Abstract
Long interspersed nucleotide element-1 (L1) is a retroelement comprising about 17% of the human genome, of which 80–100 copies are competent as mobile elements (retrotransposition: L1-RTP). Although the genetic structures modified during L1-RTP have been clarified, little is known about the cellular signaling cascades involved. Herein we found that 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct postulated as a candidate physiological ligand of the aryl hydrocarbon receptor (AhR), induces L1-RTP. Notably, RNA-interference experiments combined with back-transfection of siRNA-resistant cDNAs revealed that the induction of L1-RTP by FICZ is dependent on AhR nuclear translocator-1 (ARNT1), a binding partner of AhR, and the activation of cAMP-responsive element-binding protein. However, our extensive analyses suggested that AhR is not required for L1-RTP. FICZ stimulated the interaction of the L1-encoded open reading frame-1 (ORF1) and ARNT1, and recruited ORF1 to chromatin in a manner dependent on the activation of mitogen-activated protein kinase. Along with our additional observations that the cellular cascades for FICZ-induced L1-RTP were different from those of L1-RTP triggered by DNA damage, we propose that the presence of the cellular machinery of ARNT1 mediates L1-RTP. A possible role of ARNT1-mediated L1-RTP in the adaptation of living organisms to environmental changes is discussed.
Keywords: ARNT1, ORF1, cAMP-responsive element-binding protein, MAPK, chromatin recruitment
About 45% of the human genome is composed of transposable elements (1, 2). Long interspersed nucleotide element-1 (LINE-1; L1) is the most abundant component of retroelements, comprising about 17% of the human genome (1, 3). Its structural alignment is conserved from fish to human with two encoded proteins, open reading frames 1 and 2 (ORF1 and 2), the molecular weights of which are about 40 and 150 kDa, respectively (3, 4). ORF1 is a basic protein that binds mRNA, whereas ORF2 has dual functions of endonuclease and reverse-transcriptase activities (3). ORF1 and 2 can complete the retrotransposition of L1 (L1-RTP), which is processed by three steps: transcription, reverse transcription, and the insertion of newly synthesized L1-DNA into the host genome by target-primed reverse transcription (3). Among about 5 × 105 copies of human L1, 80–100 copies are competent as mobile elements (5), and genome shuffling by L1-RTP generates unique expression profiles depending on the integration sites of newly synthesized L1 DNA (6). DNA damage is a reported trigger of L1-RTP (7), but the cellular cascades involved or other factors responsible for the induction of L1-RTP are largely unknown.
The aryl hydrocarbon receptor (AhR), a basic helix–loop–helix/Per–Arnt–Sim (bHLH/PAS) transcription factor, is conserved in invertebrates to human (8, 9), and it recognizes various polycyclic aromatic hydrocarbons (10). For example, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a well-characterized AhR ligand, induces heterodimer formation (AhR complex; AHRC) of AhR and AhR nuclear translocator-1 (ARNT1). Depending on the nuclear localization signal (NLS) of ARNT1 (11), AHRC is translocated to the nucleus, recognizes the xenobiotic response element (XRE), and induces gene expression such as CYP1A1 mRNA (12). Sequence similarities among species and physiological functions support the idea that AHRC was an innovation in vertebrates that enabled them to metabolize xenobiotics in their environments (8). However, its real function remains elusive due to a lack of definite information about its authentic ligands (8, 12).
As a candidate physiological AhR ligand, 6-formylindolo[3,2-b]carbazole (FICZ) is of particular interest. FICZ is generated from tryptophan by ultraviolet B irradiation (13, 14), and its metabolite has been identified in human urine (15). FICZ has a high affinity for AhR (13) and induces CYP1A1 mRNA depending on the AhR (14, 16). Intriguingly, studies have proposed recently that FICZ and TCDD are differentially involved in T-cell differentiation: FICZ generates proinflammatory T cells (TH17), whereas TCDD induces regulatory T cells (Treg) (17, 18). These observations suggest that AhR ligands have novel, uncharacterized biological functions.
In the current study, we found that FICZ induced L1-RTP and that the induction of L1-RTP by FICZ depended on ARNT1, but not on AhR. Biochemical analysis revealed that FICZ activated mitogen-activated protein kinase (MAPK) and phosphorylated cAMP-responsive element-binding protein (CREB) (19), both of which were required for L1-RTP. Furthermore, FICZ induced the association of ORF1 and ARNT1, and recruited ORF1 to chromatin. These data suggest the presence of ARNT1-mediated genome shuffling by L1-RTP, and we discuss its possible involvement in the adaptation of living organisms to environmental changes.
Results
FICZ Induces L1-RTP.
