Abstract
β-turns are secondary structure elements not only exposed on protein surfaces, but also frequently found to be buried in protein–protein interfaces. Protein engineering so far considered mainly the backbone-constraining properties of synthetic β-turn mimics as parts of surface-exposed loops. A β-turn mimic, Hot═Tap, that is available in gram amounts, provides two hydroxyl groups that enhance its turn-inducing properties besides being able to form side-chain-like interactions. NMR studies on cyclic hexapeptides harboring the Hot═Tap dipeptide proved its strong β-turn-inducing capability. Crystallographic analyses of the trimeric fibritin-foldon/Hot═Tap hybrid reveal at atomic resolution how Hot═Tap replaces a βI’-turn by a βII’-type structure. Furthermore, Hot═Tap adapts to the complex protein environment by participating in several direct and water-bridged interactions across the foldon trimer interface. As building blocks, β-turn mimics capable of both backbone and side-chain mimicry may simplify the design of synthetic proteins.
Keywords: hairpin, molecular bionics, protein dynamics, synthetic miniprotein, X-ray
The β-turn is the most common nonrepetitive motif observed in folded proteins (1). Generally, proline, which does not fit many other secondary structures, and glycine, which fits any kink, are often observed in the i + 1 or i + 2 positions of β-turns (2). However, any α-amino acid can be accommodated in a β-turn given appropriate structural restraints of the global fold. In contrast to this promiscuity observed in natural proteins, synthetic and covalently constrained β-turn mimics should enforce a β-turn-like structure at any user-defined position independent of the restraints given by the protein fold. Accordingly, attempts to incorporate such turn mimics into proteins may cause destabilization of the overall fold (3). The strategy of constraining the polyamide backbone while neglecting potential side-chain interactions has restricted applicability because it may mostly work for surface-exposed peptide loops but not for β-turns embedded within the protein structure. Novel β-turn mimics providing additional side-chain interactions besides a rigid peptide backbone should therefore allow an improved fit of turn mimics within a complex protein environment.
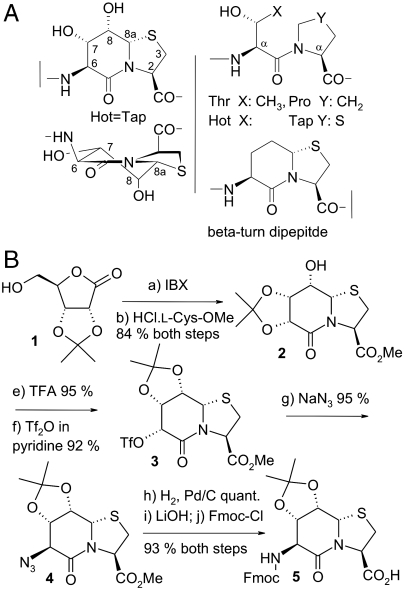
Local backbone cyclization, for restricting the local mobility of an oligopeptide, was introduced by Freidinger in order to obtain selective protein inhibitors (4). Bicyclic conformational constraints for both intervening residues i + 1 and i + 2 of a β-turn are the most common way of obtaining β-turn mimics, as established by the β-turn-dipeptide (BTD) of Nagai and Sato (5). BTD was investigated in the surface-exposed loops of several proteins like HIV-protease, (6) Zn-finger, (7), and WW domains (3) where it adapts to diverse local structural environments. Whether a β-turn mimic exclusively stabilizes a unique reverse turn conformation or, alternatively, allows other conformers as well was challenged on the basis of molecular modeling (8) and by experimental data (9). Overall, BTD can adapt to alternative conformational requirements in cyclic peptides (10, 11).
In this study the passive character of BTD is ranked against an improved turn mimic that shows additional side-chain functionalization, the bicyclic dipeptide Hot═Tap (Fig. 1A). This mimic, named for the synthetic fusion between γ-hydroxythreonine (Hot) and 4-thiaproline (Tap) with the “═” representing the two covalent ring connections (12), provides additional ring substituents, compared to BTD, that restrict alternative puckering states of the δ-valerolactam ring. We show how polar hydroxyl substituents lock the ring puckering and provide secondary interactions within the protein environment. Atomic resolution structures of the Hot═Tap turn mimic within a protein–protein interface, as represented by the fibritin–foldon trimer, show how adaptation to complex local environments is achieved. Hot═Tap can mimic a β-turn buried within the protein fold by establishing polar side-chain like interactions with neighboring structural elements.
Fig. 1.
