Abstract
Dated molecular phylogenies are the basis for understanding species diversity and for linking changes in rates of diversification with historical events such as restructuring in developmental pathways, genome doubling, or dispersal onto a new continent. Valid fossil calibration points are essential to the accurate estimation of divergence dates, but for many groups of flowering plants fossil evidence is unavailable or limited. Arabidopsis thaliana, the primary genetic model in plant biology and the first plant to have its entire genome sequenced, belongs to one such group, the plant family Brassicaceae. Thus, the timing of A. thaliana evolution and the history of its genome have been controversial. We bring previously overlooked fossil evidence to bear on these questions and find the split between A. thaliana and Arabidopsis lyrata occurred about 13 Mya, and that the split between Arabidopsis and the Brassica complex (broccoli, cabbage, canola) occurred about 43 Mya. These estimates, which are two- to threefold older than previous estimates, indicate that gene, genomic, and developmental evolution occurred much more slowly than previously hypothesized and that Arabidopsis evolved during a period of warming rather than of cooling. We detected a 2- to 10-fold shift in species diversification rates on the branch uniting Brassicaceae with its sister families. The timing of this shift suggests a possible impact of the Cretaceous–Paleogene mass extinction on their radiation and that Brassicales codiversified with pierid butterflies that specialize on mustard-oil–producing plants.
Keywords: Bayesian dating, Brassica, Brassicaceae, chronogram, fossil calibration
The most important genetic model in plant biology is Arabidopsis thaliana. It is the first plant to have its entire genome sequenced, and it serves as a key comparison point with other eukaryotic genomes. A. thaliana is diploid and has a small genome distributed on just five chromosomes, considerations in its choice as a model (1). The age of the Arabidopsis crown group (CG), previously estimated at 5.8–3 Mya (2, 3), and of splits within Brassicaceae have been used to understand the pace of evolution in genes affecting self-incompatibility (4, 5), the rate of change in signal transduction and gene expression (6, 7), the persistence of shared chromosomal rearrangements in A. thaliana and Brassica oleracea (8), the tempo of evolution of miRNA sequences (9), the evolution of pierid butterflies specializing in plants that produce mustard oils (10), and the ages of whole-genome duplication (WGD) events giving rise to gene pairs in Arabidopsis (11). As genomes of additional Brassicaceae (e.g., Capsella rubella) and other Brassicales (e.g., Carica papaya) (12) are sequenced, the importance of robust estimates of divergence dates relating these genomes to one another and to the geological record increases substantially.
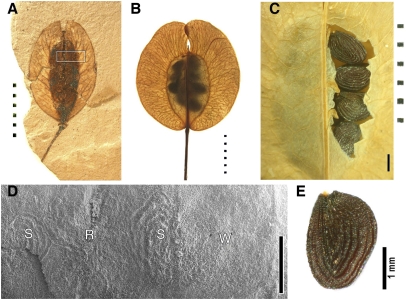
The accuracy of divergence times inferred from sequence data depends on valid, verifiable fossils to calibrate phylogenetic trees. Previous dates for the origin of Arabidopsis relied on the report of fossil pollen assigned to the genus Rorippa (Brassicaceae) (2, 3). We discovered that this report is not linked to a physical specimen, published image, or description; thus, its validity for calibration of a Brassicaceae phylogeny cannot be evaluated. Reexamination of fossils from the order Brassicales (Table S1) revealed six fossil taxa that are sufficiently documented to serve as age constraints; however, only one has been used in previous estimations of divergence times (13). Among the taxa that had been overlooked is Thlaspi primaevum (Brassicaceae) (14), an Oligocene fossil with angustiseptate winged fruits (Fig. 1A) from the Ruby Basin Flora of southwestern Montana, dated at 30.8–29.2 Mya (15). It may have been overlooked because many of the generic determinations from the Ruby Basin Flora remain to be verified, or there may have been concerns about convergence of winged fruits from unrelated angiosperm families. Although the placement of T. primaevum in Brassicaceae now has been confirmed (16), this fruit form evolved independently multiple times within the family (17). However, among extant Brassicaceae, angustiseptate fruit combined with concentrically striate seeds (Fig. 1 B, C, and E) is unique to species of Thlaspi. We examined the seed chamber of T. primaevum and found seed striations in the same pattern as those of extant Thlaspi seeds (Fig. 1D). Alliaria petiolata is the only other Brassicaceae with striated seeds (18) (Fig. S1), but it has longitudinally oriented striations, and the fruit is latiseptate; latiseptate fruits are ancestral for the clade defined by the coalescence of A. petiolata and Thlaspi arvense (Fig. S1). Thus, T. primaevum is a valid age constraint within CG Brassicaceae, and it was placed at the coalescence of T. arvense and A. petiolata as a minimum age constraint for this split (Fig. 2 and Figs. S2–S4).