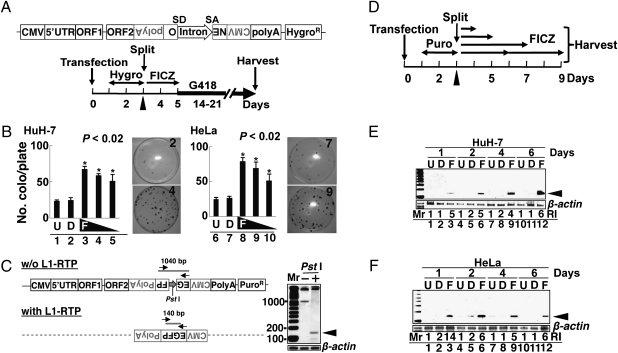
We first assessed FICZ-induced L1-RTP by a colony assay using pCEP4/L1mneoI/ColE1 (pL1-NeoR) (20); its schematic representation is depicted in Fig. 1A. pL1-NeoR contains all the components of the L1 gene and an inversely inserted transcriptional unit encoding a neomycin-resistant (NeoR) gene as a reporter. The NeoR gene had sequences of splicing donor (SD) and splicing acceptor (SA) in a sense orientation (SI Methods). The effects of FICZ on HuH-7 and HeLa cells were examined according to the experimental protocol (Fig. 1A Lower). As shown in Fig. 1B, L1-RTP was induced in both cell lines by FICZ at concentrations of 100, 10, and 1 nM (Fig. 1B, lanes 3–5 and 8–10; P < 0.02). No cytotoxic effects of the compound were detected even at 100 nM FICZ (Fig. S1A).
Fig. 1.
FICZ induces L1-RTP. (A) Schematic diagram of pL1-NeoR and the experimental protocol for the colony assay. Cells were transfected with pL1-NeoR, selected for 2 d using Hygro, and exposed for 2 d to FICZ. Selection with G418 started on day 5. In some experiments, siRNAs of target genes were transfected on day 3 (arrowhead) followed by exposure to FICZ on the next day (SI Methods). (B) Colony assay results. Cells were treated with no reagents (U, lanes 1 and 6), 0.01% dimethyl sulfoxide (DMSO; D, lanes 2 and 7), or FICZ at 100, 10, and 1 nM (F, lanes 3–5 and 8–10). The numbers of colonies were normalized by viability (Fig. S1A). The mean numbers of colonies ± standard deviation (SD) are shown. Asterisks indicate statistical significance (P < 0.02). Stained plates are also shown. Plate numbers 2 and 4 are for HuH-7 cells and plate numbers 7 and 9 are for HeLa cells. The upper and lower plates revealed NeoR colonies formed by 0.01% DMSO (2 and 7) or 10 nM FICZ (4 and 9), respectively. (C) Schematic diagram of pEF06R and the rationale for the PCR-based assay. pEF06R has a structure similar to that of pL1-NeoR except for EGFP cDNA instead of the NeoR gene. PCR primers targeting separate exons of EGFP cDNA (arrows) amplify a 1,040-bp or 140-bp fragment, depending on the induction of L1-RTP. The dotted line indicates the presence of similar structure of pEF06R. To selectively amplify the 140-bp band, DNA samples were treated with PstI (whose site is present in the intron) (Right, lane 2). (D) Experimental protocols for the PCR-based assays (SI Methods). After transfection of pEF06R, HuH-7 cells were cultured for 2 d in the presence of Puro. After exposure for 1–6 d to 10 nM FICZ, genomic DNAs were prepared and subjected to PCR-based analysis of L1-RTP. (E and F) Results of the PCR-based assays. Both HuH-7 (E) and HeLa (F) cells showed the amplified 140-bp band within 1 d after FICZ treatment (arrowheads). Relative intensities (RI) were calculated based on signal intensities of the 140-bp DNA and a DNA fragment of β-actin amplified as an internal control. D, 0.001% DMSO; F, 10 nM FICZ; U, untreated.
The induction of L1-RTP by FICZ was confirmed by a PCR-based assay with pEF06R (SI Methods) (7). As shown in Fig. 1C, PCR primers were designed in separate exons of EGFP cDNA, such that a 140-bp fragment would be amplified when L1-RTP was induced (Fig. 1C). By treating the sample DNAs with PstI (the site is present in the intron), the 140-bp band was selectively amplified (Fig. 1C Right, lane 2). According to the protocol shown in Fig. 1D, we carried out a time-course analysis of FICZ-induced L1-RTP. When 10 nM FICZ was applied, both HuH-7 cells and HeLa cells showed the 140-bp band within 1–2 d (Fig. 1 E and F). Additionally, the frequency of FICZ-induced L1-RTP was 10−4 to 10−5, when estimated by the PCR-based assay (Fig. S1B).
FICZ-Induced L1-RTP Under Down-Regulation of Endogenous AhR.