(A) Chemical formula and systematic numbering of the β-turn mimic Hot═Tap, the structurally related dipeptide motif Thr-Pro and the bicyclic dipeptide BTD (5). Constrained oligocyclic dipeptides, which fit the backbone register of oligopeptides, are named by a six-letter code (Xaa═Yaa) (12). (B) Chemical synthesis of the protected dipeptide Hot═Tap derivative 5 for solid-phase peptide synthesis.
Results and Discussion
Synthesis of the β-turn Mimic Hot═Tap.
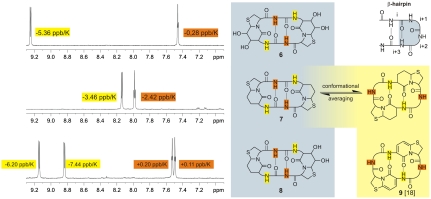
The synthesis of conformationally constrained β-turn mimics with native-like side chains is hampered by the expense of designing an oligocyclic scaffold that is adequately protected for solid-phase peptide synthesis (13). Polyhydroxylated dipeptides with diversified stereochemistries can be obtained from uronolactones in a straightforward way (14, 15). The oxidation of the primary hydroxyl group of 2,3-isopropylidene-d-ribonolactone (1) (16) yields 2,3-isopropylidene-l-riburonolactone, which reacts with l-cysteinemethylester to thiaindolizidinone 2 (Fig. 1B). Acidic isomerization of the acetonide liberates O6, which is subsequently activated to triflate 3. The crystalline azide (4, SI Text) forms under inversion of configuration. Hydrogenation, C-terminal saponification, and acylation with Fmoc-chloride yielded the Fmoc-protected Hot═Tap precursor 5 for solid-phase peptide synthesis as further described in SI Text. Evidence for the ring puckering is also given in SI Text.
Evaluation of Hot═Tap in Hexapeptide Models.
The difference between Hot═Tap, an element that has a strong tendency to induce β-turns, and BTD, which passively fits to other backbone conformations as well, becomes obvious when comparing different β-hairpin mimics in cyclic hexapeptide models. Cyclic hexapeptides comprise two antiparallel β-hairpin turns and can assume various alternative conformations (17). The cyclic hexapeptides cyclo[Hot═Tap-Gly-]2 (6), cyclo[BTD-Gly-]2 (7), and cyclo[Hot═Tap-Gly-BTD-Gly-] (8) (Fig. 2) differ in the number of hydroxyl groups on the δ-valerolactam ring. Although the hydroxyl groups are oriented outward and do not directly interfere with the intramolecular hydrogen bonds of the two β-hairpin turns, the NMR data of peptide 7 differ from peptides 6 and 8, which contain Hot═Tap. The C2-symmetric hexapeptide 6 exhibits a large chemical shift dispersion of the amide protons and a reduced solvent accessibility of the glycine amide protons in the solvent DMSO. The Hot═Tap amide proton is oriented toward the solvent, rendering its chemical shift δ (ppm) significantly temperature-dependent with a gradient of -5.36 ppb/K (SI Text and Fig. S1). The glycine NH, on the other hand, is shielded from the solvent, making its chemical shift less temperature-dependent with a gradient of only -0.28 ppb/K. The NH chemical shifts and the temperature gradients of 6 show a reversed behavior compared to other bicyclic dipeptides investigated by us, where the bicyclic dipeptide was found outside the β-turn (9, 14, 18). A pyridinone dipeptide within a cyclic hexapeptide (9) is included in Fig. 2 as an example of a peptidomimetic that prefers extended conformations with a reversed hydrogen bonding pattern resulting in a reversed temperature dependency of the NH chemical shifts (18). The averaged temperature dependencies of the amide proton of BTD (-3.46 ppb/K) and of glycine (-2.42 ppb/K) in cyclic hexapeptide 7 are explained by the averaging between conformers wherein BTD occupies the short side of the hexapeptide (similar to 6) and conformers wherein BTD occupies the long side (similar to 9). The temperature gradients of 7 are close to the empirical limit of ± 2 ppb/K for an amide that is incorporated in a hydrogen bond (17). The small chemical shift difference between the amide protons supports the notion of a flexible molecule with the peptide appearing dynamic and BTD alternating between the short side (i + 1 and i + 2 position of a β-turn) as well as in the long side (i and i + 1 position of a β-turn) of the cyclopeptide. The conformation of the cyclic hexapeptide 8 containing both BTD and Hot═Tap is again comparable with peptide 6. The glycine at the i + 3 position relative to the β-turn shows the lowest temperature gradient, but also the glycine next to BTD shows only low temperature dependency. In the mixed hexapeptide 8, Hot═Tap pushes BTD into the i + 1 and i + 2 position of the turn and a main conformation similar to 6 is observed. Cyclic hexapeptides show conclusively that Hot═Tap is different from BTD in strongly preferring the i + 1 and i + 2 position of a β-turn. NOEs further support this result, although only few NOEs are unambiguously assigned in the C2-symmetric cyclopeptides.