Fig. 1.
Fossil and extant angustiseptate winged Thlaspi fruits and striated seeds. (A) T. primaevum fossil from the Ruby Basin Flora, western Montana. Extant T. arvense fruit backlit to show placement of seeds in the two locules (wings) (B) and with a portion of the valve removed to show striated seeds (C). Locules are separated by the replum. (D) Scanning electron micrograph of the fossil seed chamber (indicated by rectangle in A) showing impressions of striated seeds. (E) T. arvense striated seed. S, seed; R, replum; W, wing. (Dotted scale bars, 5 mm; solid scale bars, 1 mm.)
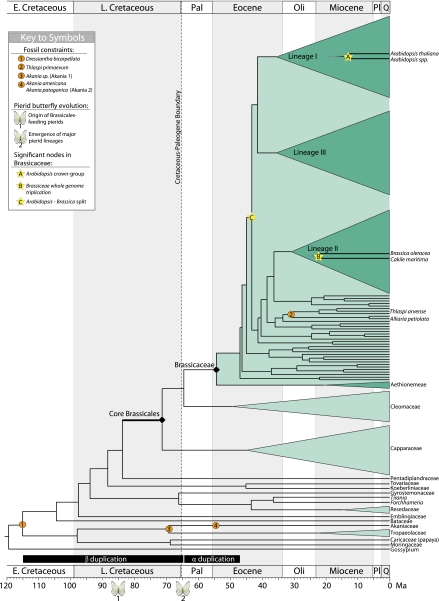
Fig. 2.
Brassicales chronogram inferred using BEAST (28). Clades with >50 species are represented by wedges proportional to species diversity. See figure for key to symbols. Thickened branch leading to Core Brassicales marks an inferred 2- to 10-fold shift in diversification rate. Putative intervals for the α and β WGD are based on refs. 12, 24, and 26. Oli, Oligocene; Pal, Paleocene; Pl, Pliocene; Q, Quaternary.
The other five potentially useful Brassicales fossils are placed outside the Brassicaceae crown group. Capparidoxylon holleisii is a Miocene wood fossil with affinities to extant Capparis, dated at 17–16.3 Mya (Table S1) (19). We explored its use as a constraint within Core Brassicales (Brassicaceae + Cleomaceae + Capparaceae) (SI Materials and Methods). Dressiantha bicarpellata is a Turonian fossil dated at 93.6–89.3 Mya (20), the oldest known putative brassicalean fossil; it provided the single age constraint used by Couvreur et al. (13). However, membership of D. bicarpellata within Brassicales is contentious (21), and thus its validity as an age constraint for estimating divergence times in the order remains unclear. To explore these issues we (i) assessed the impact of using it to constrain four nodes along the backbone of the Brassicales tree, based on results from analyses of combined nucleotide and morphological data (SI Materials and Methods) and (ii) estimated ages in Brassicales without D. bicarpellata. Finally, three species of Akania were used to constrain the ages of nodes within the Bretschneidera/Akania/Tropaeolum clade (Fig. 2). To take full advantage of these age constraints deeper in Brassicales, we expanded our sample of the plastid locus NADH dehydrogenase subunit F (ndhF) and the nuclear locus phytochrome A (PHYA), which have provided a robust phylogenetic framework for the Brassicaceae (17, 22), to include members of the Core Brassicales, giving us a matrix of 179 species (SI Material and Methods and Table S2). We estimated divergence times for trees inferred from each locus and from the combined data.