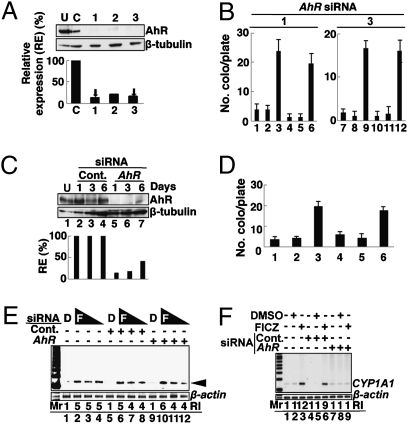
Because FICZ is an AhR ligand (14, 15), we examined whether AhR is required for FICZ-induced L1-RTP. As an initial experiment, we verified the effects of 3′-methoxy-4′-nitroflavone (MNF), an inhibitor of AhR (16). Notably, MNF did not reduce the FICZ-induced L1-RTP (Fig. S1C, lane 6), although it abolished the induction of CYP1A1 mRNA (Fig. S1D, lane 6). Data strongly suggested that AhR is not required for the induction of L1-RTP by FICZ. To prove this, we carried out RNA-interference experiments using AhR siRNA. First, we confirmed that all three AhR siRNAs prepared, when used at 10 nM, could down-regulate the endogenous AhR to a level less than 20% that of the control (Fig. 2A; see also Fig. S2A). Then, we examined the effects of AhR siRNAs on the induction of L1-RTP by FICZ. Intriguingly, the induction of L1-RTP was observed even in the presence of these siRNAs (Fig. 2B; left and right panels depict the results with AhR siRNA-1 and -3, respectively). To gain further evidence, we carried out experiments under more stringent conditions. When 50 nM AhR siRNA was transfected into HuH-7 cells, the endogenous AhR was strongly suppressed for at least 3 d (Fig. 2C, lane 6). We next examined the effects of 2-d treatment of FICZ, and G418 selection was immediately started after FICZ exposure (Fig. 1A). Even under such conditions, FICZ induced L1-RTP (Fig. 2D, lane 6). Additionally, the activities of low doses of FICZ (1 and 0.1 nM) were examined under similar conditions (50 nM AhR siRNA) and again, the PCR-based assay detected L1-RTP (Fig. 2E, lanes 11 and 12). By transfecting 10 nM AhR siRNA, the induction of CYP1A1 mRNA expression by FICZ was completely abolished (Fig. 2F, lane 9), indicating that endogenous AhR was functionally eliminated by the siRNA, although the residual amount of AhR was detectable.
Fig. 2.
AhR is dispensable for FICZ-induced L1-RTP. (A) Functional evaluation of AhR siRNAs. First, dose responses of AhR siRNAs for the suppression of endogenous AhR were verified (Fig. S2A). Then, the activities of three different AhR siRNAs (1–3) at 10 nM were examined. Relative expression (RE) of the AhR protein was calculated based on the expression levels of the proteins in the presence of control and AhR siRNAs. The RE was 11%, 19%, and 14% after transfection with AhR siRNA-1, -2, and -3, respectively. Arrows indicate the siRNAs used for the following experiments. (B) AhR siRNAs did not suppress FICZ-induced L1-RTP. Results of the colony assay performed in the presence of control siRNA (lanes 1–3 and 7–9) or AhR siRNAs (lanes 4–6 and 10–12 for siRNA-1 and -3, respectively) are shown. HuH-7 cells were treated with no reagents (lanes 1, 4, 7, and 10), 0.001% DMSO (lanes 2, 5, 8, and 11), or 10 nM FICZ (lanes 3, 6, 9, and 12). Mean numbers of colonies ± SD are shown. (C) Efficient suppression of endogenous AhR by siRNA. Western blot analysis was performed on days 1, 3, and 6 following the transfection of 50 nM AhR siRNA-1. The RE of AhR protein was calculated and plotted, indicating 13%, 16%, and 40% observed on day 1, 3, and 6 after transfection with AhR siRNA-1, respectively. Cont., control; U, untreated. (D) FICZ induced L1-RTP under stringent conditions. A colony assay was performed following the introduction of 50 nM control siRNA (lanes 1–3) or AhR siRNA-1 (lanes 4–6). HuH-7 cells were treated with no reagents (lanes 1 and 4), 0.001% DMSO (lanes 2 and 5), or 10 nM FICZ (lanes 3 and 6). G418 selection started immediately after FICZ treatment. Mean numbers of colonies ± SD are shown. (E) Low doses of FICZ induced L1-RTP under stringent conditions. On day 2 following the introduction of 50 nM control siRNA (lanes 5–8) or AhR siRNA-1 (lanes 9–12), HuH-7 cells were treated for 2 d with FICZ at 10 nM (lanes 2, 6, and 10), 1 nM (lanes 3, 7, and 11), or 0.1 nM (lanes 4, 8, and 12). Results of the PCR-based assay performed on the second day of FICZ treatment are shown. The arrowhead indicates L1-RTP. The RI of the 140-bp band was also calculated. (F) AhR siRNA suppressed CYP1A1 mRNA expression. RT-PCR done on day 2 after introduction of 10 nM control siRNA (lanes 4–6) or AhR siRNA-1 (lanes 7–9) is shown. Cells were treated for 6 h with 10 nM FICZ and subjected to analysis. The RI was calculated based on the signal intensities of the transcript by the control and AhR siRNAs.
Based on these data, we concluded that the induction of L1-RTP by FICZ is independent of AhR.
FICZ-Induced L1-RTP Is Dependent on ARNT1.