Fig. 2.
1H NMR (600 MHz, DMSO-d6). The amide region from the 1H NMR (upper left) of cyclo[Hot═Tap-Gly-]2 (6) identifies a solvent-exposed amide proton at 9.25 ppm (yellow) exhibiting a strong temperature dependence (-5.36 ppb/K). The triplet at 7.45 ppm (Gly-NH, brown) on the contrary is hardly influenced by the temperature (-0.28 ppb/K) as expected for a proton engaged in an intramolecular hydrogen bond. Based on these data, Hot═Tap is expected to occupy the turn position in hexapeptide 6 (gray background, top). The amide region from the 1H NMR (middle left) of cyclo[BTD-Gly-]2 (7) shows averaged chemical shifts and temperature gradients caused by the conformational adaptability of BTD, which fits at least two different positions in the hexapeptide. The lower left 1H NMR of the mixed hexapeptide cyclo[Hot═Tap-Gly-BTD-Gly-]2 (8) is again similar to the 1H NMR of 6 with a doubled signal caused by the broken C2 symmetry. The proposed alternative conformation of 7 is shown with a yellow background similar to 9 (18) as an example of a cyclic hexapeptide populating an alternative conformation (yellow background).
Overall, Hot═Tap surpasses the well-known BTD because of the restrained pucker of its six-membered δ-valerolactam ring (SI Text and Table S1). Although the bicyclic dipeptide BTD efficiently kinks the peptide backbone, neither this nor related turn mimics can completely restrict a given oligopeptide into a β-hairpin turn conformation. Hot═Tap not only kinks the peptide backbone as any other related ring structure would do but constricts a complete reverse turn.
Design of Hot═Tap-Foldon Hybrids.
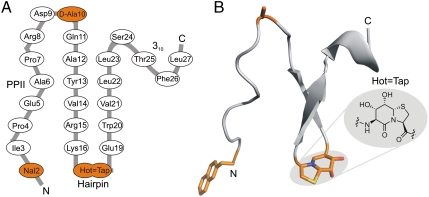
The foldon protein was first described as the C-terminal domain (G457-A486) of the T4 phage fibritin (19, 20), a trimeric protein that serves as fibrous “whiskers” during late stages of the assembly of T4 phage particles. Foldon has a small size of 30 amino acids (here Gly1-Ala30) with a strong tendency to trimerize in a simple fold comprising a β-hairpin that is preceded by a type-II polyproline helix (Fig. 3). Hence, this miniprotein has often been used in biophysical studies, in cooperative protein folding and oligomerization studies (21), and in analyzing the structural features of similar fibrous proteins fused to the foldon domain (22).
Fig. 3.
(A) Schematic representation of the secondary structure of the FV-1. Amino acid exchanges are highlighted in orange. The bicyclic dipeptide is abbreviated with the six-letter code Hot═Tap. (B) Backbone of one subunit of the crystalline trimeric FV-1 with the side-chain and backbone modifications highlighted in orange. The chemical structure of the dipeptide mimic Hot═Tap is shown in the inlay (PPII: polyproline II helix).
This fibritin–foldon was chosen as a template to replace its β-turn, which is embedded within the trimer interfaces, by the turn mimic Hot═Tap. Foldon incorporates two Asp-Gly motifs, of which the first Asp9-Gly10 terminates the N-terminal type-II polyproline helix and the second Asp17-Gly18 forms the β-turn. Asp9-Gly10 is part of a left-handed loop with the Φ-torsion of Gly10 adopting a positive value. In order to avoid aspartimide formation during SPPS (23), we replaced Gly10 with the glycine surrogate d-Ala, which adopts main chain conformations otherwise only accessible to glycine (24). The second Asp-Gly motif forms the central amino acids (i + 1 and i + 2) of the βI’-type turn (Lys16-Glu19). This loop is expected to be highly sensitive against structural distortion because it not only holds together the antiparallel β-strands 1 and 2 (Ala12-Leu23), making up the majority of the foldon trimer, but also packs against the loop of a neighboring monomer connecting its N-terminal type-II polyproline helix with β-strand 1. Apart from the high stability of the antiparallel β-sheet, temperature-dependent NMR-experiments of foldons indicated an extraordinary stability of its βI’ turn (25).
Synthetic Fibritin–Foldon Variants.