Most estimates of divergence times in Brassicaceae have assumed that rates of nucleotide evolution are equal across the tree and have been based on either the mutation rate at synonymous sites (2, 3) or the estimated times of genome duplications (23–26). Two recent analyses have allowed rates to be uncorrelated across the tree, but one of these analyses relied on D. bicarpellata as a single age constraint (13), and the other relied on a single secondary age constraint (27). Neither of these analyses took into account uncertainties in the topology of the tree (13, 27). Our analyses used multiple age constraints, allowed rates of nucleotide evolution to be uncorrelated across the tree, and accounted for uncertainty in the phylogenetic hypothesis and in the placement of the fossil age constraints.
Results and Discussion
Using Bayesian approaches (28), we estimated the origin of CG Arabidopsis at ≈13.0 Mya [95% highest probability density (HPD): 17.9–8.0] (Fig. 2 and Table 1), considerably older than the frequently cited estimate of 5.8–3 Mya (Table 1) (2, 3). When a penalized likelihood approach (29, 30) was used, the estimate for this node was even older (Table S3). In the subsequent discussion, we focus on estimates from the Bayesian analyses (Table 1 and Table S3). The placement or exclusion of D. bicarpellata had little impact on the age of this node and other nodes in the tree (Table S3). Conversely, the T. primaevum constraint did lead to slightly older ages for most nodes in the tree (Table S4). Nonetheless, even without this constraint within CG Brassicaceae, age estimates were substantially older than previous estimates (Table 1), possibly resulting from our use of multiple constraints, dense taxon sampling (but see ref. 13), from our allowing for phylogenetic uncertainty, alternative placement of fossil calibrations, from the relative completeness of our molecular datasets (cf. ref. 13), or from a combination of these factors. The previous estimate for CG Arabidopsis is primarily Pliocene (5.3–1.8 Mya), whereas the revised age falls within the Miocene (23.03–5.3 Mya) and spans a particularly warm period in recent earth history that included the Middle Miocene climatic optimum (31). Thus, warming may have played a role in the divergence of A. thaliana from other Arabidopsis. Moreover, the much older optimal age estimate of 13.0 Mya suggests that the pace of chromosomal rearrangements (1), divergent gene regulation (6), and the breakdown of self-incompatibility (4) may have proceeded more slowly than has been appreciated.
Table 1.
Comparison of published dates with those estimated in this study using Bayesian inference in BEAST on the ndhF + PHYA Brassicales chronogram
| Node age | Koch et al. 2000 (2), 2001 (3) synonymous substitution rate, single fossil constraint (Mya) | Franzke et al. 2009 (27) BEAST, nad4, single secondary constraint (Mya) | Couvreur et al. 2010 (13) BEAST, supermatrix, single fossil constraint (Mya) | This study BEAST, ndhF + PHYA, four fossil constraints (Mya) |
| Arabidopsis CG | 5.8–3 | ND | ND | 17.9 – 13.0 – 8.0 |
| Brassiceae CG (whole genome triplication) | 14 | ND | ND | 28.3 – 22.5 – 15.6 |
| Lineage I CG | 19–13 | 19.0 – 8.0 – 0.5 | 36.1 – 27.3 – 18.2 | 42.8 – 35.6 – 28.5 |
| Lineage II CG | 18.1 | ND | 37.2 – 28.2 – 18.1 | 37.8 – 30.8 – 23.7 |
| Lineage III CG | ND | ND | 29.3 – 21.4 – 14.8 | 42.7 – 35.5 – 28.3 |
| Arabidopsis–Brassica MRCA | 21–16 | ND | ≈34* | 50.7 – 43.2 – 36.6 |
| Core Brassicaceae CG | ND | 28.0 – 11.0 – 1.0 | 42.8 – 32.3 – 20.9 | 54.3 – 46.9 – 39.4 |
| Brassicaceae CG | 60–30 | 35.0 – 15.0 – 1.0 | 49.4 – 37.6 – 24.2 | 64.2 – 54.3 – 45.2 |
| Brassicaceae–Cleomaceae MRCA | ND | 35.0 – 19.0 – 1.0 | ND | 76.5 – 64.5 – 54.4 |
| Core Brassicales CG | ND | ND | ND | 83.2 – 71.3 – 59.7 |
Ages in bold are optimal reconstructions and are bracketed by upper and lower bounds of the 95% HPD interval. CG, crown group; MRCA, most recent common ancestor; ND, not determined.