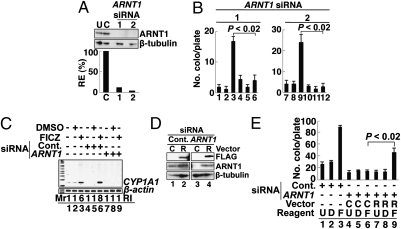
Next, we examined the involvement of ARNT1 and observed that two different ARNT1 siRNAs (1 and 2) efficiently suppressed the expression of endogenous ARNT1 (Fig. 3A; data with siRNA-1 and -2 are shown; see also Fig. S2B), and both siRNAs completely abolished FICZ-induced L1-RTP (Fig. 3B). The PCR-based assay also detected the inhibitory effects of the siRNA (Fig. S3A, lane 9), and the expression of CYP1A1 mRNA was also inhibited by the siRNA (Fig. 3C, lane 9). Data suggested that the siRNA abrogated the function of endogenous ARNT1 and inhibited L1-RTP. To confirm this, we carried out a back-transfection experiment using siRNA-resistant ARNT1 cDNA (21). First, we confirmed that the introduction of a plasmid DNA expressing an siRNA-resistant Flag-tagged ARNT1 mRNA (pSiR-ARNT1; SI Methods) could restore the protein expression that had been reduced by the siRNA (Fig. 3D, lane 4). Then, we observed that pSiR-ARNT1 recovered the formation of NeoR colonies, which had been suppressed by the siRNA (Fig. 3E, compare lanes 6 and 9; SI Methods). These data indicated that ARNT1 is involved in FICZ-induced L1-RTP.
Fig. 3.
ARNT1 is required for FICZ-induced L1-RTP. (A) Functional evaluation of ARNT1 siRNAs. First, dose responses of ARNT1 siRNAs for the suppression of endogenous ARNT1 were verified (Fig. S2B). Then, the activities of two different ARNT1 siRNAs (1 and 2) at 10 nM were examined. The RE of endogenous ARNT1 protein was 12% and 4% when ARNT1 siRNA-1 and -2 were introduced, respectively. (B) ARNT1 siRNAs abrogated L1-RTP. A colony assay was performed on HuH-7 cells following the introduction of control siRNA (lanes 1–3 and 7–9) or ARNT1 siRNA-1 and -2 (lanes 4–6 and 10–12). Cells were treated with no reagents (lanes 1, 4, 7, and 10), 0.001% DMSO (lanes 2, 5, 8, and 11), or 10 nM FICZ (lanes 3, 6, 9, and 12). Mean numbers of colonies ± SD are shown. The effects of ARNT1 siRNAs were significant (P < 0.02). (C) Inhibitory effects of ARNT1 siRNA on the expression of CYP1A1 mRNA. Results of RT-PCR analysis with 10 nM control siRNA (lanes 4–6) or ARNT siRNA-1 (lanes 7–9) are shown. The RI of ARNT1 mRNA is also given. (D) pSiR-ARNT1 restored protein expression. HuH-7 cells were introduced with control siRNA (lanes 1 and 2) or ARNT1 siRNA-1 (lanes 3 and 4). On the next day, a control vector (C, lanes 1 and 3) or pSiR-ARNT1 (R, lanes 2 and 4) was transfected. Protein expression was detected by Western blot analysis on day 2 after the second transfection. (E) pSiR-ARNT1 recovered the L1-RTP suppressed by the siRNA. HuH-7 cells were transfected with ARNT1 siRNA, followed by the introduction of a control vector (lanes 4–6) or pSiR-ARNT1 (lanes 7–9) on the following day. Then, cells were treated with 10 nM FICZ for 2 d followed by G418 selection. Mean numbers of colonies ± SD are shown. The difference between the number of NeoR colonies obtained by pSiR-ARNT1 and by a control vector was significant (P < 0.02; compare lanes 6 and 9). C, control vector; D, 0.001% DMSO; F, 10 nM FICZ; R, pSiR-ARNT1; U, untreated.
To exclude the possibility that other cellular proteins related to the activity of AHRC are involved in FICZ-induced L1-RTP, we examined the effects of 10 nM siRNAs of AhRR (22) and ARNT2 (23) on L1-RTP (Fig. S2 C and D). The results revealed that these molecules are not required for FICZ-induced L1-RTP (Fig. S3B, lanes 9 and 12).
Involvement of MAPK and CREB in the Induction of L1-RTP by FICZ.
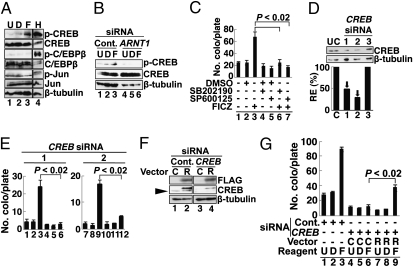
As TCDD activates MAPK (24), we tested the phosphorylation of MAPK substrates possibly related to the induction of L1-RTP. Among the three MAPK substrates examined, CREB was phosphorylated by FICZ (Fig. 4A, lane 3), whereas C/EBP-β and c-Jun were not (Fig. 4A). Notably, the FICZ-induced phosphorylation of CREB was abolished by the down-regulation of ARNT1 by siRNA (Fig. 4B, lane 6). We then examined the effects of MAPK inhibitors on FICZ-induced L1-RTP. As shown in Fig. 4C, the MAPK inhibitors, SB202190 for p38 (25) and SP600125 for JNK (26), efficiently suppressed L1-RTP (compare lanes 3, 6, and 7). The PCR-based assay revealed similar results, indicating the involvement of MAPK in FICZ-induced L1-RTP (Fig. S3C, lanes 6, 8, and 10).
Fig. 4.