Besides wild type, eleven fibritin–foldon variants, FV-1 to FV-11, were generated incorporating either d-amino acids or β-turn mimics or both (Table 1 and SI Text). NMR analyses of these foldon variants including the wild-type peptide verified a folded state for FV-1 to FV-7. Three of them, FV-1 to FV-3, formed crystals and high-resolution structures could be obtained of FV-1 and FV-2. Both variants harbor the d-Ala10 mutation, although this mutation is per se not necessary for cooperative folding, as proven for the other foldon variants (Table 1). Whereas two of the three crystalizable foldon variants contain Hot═Tap in the turn position, no crystals could be obtained of the BTD-foldon. As predicted from the structure, no hybrid foldon variant containing Hot═Tap at positions other than Asp17-Gly18 demonstrated cooperative folding, as shown by FV-9 to FV-11. The stability and tertiary structure of the synthetic foldon trimers were found to depend on the preservation of long-range interactions via side chains, as exemplified by the intermolecular salt bridge Glu5-Arg15 (21, 25). Changes in synthetic fibritin–foldon variants that appeared minor, like the Asp9 → Glu mutation (FV-8) where only a single hydrogen bond between Asp9 and Tyr13 is disturbed, completely disrupted cooperative folding, whereas minor modifications as exemplified by the [d-Ala10, d-Phe17] foldon (FV-2) are structurally tolerated. Likewise, the deletion of the N-terminal glycine does not significantly affect stability, whereas a Tyr2 → Nal (naphthylalanine) mutation was subsequently found to be beneficial for crystallization.
Table 1.
Synthetic foldon variants
| Foldon variant | X-ray | Fold * | |
| Wild-type | no | yes | |
| FV-1 | [ΔGly1, Nal2, d-Ala10, Hot17═Tap18] | 1.06 Å | yes |
| FV-2 | [d-Ala10, d-Phe17] | 0.98 Å | yes |
| FV-3 | [d-Ala10, Hot17═Tap18] | 1.50 Å | yes |
| FV-4 | [d-Ala10] | no | yes |
| FV-5 | [d-Ala10, d-Phe17, Pro18] | no | yes |
| FV-6 | [Hot17═Tap18] | no | yes |
| FV-7 | [d-Ala10, Btd17,18] | no | yes |
| FV-8 | [Glu9, Hot17═Tap18] | no | no |
| FV-9 | [Hot6═Tap7, d-Ala10, Hot17═Tap18] | no | no |
| FV-10 | [d-Ala10, Hot17═Tap18, Hot23═Tap24] | no | no |
| FV-11 | [d-Ala10, Hot17═Tap18,Hot25═Tap26] | no | no |
1H NMR spectra are shown in SI Text.
*Determined by 1H NMR.
Overall Structure of a Fibritin–Foldon–Hot═Tap Hybrid.
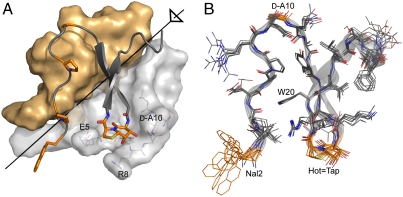
The synthetic [ΔGly1, Nal2, d-Ala10, Hot17═Tap18] foldon (FV-1) (Fig. 3A) yielded crystals diffracting to 1.06 Å resolution (Table S2 and Fig. S2). Relative to the wild-type protein this variant has been chemically modified at four out of its 26 residues and misses the N-terminal Gly1 (Fig. 3B). As expected, the FV-1 (Fig. 4A) adopts a right-hand twisted β-sheet conformation for Ala12-Leu23 that is preceded by an extended N-terminal region (Nal2-Gln11) and followed by a short 310 helix (Leu23-Leu27). The trimer of the FV-1 is stabilized by several hydrophobic interactions, the Glu5-Arg15 salt bridge and backbone-to-backbone interactions between neighboring β-hairpins involving Tyr13 and Arg15. It can thus be best regarded as a six-stranded β-sheet that comprises the antiparallel β-strands 1 and 2 from each subunit. The N-terminal modification Tyr2 → Nal increases the overall hydrophobicity of the synthetic foldon but does not contribute detectably to the overall folding/trimerization tendency, because there is a lack of hydrophobic interactions between the naphthyl side chains in the FV-1 trimer. The crystal form obtained for the FV-1 harbors two independent foldon trimers within the asymmetric unit whose monomers are overlaid in Fig. 4B. Due to atomic resolution their constituting monomers could be independently refined, implying that even minor conformational differences between foldon monomers, which are caused by their intrinsic mobility or the influence of the chemical environment, become visible (26). Accordingly, the superposition of the six foldon monomers shows, apart from conformational variability at the N and C termini, significant deviations only for the solvent-exposed side chains of Arg8, Lys16, Glu19, and Phe26. In contrast, the hydrophobic core of the fibritin–foldon—Ala6, Pro7, Tyr13, Trp20, and Leu22—is almost invariant; the rmsd values for its 26 Cα-positions range between 0.32 and 0.71 Å (Fig. 4B). Furthermore, the conformation of the FV-1 closely corresponds to chemically unmodified foldon units (27–30). For example, compared to the foldon unit within a genetically engineered fusion to a viral nucleocapsid protein (27) the rmsd of the Cα-positions is only 0.65 Å (Fig. S3).