*Approximate mean date inferred from chronogram and for which HPD data were not available (13).
The first split within CG Brassicaceae occurred in the early Eocene, ≈54.3 Mya (95% HPD: 64.2–45.2) (Fig. 2 and Figs. S2–S4). CG Lineages I, II, and III, which contain the majority of Brassicaceae (17, 22), radiated in the mid to late Eocene, from 43.4 to 33.3 Mya (Table 1). Because A. thaliana and B. oleracea (broccoli and related plants) occur in Lineages I and II, respectively, the relatively deep coalescence of these scientifically and economically important species dates to ≈43.2 Mya (95% HPD: 50.7–36.6; Table 1), indicating that conservation of the large chromosomal blocks shared by these two species (8) has persisted for longer than previously thought (2, 3, 13). The B. oleracea genome, along with several close relatives, including Brassica rapa (turnip) and Brassica napus (canola), has been the subject of intense study because of the agricultural importance of the group. The whole-genome triplication event that likely facilitated the origin of these crops was dated previously at 14 Mya (32). Our age estimate for the triplication is centered at 22.5 Mya (95% HPD: 28.3–15.6; Table 1).
Our data suggest that the Core Brassicales stem group (SG) originated around 71.3 Mya (95% HPD: 83.2–59.7), shortly before the end of the Cretaceous, when SG Capparaceae split from SG Brassicaceae and SG Cleomaceae (Fig. 2 and Table 1). Furthermore, using LASER (33), we detected a twofold shift in net diversification rate along the branch to core Brassicales when extinction rates are low or a 10-fold shift when extinction rates are high (Table S5). SG Brassicaceae split from SG Cleomaceae at the Cretaceous–Paleogene boundary (KPB; ≈65.5 Mya) (Table 1 and Fig. 2). Thus, stem members of all three families may have survived the KPB mass extinction event before their subsequent radiations in the Eocene (Fig. 2 and Fig. S4). A pattern of origin before the KPB and radiation afterward has been noted in animal (34) and other plant lineages (35), although the lag time before recovery is greater in Brassicaceae than has been inferred in the other groups (35).
A great deal of interest has centered on the timing of WGD events in the history of A. thaliana, particularly on the most recent of these events, the α WGD, estimated to have occurred between 100 and 20 Mya (11, 36–39), and the β WGD, estimated to have occurred between 235 and 112 Mya (36, 38, 39) (Fig. 2). Recent lines of evidence place the α WGD within Brassicales (12), perhaps within Brassicaceae (26, 27, 36, 40), and place the β WGD within Brassicales (12, 40). These placements have fueled speculations that genome-doubling events are linked with diversification in Brassicaceae (13, 41) and with survival of the KBP mass extinction event (24). These hypotheses are attractive, because WGD may result in the colonization of new habitats (42), major ecological transitions (43), invasiveness (44), and the generation of morphological novelty (45). Our chronogram (Fig. 2 and Fig. S4) provides the framework for testing these hypotheses. For example, if the β WGD occurred around 70 Mya, placing it on the stem lineage of Core Brassicales (Fig. 2), we would expect to find that paralogs stemming from this event are shared by members of Brassicaceae, Cleomaceae, and Capparaceae but not by other families in Brassicales. If a WGD maps to this branch, the locus of the shift in net diversification rate is consistent with a link between genome doubling and survival across the KPB mass extinction and perhaps to subsequent diversification.