MAPK and CREB are involved in L1-RTP. (A) Phosphorylation of CREB by FICZ. HuH-7 cells were treated with no reagents (U, lane 1), 0.001% DMSO (D, lane 2), 10 nM FICZ (F, lane 3), and 1 mM H2O2 (H, lane 4) as a positive control. Antibodies to nonphosphorylated or phosphorylated forms of CREB, C/EBP-β, and c-Jun were used. As a loading control, β-tubulin was detected. (B) FICZ-induced CREB phosphorylation depended on ARNT1. The FICZ-induced CREB phosphorylation was examined after introducing control siRNA (lanes 1–3) or ARNT1 siRNA-1 (lanes 4–6). U, untreated (lanes 1 and 4); D, 0.001% DMSO (lanes 2 and 5); F, 10 nM FICZ (lanes 3 and 6). (C) Effects of MAPK inhibitors on L1-RTP. SB202190 (1 μM) and SP600125 (100 μM), which inhibit p38 and JNK, respectively, were added 30 min before the addition of 10 nM FICZ. Mean numbers of colonies ± SD are shown. The reductions in the numbers of NeoR colonies by inhibitors were significant (P < 0.02; compare lanes 3, 6, and 7). (D) Functional evaluation of CREB siRNAs. First, dose responses of CREB siRNAs for the suppression of endogenous CREB were verified (Fig. S2C). Then, the activities of three different siRNAs (1–3) at 10 nM were examined. The RE of endogenous CREB protein was 48% and 28% when CREB siRNA-1 and -2 were introduced. CREB siRNA-3 was not effective. Arrows indicate the siRNAs used for the next experiments. (E) CREB siRNAs abrogated the L1-RTP. HuH-7 cells were transfected with 10 nM CREB siRNA-1 or -2, and treated with no reagents (lanes 1, 4, 7, and 10), 0.001% DMSO (lanes 2, 5, 8, and 11), or 10 nM FICZ (lanes 3, 6, 9, and 12). Then, a colony assay was carried out. Mean numbers of colonies ± SD are shown. CREB siRNA-1 (Left) and -2 (Right) significantly suppressed L1-RTP (P < 0.02). (F) pSiR-CREB restored protein expression. HuH-7 cells were introduced with control siRNA (lanes 1 and 2) or CREB siRNA-1 (lanes 3 and 4) followed by transfection of a control vector (C) or pSiR-CREB (R). CREB expression was checked on day 2 after transfection. Note that the band with a lower molecular weight is an endogenous CREB protein (lanes 1 and 2, arrowhead), which was abolished by CREB siRNA-1. (G) pSiR-CREB recovered the L1-RTP suppressed by the siRNA. Experiments were performed as described (Fig. 3E). Mean numbers of colonies ± SD are shown. The difference between the number of NeoR colonies recovered by pSiR-CREB and a control vector was significant (P < 0.02; compare lanes 6 and 9). C, control vector; D, 0.001% DMSO; F, 10 nM FICZ; R, pSiR-CREB; U, untreated.
To obtain direct evidence of the requirement of CREB for L1-RTP, the effects of CREB siRNAs were examined. Two different CREB siRNAs, which efficiently suppressed the expression of endogenous CREB protein (Fig. 3D, lanes 1 and 2), blocked L1-RTP (Fig. 4E; data with siRNA-1 and -2 are shown; see also Fig. S2C). Again, the PCR-based assay gave similar results (Fig. S3D, lane 9). Moreover, a plasmid DNA expressing an siRNA-resistant Flag-tagged CREB mRNA (pSiR-CREB) successfully restored both CREB expression (Fig. 4F, lane 4) and L1-RTP (Fig. 4G, compare lanes 6 and 9). These data indicated that in response to FICZ, ARNT1 modulates the activity of MAPK, phosphorylates CREB, and induces L1-RTP.
Interestingly, SP600125 abrogated the induction of L1-RTP by FICZ, but we did not observe that FICZ induced the phosphorylation of c-Jun. Although SP600125 was originally proposed as a specific inhibitor of JNK (26), it was ascertained by subsequent studies that the compound could inhibit activities of several other protein kinases such as SGK, p70 ribosomal protein S6 kinase, AMPK, CDK2, and CK1δ (27). Data suggest the presence of an uncharacterized substrate(s), the activation of which is critical for L1-RTP by FICZ and sensitive to SP600125.
Chromatin Recruitment of ORF1 by FICZ-Dependent Activation of MAPK.
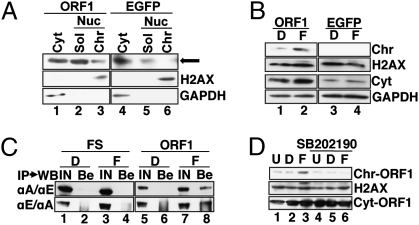
Recently, Goodier et al. reported that ORF1, originally localized in cytoplasmic stress granules, is translocated to chromatin in response to stress stimuli (28). This led us to hypothesize that FICZ induces the nuclear trafficking of ORF1. To demonstrate this, we introduced a plasmid DNA encoding a chimeric protein composed of a codon-optimized ORF1 (29) tagged with a motif for tandem affinity purification (TAP) (30) into HuH-7 cells (SI Methods) and examined the FICZ-induced chromatin recruitment of ORF1. Western blot analysis revealed that FICZ increased ORF1 in the chromatin fraction (Fig. 5B, lane 2), although a slight amount of ORF1 was originally present in the chromatin fraction (Fig. 5A, lane 3). FICZ induced no increase in cellular ORF1 (Fig. S4 A and B) or ORF2 (Fig. S4 C and D). Immunoprecipitation followed by Western blot analysis revealed that FICZ triggered the complex formation of ORF1 and ARNT1 (Fig. 5C, compare lanes 4 and 8). Moreover, the MAPK inhibitor attenuated the FICZ-induced chromatin recruitment of ORF1 (Fig. 5D, compare lanes 3 and 6). Along with observations that the level of endogenous ARNT1 was not changed by FICZ (Fig. S4E), data indicated that FICZ mobilizes the L1 component to chromatin in a manner dependent on the activation of MAPK and ARNT1.