Fig. 4.
(A) Crystal structure of the trimeric FV-1. Two subunits are shown as surface representations (gray/yellow), one including three side chains (light gray) that embrace the Hot═Tap mimic of the third subunit (peptide backbone with selected side chains shown). The threefold axis is indicated in black. (B) Superposition of the backbones of all six different monomeric subunits in the asymmetric unit of the crystal structure of FV-1. Selected side chains are highlighted in orange.
Adaption of the Hot═Tap Turn Mimic into a Protein Interface.
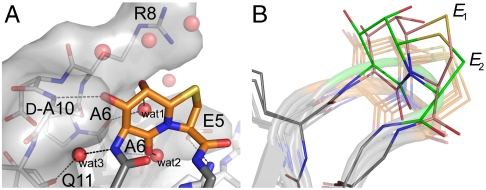
The β-turn mimic Hot═Tap occupies the anticipated i + 1 and i + 2 position within the β-turn loop Lys16-Glu19. The six-membered δ-valerolactam ring of the β-turn mimic Hot═Tap uniformly adopts an envelope conformation (E8) thus directing the C8-hydroxyl group into an axial orientation (Fig. 5). In contrast, the five-membered thiazolidine ring of Hot═Tap occurs in at least two pucker conformations as shown in Fig. 5B. Five out of six monomers have an E2 envelope conformation, but in the sixth monomer the thiazolidine ring in Hot═Tap adopts two alternative conformations. In the second conformer the sulfur atom of the thiazolidine flips out of the ring plane, thus adopting an E1 conformation. Obviously, puckering of the thiazolidine allows adaptability of the Hot═Tap turn mimic to accommodate local structural variations of the FV-1. Due to the conformation of the bicyclic Hot═Tap mimic, its amino group projects equatorially from the six-membered ring, whereas its carboxyl group protrudes axially from the thiazolidine’s ring plane. In the FV-1 the β-hairpin loop conformation can thus be best described as belonging to the βII’ type, instead of the βI’ type that is found in unmodified foldon units. A second difference, compared with the wild-type foldon structure, is given by the alternative trace taken by the Hot═Tap moiety. Whereas the thiazolidine occupies almost exactly the same position as Gly18 in the wild-type fibritin–foldon (deviation between C3 atom and corresponding Cα atom: ∼0.5 Å, Fig. S3), the δ-valerolactam ring is significantly shifted from the position occupied by Asp17 (deviation between C6 and Cα atom: ∼1.6 Å). As the backbone cyclization that is implemented in Hot═Tap should restrict only rotary degrees of freedom along β-turn positions i + 1 and i + 2 without shifting the register of β-hairpins, the overall structural effects on the conformation of the Ala12-Leu23 β-hairpin are minor. Accordingly, the hydrogen bond between positions i and i + 3 (Lys16, Glu19) is almost unaffected (3.0 Å) due to the structure-inducing properties of the Hot═Tap mimic. The expected function of Hot═Tap as a generally strong β-turn inducer unable to adopt largely differing conformations is underlined by the fact that this building block is not tolerated in other positions of the foldon trimer (Table 1, FV-9 to FV-11).
Fig. 5.
(A) Detailed view of Hot═Tap turn mimic coordinating directly to the neighboring subunit and indirectly via three water molecules (wat1, wat2, and wat3). (B) Detailed view of the Hot═Tap dipeptides in the six overlayed structures of Fig. 4B. Five chains (shown transparently) adopt an E1 envelope conformation for the thiazolidine ring of Hot═Tap, whereas the thiazolidine ring of chain C shows two alternative puckers (salmon: E1; green: E2).