Despite the interest in genome doubling and its effects, it is important to consider other factors that might promote diversification. Ehrlich and Raven (46) cited the interaction between Brassicales and pierid butterflies as a prime example of coevolution between plants and insects. Pierids specializing on Brassicales produce nitrile-specifier protein essential for detoxification of glucosinolates, a chemical defense found almost exclusively in Brassicales (10). This key innovation facilitated a host switch onto Brassicales, dated at ≈85 Mya (10, 47), and was followed by the subsequent diversification of pierids (10). The locus of the shift in Brassicales net diversification rate and the timing of the pierid butterfly radiation suggest that they codiversified (Fig. 2 and Fig. S4), supporting a central tenet of coevolutionary theory (46). However, the extent to which diversification in Brassicales may have driven pierid diversification, and vice-versa, requires further study.
Carefully designed comparative studies reveal the processes that generate and maintain biodiversity. In this study we identify a major shift in diversification rate that may correlate with survival across and diversification following the KPB mass extinction event, WGD events, and the evolution of pierid butterflies. Our results suggest a number of ways in which our understanding of the history of CG Arabidopsis should be revised, notably that it evolved during a period of warming rather than cooling and that genome structure and developmental processes have been slower to evolve than has been appreciated. The evolutionary history of A. thaliana and its neighborhood described here will allow more precise application of our understanding of this model organism to other flowering plants, land plants, and all eukaryotes.
Materials and Methods
GenBank accession numbers, specifics of the evaluation of published fossils, and detailed methods can be found in SI Materials and Methods.
Phylogenetic Inference.
We used RAxML (48) to infer phylogeny in Brassicales from three alignments. The plastid ndhF alignment comprised 2,067 nucleotide sites representing 170 species; the nuclear PHYA alignment comprised 1,824 nucleotide sites representing 139 species; the combined alignment comprised 3,798 nucleotide sites from 177 species (Table S2). Analyses assumed a general time-reversible model of sequence evolution, with γ-distributed rate heterogeneity; partitions in the combined matrix were allowed to evolve independently. The topology and clade support in the resulting trees are consistent with previously published phylogenetic estimates using ndhF (17) and other plastid (49) and nuclear (3, 22, 50, 51) markers.
Ultrametric Tree and Divergence Date Estimation.
We calculated divergence dates for Brassicales from all three alignments using r8s v. 1.71 (30) and BEAST v. 1.5.3 (28). These methods account in different ways for variation in substitution rates among branches on the tree. We used r8s to explore the impact of placing the Dressiantha constraint at different nodes along the Brassicales backbone; potential placements were inferred in analyses of combined ndhF and morphological data (details are given in SI Materials and Methods). Because the placement of Dressiantha had little impact on age estimates within Core Brassicales (Table S3), we used a single placement in our BEAST analyses.
We allowed BEAST to infer topology, branch lengths, and dates for ndhF and combined data. For the PHYA data, we fixed the tree to that used in the r8s analyses and allowed BEAST to alter branch lengths while inferring dates (SI Materials and Methods). We used a uniform distribution for all three fossil calibrations with the lower hard bound of the distribution set to the youngest age of the fossil (see text, SI Materials and Methods, and Table S1) and the upper hard bound set to the first fossil record for eudicot pollen (125 Mya) (52) for D. bicarpellata and Akania sp. For other fossil calibrations the upper hard bound was determined by the age of other fossils used in the analysis (SI Materials and Methods). BEAST runs of 3 × 107 generations, saving data every 1,000 generations, produced 30,000 estimates of dates under a Yule speciation prior and an uncorrelated relaxed clock (28) for the single-gene datasets. Convergence statistics for each single-gene run were analyzed in Tracer, resulting in 27,000 post–burn-in trees. BEAST runs of 6 × 107 generations, saving data every 1,000 generations, produced 60,000 estimates under a Yule speciation (53) prior and uncorrelated relaxed clock (28) for the combined data. Also, for these data, we resampled at a lower frequency using LogCombiner v. 1.5.3 (28), resulting in a tree file with 30,000 trees. We used TreeAnnotator v. 1.5.3 (28) to produce maximum clade credibility trees from the post–burn-in trees and to determine the 95% probability density of ages for all nodes in the tree (Figs. S2–S4).