Fig. 5.
The FICZ-induced chromatin recruitment of ORF1 depends on MAPK activity. (A) ORF1 present in the chromatin fraction. HuH-7 cells were transfected with plasmid DNAs encoding ORF1-TAP (lanes 1–3) or EGFP-TAP (lanes 4–6). On day 2 after transfection, cytoplasmic (Cyt, lanes 1 and 3), nuclear soluble (Sol, lanes 2 and 5), and chromatin (Chr, lanes 3 and 6) fractions were prepared and analyzed (SI Methods). H2AX and GAPDH were also detected to specify the subcellular localizations of the proteins. The arrow indicates the position of ORF1 in the chromatin fraction detected in lane 3. (B) Chromatin recruitment of ORF1 by FICZ. Similar experiments were done as described in A, and a chromatin fraction was prepared and analyzed 24 h after the addition of FICZ; 0.001% DMSO (D, lanes and 3); 10 nM FICZ (F, lanes 2 and 4). (C) FICZ stimulated the interaction of ARNT1 and ORF1. HuH-7 cells were transfected with pFS-EGFP (FS) (lanes 1–4) or pORF1-EGFP (ORF1) (lanes 5–8). At 24 h after the addition of 0.001% DMSO (D, lanes 1, 2, 5, and 6) or 10 nM FICZ (F, lanes 3, 4, 7, and 8), cellular extracts were prepared and subjected to reciprocal experiments of immunoprecipitation (IP) followed by Western blot analysis (WB) (IP/WB). Upper row, IP with αARNT1 (αA) followed by WB using αE (αA/αE); lower row, IP with αEGFP (αE) followed by WB using αA (αE/αA). Both input sample (IN) (lanes 1, 3, 5, and 7) and protein recovered by protein G beads (Be) (lanes 2, 4, 6, and 8) were analyzed. As an IN sample, about one-tenth of the cell extract used for the IP was loaded. (D) MAPK inhibitor attenuated the chromatin recruitment of ORF1. SB202190 (1 μM) (lanes 4–6) was added 30 min before the addition of FICZ. U, untreated (lanes 1 and 4); D, 0.001% DMSO (lanes 2 and 5); F, 10 nM FICZ (lanes 3 and 6). ORF1 present in the chromatin fraction (Chr-ORF1 in top row) and cytoplasmic fraction (Cyt-ORF1 in bottom row) is shown. H2AX was also detected to specify the chromatin fraction (middle row). A similar result was obtained by an independent experiment.
Discussion
In this study, we found that FICZ, a candidate physiological AhR ligand of a tryptophan photoproduct, induced L1-RTP. RNA-interference experiments conducted under strongly repressed expression of endogenous AhR suggested that AhR was not required for L1-RTP. In contrast, ARNT1, MAPK, and CREB were all involved in FICZ-induced L1-RTP. FICZ induced the association of ARNT1 and ORF1, and promoted the chromatin recruitment of ORF1 depending on the activation of MAPK. As the target of MAPK, CREB was pivotal for FICZ-induced L1-RTP, but our additional experiments revealed that the simple overexpression of a constitutively active form of CREB cDNA (pCREBY134F) (31) did not induce L1-RTP (Fig. S5). Additional functions of ARNT1 or MAPK are likely required for FICZ-induced L1-RTP.
We found that FICZ-induced L1-RTP was regulated in a fashion different from that induced by DNA double-strand breaks (DSB) (7). First, we observed that the time points of L1-RTP induced by FICZ and X-ray irradiation were different. In contrast, FICZ-induced L1-RTP was observed within 1–2 d (Fig. 1 E and F), but L1-RTP induced by DNA damage was observed only after 12 d of X-ray irradiation (Fig. S6A). Moreover, MAPK inhibitors completely blocked FICZ-induced L1-RTP (Fig. 4C), but not DSB-induced L1-RTP (Fig. S6B, lanes 7–9). Additionally, we found that TCDD was genotoxic, as judged by the increase of γ-H2AX (H2AX phosphorylated at serine 139), which is a sensitive cellular marker of DSB (32) (Fig. S6C, lanes 4 and 5), but it did not induce L1-RTP in HuH-7 cells (Fig. S6D), even though the same dose of TCDD induced the expression of CYP1A1 mRNA (Fig. S6E).
We carefully evaluated whether ARNT1 is a primary target involved in the rate-limiting process of FICZ-induced L1-RTP. Because XRE is present in the region of the 5′ UTR of human L1 (33) and mouse retroposons (34), we first excluded the possibility that FICZ induces L1-RTP by up-regulating the expression level of L1 mRNA. Quantitative RT-PCR analysis revealed that FICZ did not increase the expression of CMV-driven L1 mRNA (Fig. S7 A and B). Furthermore, MAPK inhibitors or siRNAs of ARNT1 and CREB did not change the level of CMV-driven L1 mRNA (Fig. S7B, lanes 5–9). Additionally, FICZ did not increase the splicing efficiency of the immature L1 transcript (Fig. S7 C and D), ORF2 mRNA (Fig. S7E), or the expression levels of ORF1/ORF2 proteins (Fig. S4 B and D). These findings strongly suggest that ARNT1 functions as the mediator for FICZ-induced L1-RTP.