In the wild-type fibritin–foldon the β-hairpin Lys16-Glu19 participates in trimer stabilization by forming several hydrogen bonds: first, between the side chain of Asp17 and the amide of Gly10 of another monomer within the trimer, and secondly, by several water-bridged interactions between the amides deriving from Asp17 and Gly18 with the carbonyl group of Pro7 and the side chain of Glu5. Despite some differences for the conformation of the β-hairpin within the FV-1 as compared to the unmodified foldon, the functionalized Hot═Tap group fits to the surface of the neighboring monomer and is well nestled among side chains derived from residues Glu5, Arg8, and d-Ala10 (Fig. 4A). Here, Hot═Tap emulates the aforementioned interactions between the β-hairpin turn and the neighboring monomer. Instead of Asp17 in the unmodified foldon, the C7-hydroxyl group forms a hydrogen bond (3.0 Å) with the amide of the glycine surrogate d-Ala10 (Fig. 5A). One water molecule (wat1) corresponds to a water molecule found in the wild-type structure by bridging between the axial C8-hydroxyl and the Pro7 carbonyl; two additional water molecules are trapped in the trimer interface. One of them (wat2) forms a hydrogen bond between the C5-carbonyl of the Hot═Tap mimic and the amide of Ala6, whereas the other water molecule (wat3) invades the interface by bridging the amino group of Hot═Tap to the backbone carbonyl of Gln11.
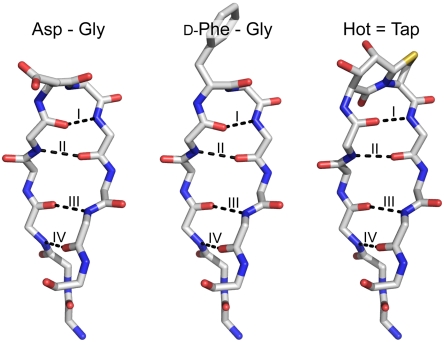
Despite the different β-turn types adopted, the Hot═Tap turn mimic neither affects the intermolecular salt bridge between Glu5 and Arg15 (2.8 Å), which is crucial for trimer formation, (21) nor distorts the attached antiparallel β-sheet. The β-turns of three crystalline fibritin-foldon variants are shown in Fig. 6 together with the four hydrogen bonds formed by their constituting antiparallel β-sheets. Like the peptide backbones shown for the related FV-2 foldon variant that misses a turn mimic, the Hot═Tap/foldon hybrid is indistinguishable from the best available crystal structure of a recombinant foldon. Apparently, the FV-1 is characterized only by a 180° flip of its central amide bond causing the βI' → βII conversion, combined with a progression of hydrogen bonds that are almost identical to the native protein.
Fig. 6.
Antiparallel β-sheet with a central Asp17-Gly18 βI’-turn as observed in a recombinant fibritin–foldon fusion protein [PDB code 2IBL (27)]. Hydrogen bonds are indicated by black dashed lines. The same region with d-Phe-Gly turn in the FV-2. Hot═Tap β-turn of FV-1 forming a nearly identical β-sheet that has a flipped amide bond orientation restricting the backbone torsions of a βII’-turn.
Conclusion
We replaced the two central amino acids of the β-turn in the miniprotein fibritin–foldon with the bicyclic dipeptide mimic Hot═Tap, which constrains the peptide backbone and forms polar contacts to neighboring structural elements within a quaternary protein fold. Residual mobility of the bicyclic ring system and several noncovalent side-chain-like interactions allow the assembly of the intact protein oligomer and trapping of the turn mimic within a protein–protein interface.
β-turn mimics are of general interest not only in the design of enzyme inhibitors but also as building blocks for synthetic proteins. Design of the latter may involve different strategies of simplification, e.g., the usage of reduced amino acid alphabets (31) or conformationally restricted building blocks like the synthetic turn mimic Hot═Tap. Accordingly, the engineering of diversity-minimized biopolymers may pave the way toward a “molecular bionics” of proteins and be in the long-term a new strategy to construct therapeutic proteins.
Materials and Methods
Synthesis of the Hexapeptide Models.
Fragment condensation of two adequately protected Hot═Tap-Gly tripeptides yielded the linear urethane protected Hot═Tap-Gly-Hot═Tap-Gly methyl ester. The linear peptides were deprotected under high dilution conditions and cyclized to 5, 6, and 7, respectively (4).
Polypeptide Synthesis.
Compound 4 is a storable white powder convenient for the automated solid-phase peptide synthesis. Fibritin–foldon miniproteins were produced on the 0.1 mM scale using a modified standard procedure as given in SI Text.
Crystallization and Data Collection of Fibritin–Foldon Peptides.