Shifts in Net Diversification Rate.
We used LASER v. 2.2 (33) to test for significant shifts in net diversification rates over the history of Brassicales. Clade diversity for each group was determined from the Angiosperm Phylogeny Website (54). We calculated the likelihoods of our phylogenetic data under a model of constant diversification rate over time and under a two-rate model in which the diversification rate is permitted to change (Table S5). Because analyses of diversification rate can be sensitive to extinction, we determined whether our results were sensitive to differences in extinction fraction by reanalyzing the data under low (a = 0) and high (a = 0.99) extinction (Table S5).
Supplementary Material
Acknowledgments
We thank Elisabeth Wheeler for her expertise in evaluating Capparidoxylon holleisii using Inside Wood (http://insidewood.lib.ncsu.edu), Aleksej Hvalj (Komorov Botanical Institute, Russian Academy of Sciences) for help identifying paleobotanical references from Russian scientific literature, Jocelyn Hall (University of Alberta, Edmonton, AB, Canada) for DNA material of Brassicales species, and Paul Forster (Queensland Herbarium, Brisbane, QLD, Australia) and David Woodlee (Burringbar Rainforest Nursery, Burringbar, NSW, Australia) for plant material of Akania bidwillii. This work was supported by a Mercer Postdoctoral Fellowship (to M.A.B.) awarded by the Arnold Arboretum of Harvard University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: All accession numbers for sequences downloaded from GenBank and sequences deposited in GenBank as part of this study appear in Table S2.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0909766107/-/DCSupplemental.
References
- 1.Berr A, et al. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 2006;48:771–783. doi: 10.1111/j.1365-313X.2006.02912.x. [DOI] [PubMed] [Google Scholar]
- 2.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 3.Koch M, Haubold B, Mitchell-Olds T. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 2001;88:534–544. [PubMed] [Google Scholar]
- 4.Sherman-Broyles S, et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2009;106:5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha M, Kim ED, Chen ZJ. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci USA. 2009;106:2295–2300. doi: 10.1073/pnas.0807350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Ziolkowski PA, Kaczmarek M, Babula D, Sadowski J. Genome evolution in Arabidopsis/Brassica: Conservation and divergence of ancient rearranged segments and their breakpoints. Plant J. 2006;47:63–74. doi: 10.1111/j.1365-313X.2006.02762.x. [DOI] [PubMed] [Google Scholar]
- 9.Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14:2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheat CW, et al. The genetic basis of a plant-insect coevolutionary key innovation. Proc Natl Acad Sci USA. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couvreur TLP, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- 14.Becker HF. Oligocene Plants from the Upper Ruby River Basin, Southwestern Montana. Vol. 82. Geological Society of America Memoir (Geological Society of America); 1961. pp. 1–127. [Google Scholar]
- 15.Wing SL. Eocene and Oligocene floras and vegetation of the Rocky Mountains. Ann Mo Bot Gard. 1987;74:748–784. [Google Scholar]
- 16.Manchester S, O'Leary E. Phylogenetic distribution and identification of fin-winged fruits. Bot Rev. 2010;76:1–82. [Google Scholar]
- 17.Beilstein MA, Al-Shehbaz IA, Kellogg EA. Brassicaceae phylogeny and trichome evolution. Am J Bot. 2006;93:607–619. doi: 10.3732/ajb.93.4.607. [DOI] [PubMed] [Google Scholar]
- 18.Appel O, Al-Shehbaz IA. Cruciferae. In: Kubitzki K, editor. Families and Genera of Vascular Plants. Vol. 5. Berlin: Springer; 2002. pp. 75–174. [Google Scholar]