ARNT1, as a central molecule of the class II bHLH/PAS proteins, either homodimerizes or promiscuously heterodimerizes with various bHLH/PAS members (9). As the best-characterized function, the NLS of ARNT1 contributes to nuclear trafficking of the ligand-bound AhR (11). Because ARNT1 has not been reported to function as a receptor to ligands, ARNT1 would likely associate with a novel molecule that binds FICZ. Interestingly, FICZ increases the mRNA expression of Cry1 and Cry2 genes, members of the bHLH/PAS family that are involved in the regulation of circadian rhythm (35). Furthermore, FICZ has been shown to change the electric activities of cells in the suprachiasmatic nucleus, where the master clock of circadian rhythm is maintained and controlled in response to light stimuli (36). These observations make it tempting to speculate that certain gene products involved in circadian rhythm can recognize FICZ, function as its receptor, and cooperate with ARNT1 for the induction of L1-RTP.
Our PCR-based assay revealed that picomolar levels of FICZ (3 pM) can induce L1-RTP (Fig. S8). About 8 pM FICZ was reported to be generated after a 24-h exposure of tissue-culture medium to ordinary laboratory light (14). Given that the concentration of tryptophan in human blood (about 70 μM) (37) is comparable to that present in tissue-culture medium (about 80 μM), FICZ may be generated in vivo and triggers L1-RTP. L1 is conserved in organisms from zebrafish to human (4), and a human L1 homolog of Candida albicans is competent for retrotransposition in Saccharomyces cerevisiae (38). Furthermore, cellular responses to FICZ have been reported in both a Xenopus laevis cell line (39) and zebrafish embryos (40), implying that FICZ can induce L1-RTP in various living organisms. Although further study is required, our current observations suggest the possibility that a member(s) of the bHLH/PAS family is involved in the epigenetic mode of L1-RTP. One possibility is that the genome shuffling by ARNT1-mediated L1-RTP provides living organisms the opportunity to acquire novel characteristics in response to environmental changes such as daylight.
Methods
Cells.
HuH-7 cells (a human hepatocellular carcinoma cell line) and HeLa cells (a cervical carcinoma cell line) were used. The transfection efficiency of these cell lines, when judged by EGFP-positive cells after transfection of an EGFP expression vector, was about 70% and 30%, respectively.
L1-RTP Assays.
Two different systems were used: a colony assay using pL1-NeoR and a PCR-based assay using pEF06R. Each assay was performed at least twice, and representative results are presented. FICZ was used at 10 nM, unless stated otherwise.
RNA-Interference and Back-Transfection Experiments Using siRNA-Resistant Clones.
Knockdown experiments were conducted using two different siRNAs for each gene. Each siRNA was used at 10 nM. To generate stringent conditions for down-regulated AhR expression, 50 nM AhR siRNA was used. Nucleotide sequences of used siRNAs were summarized in Table S1. The expression vectors for siRNA-resistant ARNT1 and CREB cDNAs (pSiR-ARNT1 and pSiR-CREB) and constitutively active CREB (pCREBY134F) were constructed based on pcDNA3.1 Zeo (+) (Invitrogen). Their quality was verified by restriction mapping and nucleotide sequencing.
Chromatin Recruitment of ORF1.
Plasmid DNAs encoding ORF1-TAP (pORF1-TAP) and EGFP-TAP (pEGFP-TAP) were generated using pcDNA3.1 Zeo. To prepare the DNA fragments, pBudORF1syn expressing a codon-optimized ORF1 cDNA (29), pZome-1-C vector for TAP-tag cDNA (30), and pBOSH2BGFP-N1 for EGFP cDNA (41) were used.
Association of ORF1 and ARNT1.
Plasmid DNAs encoding ORF1-EGFP (pORF1-EGFP) and Flag-streptag EGFP (pFS-EGFP) were produced based on pcDNA3.1 Zeo. cDNA fragments were prepared from pBudORF1syn and pBOSH2BGFP-N1. pFS-EGFP was prepared by inserting an oligonucleotide cassette encoding -WSHPQFEK-WSHPQFEK-M- (amino acid sequence for 2xstreptag peptide is depicted by single letters, with spacers indicated by “-”) into 5′ to EGFP cDNA that had been inserted in pFlag-CMV2 (Sigma).
Statistics.
Statistical significance was evaluated using the Mann–Whitney U test. A P value of <0.05 was considered to be statistically significant.
Experimental procedures and construction of the plasmid DNAs are described in detail in SI Methods.