Purified protein was solved in low salt buffer (9 mM potassium phosphate, pH 7.0, 10% (v/v) D2O) at a concentration of 2 mg/mL. After gentle heating the NMR tube was left in a warm water bath to cool to room temperature overnight, where spontaneous crystallization occurred. Crystals were shortly soaked in cryo buffer (9 mM potassium phosphate, pH 7.0, 10% (v/v) D2O, 40% PEG400) and flash frozen in liquid nitrogen. Data collection was performed at ID23-1 at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) yielding a low- and high-resolution dataset with 2.2 Å and 1.06 Å resolution in space group P1 for the FV-1 and a low- and high-resolution dataset at 2.25 Å and 0.98 Å in space group P32 for the FV-2 foldon.
Structure Solution of Fibritin–Foldon Peptides.
Diffraction data were integrated and merged using imosflm (32) in space groups P1 and P32 for FV-1 and FV-2, respectively. Scaling of data was performed using SCALA 6.0 in the CCP4 package (33) and structure solution was carried out with the program PHASER (34). Residues 135–161 of the structure of a fibritin–foldon fusion protein (PDB code: 1NAY) (29) were used as search model assuming two trimers per asymmetric unit for the FV-1 and one trimer per asymmetric unit for the FV-2, respectively. The graphics program Coot (35) was used for manual model building and refinement was carried out with REFMAC5 from the CCP4i package (33, 36). Restraints used for refinement of the synthetic turn mimic were created using the MOE v2008.1 package (Chemical Computing Group Inc.) and MMFF94 as force field. Figures were made using the program PYMOL 0.99 (37).
Supplementary Material
Acknowledgments.
The authors thank the Deutsche Forschungsgemeinschaft (DFG), Uwe Linne for his assistance with HPLC-MS analyses, Tobias Craan for discussion, Sandor Brockhauser for support at ESRF beamline ID23-1, Grenoble, and Michael Marsch and Klaus Harms for the crystal structures of the synthetic intermediates discussed in SI Text. B.E. thanks the Evonik foundation for the Werner–Schwarze scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004187107/-/DCSupplemental.
Data deposition: X-ray crystallographic structure factors and coordinates of [ΔGly1, Nal2, d-Ala10, Hot17 = Tap18] foldon (FV-1) and of [d-Ala10, d-Phe17] foldon (FV-2) have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, www.pdb.org (PDB ID codes 2WW7 and 2WW6).
References
- 1.Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller AA, et al. Evaluating beta-turn mimics as beta-sheet folding nucleators. Proc Natl Acad Sci USA. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freidinger RM, Veber DF, Perlow DS, Brooks JR, Saperstein R. Bioactive conformation of luteinizing-hormone-releasing hormone—evidence from a conformationally constrained analog. Science. 1980;210:656–658. doi: 10.1126/science.7001627. [DOI] [PubMed] [Google Scholar]
- 5.Nagai U, Sato K. Synthesis of a bicyclic dipeptide with the shape of [beta]-turn central part. Tetrahedron Lett. 1985;26:647–650. [Google Scholar]
- 6.Baca M, Kent SBH, Alewood PF. Structural engineering of the HIV-1 protease molecule with a beta-turn mimic of fixed geometry. Protein Sci. 1993;2:1085–1091. doi: 10.1002/pro.5560020702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viles JH, et al. Design, synthesis and structure of a zinc finger with an artificial β-turn. J Mol Biol. 1998;279:973–986. doi: 10.1006/jmbi.1998.1764. [DOI] [PubMed] [Google Scholar]
- 8.Müller G, Hessler G, Decornez HY. Are beta-turn mimetics mimics of beta-turns? Angew Chem Int Edit. 2000;39:894–896. doi: 10.1002/(sici)1521-3773(20000303)39:5<894::aid-anie894>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Tremmel P, Geyer A. Coupled Hydrogen-Bonding Networks in Polyhydroxylated Peptides. Angew Chem Int Edit. 2004;43:5789–5791. doi: 10.1002/anie.200461099. [DOI] [PubMed] [Google Scholar]
- 10.Cluzeau J, Lubell WD. Design, synthesis, and application of azabicyclo[XYO]alkanone amino acids as constrained dipeptide surrogates and peptide mimics. Biopolymers. 2005;80:98–150. doi: 10.1002/bip.20213. [DOI] [PubMed] [Google Scholar]
- 11.Etzkorn FA, Guo T, Lipton MA, Goldberg SD, Bartlett PA. Cyclic hexapeptides and chimeric peptides as mimics of tendamistat. J Am Chem Soc. 1994;116:10412–10425. [Google Scholar]