- 19.Selmeier A. Capparidoxylon holleisii nov. spec., a silicified Capparis (Capparaceae) wood with insect coprolites from the Neogene of southern Germany. Zitteliana. 2005;45:199–209. [Google Scholar]
- 20.Gandolfo MA, Nixon KC, Crepet WL. A new fossil flower from the Turonian of New Jersey: Dressiantha bicarpellata gen. et sp. nov. (Capparales) Am J Bot. 1998;85:964–974. [PubMed] [Google Scholar]
- 21.De Craene LPR, Haston E. The systematic relationships of glucosinolate-producing plants and related families: A cladistic investigation based on morphological and molecular characters. Bot J Linn Soc. 2006;151:453–494. [Google Scholar]
- 22.Beilstein MA, Al-Shehbaz I, Mathews S, Kellogg E. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: Tribes and trichomes revisited. Am J Bot. 2008;95:1307–1327. doi: 10.3732/ajb.0800065. [DOI] [PubMed] [Google Scholar]
- 23.Ermolaeva MD, Wu M, Eisen JA, Salzberg SL. The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol. 2003;51:859–866. doi: 10.1023/a:1023001130337. [DOI] [PubMed] [Google Scholar]
- 24.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry Y, Bedhomme M, Blanc G. History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends Plant Sci. 2006;11:267–273. doi: 10.1016/j.tplants.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzke A, German D, Al-Shehbaz IA, Mummenhoff K. Arabidopsis family ties: Molecular phylogeny and age estimates in Brassicaceae. Taxon. 2009;58:425–437. [Google Scholar]
- 28.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 30.Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 31.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 32.Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinf Online. 2006:257–260. [PMC free article] [PubMed] [Google Scholar]
- 34.Montgelard C, Forty E, Arnal V, Matthee CA. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 2008;8:321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McElwain JC, Punyasena SW. Mass extinction events and the plant fossil record. Trends Ecol Evol. 2007;22:548–557. doi: 10.1016/j.tree.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Bowers JE, Chapman BA, Rong JK, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 37.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 38.Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 40.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 42.Rieseberg LH, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 44.Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin DA. Polyploidy and novelty in flowering plants. Am Nat. 1983;122:1–25. [Google Scholar]
- 46.Ehrlich P, Raven P. Butterflies and plants: A study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 47.Braby MF, Vila R, Pierce NE. Molecular phylogeny and systematics of the Pieridae (Lepidoptera: Papilionoidea): Higher classification and biogeography. Zool J Linn Soc. 2006;147:238–275. [Google Scholar]
- 48.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 49.Koch MA, et al. Supernetwork identifies multiple events of plastid trnF(GAA) pseudogene evolution in the Brassicaceae. Mol Biol Evol. 2007;24:63–73. doi: 10.1093/molbev/msl130. [DOI] [PubMed] [Google Scholar]
- 50.Bailey CD, et al. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- 51.Galloway GL, Malmberg RL, Price RA. Phylogenetic utility of the nuclear gene arginine decarboxylase: An example from Brassicaceae. Mol Biol Evol. 1998;15:1312–1320. doi: 10.1093/oxfordjournals.molbev.a025859. [DOI] [PubMed] [Google Scholar]
- 52.Brenner GJ. Evidence of the earliest stage of angiosperm pollen evolution: A paleoequatorial section from Israel. In: Taylor DW, Hickey LJ, editors. Flowering Plant Origin, Evolution and Phylogeny. New York: Chapman and Hall; 1996. pp. 91–115. [Google Scholar]
- 53.Yule GU. A mathematical theory of evolution based on the conclusions of Dr. J.C. Willis. Philos Trans R Soc Lond B Biol Sci. 1924;213:21–87. [Google Scholar]
- 54.The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.