Supplementary Material
Acknowledgments
We are grateful to Drs. Nicolas Gilbert (University of Michigan Medical School, Ann Arbor, MI), Eline T. Luning Prak (University of Pennsylvania School of Medicine, Philadelphia, PA), John Goodier (University of Pennsylvania School of Medicine, Philadelphia, PA), Gabriele Vielhaber (Symrise, Holzminden, Germany), Mark Hahn (Woods Hole Oceanographic Institution, Woods Hole, MA), Oliver Hankinson (Jonsson Comprehensive Cancer Center, University of California, Los Angeles, CA), Kumiko Saeki (National Center for Global Health and Medicine), Astrid M. Roy-Engel (Tulane University Health Sciences Center, New Orleans, LA), and Geoffrey M. Wahl (Salk Institute for Biological Studies, La Jolla, CA) for providing us with pCEP4/L1mneoI/ColE1, pEF06R, an antibody to ORF2, MNF, an antibody to AhRR, ARNT1 cDNA, CREB cDNA, pBudORF1syn/ORF2syn, and pBOSH2BGFP-N1. Dr. Takuro Shimbo (National Center for Global Health and Medicine) kindly carried out statistical analysis of experimental data. This work was supported by a grant from the National Center for Global Health and Medicine (21A-104) and in part by a research grant for the Long-Range Research Initiative from the Japan Chemical Industry Association.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001252107/-/DCSupplemental.
See Commentary on page 18239.
References
- 1.Bannert N, Kurth R. Retroelements and the human genome: New perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Furano AV, Duvernell DD, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 7.Farkash EA, Kao GD, Horman SR, Luning Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn ME. Aryl hydrocarbon receptors: Diversity and evolution. Chem Biol Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 9.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 10.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi H, Ikuta T, Tachibana T, Yoneda Y, Kawajiri K. A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1β is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J Biol Chem. 1997;272:17640–17647. doi: 10.1074/jbc.272.28.17640. [DOI] [PubMed] [Google Scholar]
- 12.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rannug A, et al. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J Biol Chem. 1987;262:15422–15427. [PubMed] [Google Scholar]
- 14.Oberg M, Bergander L, Håkansson H, Rannug U, Rannug A. Identification of the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole, in cell culture medium, as a factor that controls the background aryl hydrocarbon receptor activity. Toxicol Sci. 2005;85:935–943. doi: 10.1093/toxsci/kfi154. [DOI] [PubMed] [Google Scholar]
- 15.Wincent E, et al. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem. 2009;284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 16.Fritsche E, et al. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc Natl Acad Sci USA. 2007;104:8851–8856. doi: 10.1073/pnas.0701764104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 18.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 19.Saeki K, Saeki K, Yuo A. Distinct involvement of cAMP-response element-dependent transcriptions in functional and morphological maturation during retinoid-mediated human myeloid differentiation. J Leukoc Biol. 2003;73:673–681. doi: 10.1189/jlb.1002512. [DOI] [PubMed] [Google Scholar]
- 20.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman EC, et al. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 22.Baba T, et al. Structure and expression of the Ah receptor repressor gene. J Biol Chem. 2001;276:33101–33110. doi: 10.1074/jbc.M011497200. [DOI] [PubMed] [Google Scholar]
- 23.Dougherty EJ, Pollenz RS. Analysis of Ah receptor-ARNT and Ah receptor-ARNT2 complexes in vitro and in cell culture. Toxicol Sci. 2008;103:191–206. doi: 10.1093/toxsci/kfm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon M-J, Jeong K-S, Choi EJ, Lee BH. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-induced activation of mitogen-activated protein kinase signaling pathway in Jurkat T cells. Pharmacol Toxicol. 2003;93:186–190. doi: 10.1034/j.1600-0773.2003.930406.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee HE, et al. Biphasic activation of p38MAPK suggests that apoptosis is a downstream event in pemphigus acantholysis. J Biol Chem. 2009;284:12524–12532. doi: 10.1074/jbc.M808204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: An update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 31.Du K, Asahara H, Jhala US, Wagner BL, Montminy M. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol Cell Biol. 2000;20:4320–4327. doi: 10.1128/mcb.20.12.4320-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 33.Stribinskis V, Ramos KS. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 2006;66:2616–2620. doi: 10.1158/0008-5472.CAN-05-3478. [DOI] [PubMed] [Google Scholar]
- 34.Roman AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci USA. 2008;105:1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukai M, Tischkau SA. Effects of tryptophan photoproducts in the circadian timing system: Searching for a physiological role for aryl hydrocarbon receptor. Toxicol Sci. 2007;95:172–181. doi: 10.1093/toxsci/kfl126. [DOI] [PubMed] [Google Scholar]
- 36.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki Y, et al. Increased serum kynurenine/tryptophan ratio correlates with disease progression in lung cancer. Lung Cancer. 2010;67:361–365. doi: 10.1016/j.lungcan.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Dong C, Poulter RT, Han JS. LINE-like retrotransposition in Saccharomyces cerevisiae. Genetics. 2009;181:301–311. doi: 10.1534/genetics.108.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laub LB, Jones BD, Powell WH. Responsiveness of a Xenopus laevis cell line to the aryl hydrocarbon receptor ligands 6-formylindolo[3,2-b]carbazole (FICZ) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Chem Biol Interact. 2010;183:202–211. doi: 10.1016/j.cbi.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jönsson ME, et al. The tryptophan photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) binds multiple AHRs and induces multiple CYP1 genes via AHR2 in zebrafish. Chem Biol Interact. 2009;181:447–454. doi: 10.1016/j.cbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.