- 12.Haack M, Enck S, Seger H, Geyer A, Beck-Sickinger AG. Pyridone dipeptide backbone scan to elucidate structural properties of a flexible peptide segment. J Am Chem Soc. 2008;130:8326–8336. doi: 10.1021/ja8004495. [DOI] [PubMed] [Google Scholar]
- 13.Hanessian S, McNaughton-Smith G, Lombart H-G, Lubell WD. Design and synthesis of conformationally constrained amino acids as versatile scaffolds and peptide mimetics. Tetrahedron. 1997;53:12789–12854. [Google Scholar]
- 14.Hörger R, Geyer A. A shape-persistent D,L-dipeptide building block for the assembly of rigidified oligopeptides. Org Biomol Chem. 2006;4:4491–4496. doi: 10.1039/b613172g. [DOI] [PubMed] [Google Scholar]
- 15.Tremmel P, Geyer A. An oligomeric Ser-Pro dipeptide mimetic assuming the polyproline II helix conformation. J Am Chem Soc. 2002;124:8548–8549. doi: 10.1021/ja026098u. [DOI] [PubMed] [Google Scholar]
- 16.Williams JD, Kamath VP, Morris PE, Townsend LB. D-Ribolactone and 2,3-Isopropylidene(D-Ribolactone) Org Synth. 2005;82:75–79. [Google Scholar]
- 17.Kessler H. Conformation and biological activity of cyclic peptides. Angew Chem Int Edit. 1982;21:512–523. [Google Scholar]
- 18.Seger H, Geyer A. Synthesis and diversification of pyridone dipeptide chromophores. Synthesis. 2006;19:3224–3230. [Google Scholar]
- 19.Letarov AV, Londer YY, Boudko SP, Mesyanzhinov VV. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry-Moscow. 1999;64:817–823. [PubMed] [Google Scholar]
- 20.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: A segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 21.Habazettl J, Reiner A, Kiefhaber T. NMR structure of a monomeric intermediate on the evolutionarily optimized assembly pathway of a small trimerization domain. J Mol Biol. 2009;389:103–114. doi: 10.1016/j.jmb.2009.03.073. [DOI] [PubMed] [Google Scholar]
- 22.Boudko S, et al. Nucleation and propagation of the collagen triple helix in single-chain and trimerized peptides: Transition from third to first order kinetics. J Mol Biol. 2002;317:459–470. doi: 10.1006/jmbi.2002.5439. [DOI] [PubMed] [Google Scholar]
- 23.Mergler M, Dick F, Sax B, Weiler P, Vorherr T. The aspartimide problem in Fmoc-based SPPS. Part I. J Pept Sci. 2003;9:36–46. doi: 10.1002/psc.430. [DOI] [PubMed] [Google Scholar]
- 24.Valiyaveetil FI, Sekedat M, MacKinnon R, Muir TW. Glycine as a D-amino acid surrogate in the K+-selectivity filter. Proc Natl Acad Sci USA. 2004;101:17045–17049. doi: 10.1073/pnas.0407820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier S, Guthe S, Kiefhaber T, Grzesiek S. Foldon, the natural trimerization domain of T4 fibritin, dissociates into a monomeric A-state form containing a stable beta-hairpin: Atomic details of trimer dissociation and local beta-hairpin stability from residual dipolar couplings. J Mol Biol. 2004;344:1051–1069. doi: 10.1016/j.jmb.2004.09.079. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt A, Lamzin VS. Veni, vidi, vici—atomic resolution unravelling the mysteries of protein function. Curr Opin Struct Biol. 2002;12:698–703. doi: 10.1016/s0959-440x(02)00394-9. [DOI] [PubMed] [Google Scholar]
- 27.Boudko SP, Kuhn RJ, Rossmann MG. The coiled-coil domain structure of the sin nombre virus nucleocapsid protein. J Mol Biol. 2007;366:1538–1544. doi: 10.1016/j.jmb.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudko SP, Strelkov SV, Engel J, Stetefeld J. Design and crystal structure of bacteriophage T4 mini-fibritin NCCF. J Mol Biol. 2004;339:927–935. doi: 10.1016/j.jmb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Stetefeld J, et al. Collagen stabilization at atomic level: Crystal structure of designed (GlyProPro)10foldon. Structure. 2003;11:339–346. doi: 10.1016/s0969-2126(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 30.Strelkov SV, Tao T, Shneider MM, Mesyanzhinov VV, Rossmann MG. Structure of Bacteriophage T4 Fibritin M: a Troublesome Packing Arrangement. Acta Crystallogr D. 1998;54:805–816. doi: 10.1107/s0907444997018878. [DOI] [PubMed] [Google Scholar]
- 31.Fan K, Wang W. What is the minimum number of letters required to fold a protein? J Mol Biol. 2003;328:921–926. doi: 10.1016/s0022-2836(03)00324-3. [DOI] [PubMed] [Google Scholar]
- 32.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 33.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.