000015432. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt C. Assays that predict outcomes make slow progress toward prime time. J Natl Cancer Inst. 2010;102:677–679. doi: 10.1093/jnci/djq185. [DOI] [PubMed] [Google Scholar]
- 25.Marrinucci D, et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007;38:514–519. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 26.Marrinucci D, et al. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Arch Pathol Lab Med. 2009;133:1468–1471. doi: 10.1043/1543-2165-133.9.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Mehdi AB, et al. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: a new model for metastasis. Nat Med. 2000;6:100–102. doi: 10.1038/71429. [DOI] [PubMed] [Google Scholar]
- 29.Xia YN, Whitesides GM. Soft lithography. Ann Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 30.Murthy SK, Sin A, Tompkins RG, Toner M. Effect of flow and surface conditions on human lymphocyte isolation using microfluidic chambers. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.