Abstract
Platelet-derived growth factor (PDGF) is a broadly expressed mitogenic and chemotactic factor with diverse roles in a number of physiologic and pathologic settings. The zinc finger transcription factors Sp1, Sp3 and Egr-1 bind to overlapping elements in the proximal PDGF B-chain promoter and activate transcription of this gene. The anthracycline nogalamycin has previously been reported to inhibit the capacity of Egr-1 to bind DNA in vitro. Here we used electrophoretic mobility shift assays to show that nogalamycin added to cells in culture did not alter the interaction of Egr-1 with the PDGF-B promoter. Instead, it enhanced the capacity of Sp1 to bind DNA. Nogalamycin increased PDGF-B mRNA expression at the level of transcription, which was abrogated by mutation of the Sp1 binding site in the PDGF-B promoter or overexpression of mutant Sp1. Rather than increasing total levels of Sp1, nogalamycin altered the phosphorylation state of the transcription factor. Overexpression of dominant-negative PKC-ζ blocked nogalamycin-inducible Sp1 phosphorylation and PDGF-B promoter-dependent expression. Nogalamycin stimulated the phosphorylation of PKC-ζ (on residue Thr410). These findings demonstrate for the first time that PKC-ζ and Sp1 phosphorylation mediate the inducible expression of this growth factor.
INTRODUCTION
Platelet-derived growth factor (PDGF) is a 30 kDa dimer composed of an A- and/or B-chain, which are encoded by separate genes and regulated independently (1,2). PDGF has both mitogenic and chemoattractant properties and plays a functional role in both normal and pathological settings. PDGF has been implicated in wound healing (3), new bone formation (4), angiogenesis (5) and apoptosis (6). PDGF is also involved in neoplasia, atherosclerosis and inflammatory disease (7,8). Understanding the mechanisms regulating the expression of PDGF will provide valuable insights into strategies to control expression of the growth factor in these pathologic settings.
The minimal promoter for the human PDGF-B gene comprises ∼100 bp (9–11). A number of functionally important transcription factors have been shown to interact with distinct sites in this region of the PDGF-B promoter, including Sp1, Sp3 and Egr-1 (12). Sp1 was the first endogenous nuclear factor shown to bind the PDGF-B promoter ∼30 bp upstream of the TATA box. This specific interaction mediates basal PDGF-B gene expression in numerous cell types (13), including vascular endothelial cells (10) and smooth muscle cells (SMCs) (14). To date, there have been no reports implicating the structural modification of Sp1 as a mediator of increased PDGF expression.
Nogalamycin, an anthracycline isolated from Streptomyces nogalator var, binds to DNA and selectively inhibits DNA-directed RNA synthesis in vivo (15). Nogalamycin is composed of a planar aglycone, with a methyl ester and nalgose sugar at one end, and a positively charged bicyclo amino sugar at the other. These bulky substitutes on the aglycone confer a dumbell shape (16). Nogalamycin has been shown to penetrate the DNA double helix and form a stable intercalative complex (17) with slow association and dissociation rates (18). Anthracyclines readily localise in cytosol and nuclei after incubation with cultured cells (19). Nogalamycin has, in recent years, been employed as a DNA binding drug. Chiang et al. (20) used the Herpes simplex virus latency promoter to demonstrate that nogalamycin binds to GC-rich nucleotide sequences and prevents Egr-1 nucleoprotein complex formation when added to nuclear extracts. In this paper, we report that rather than down-regulating PDGF-B expression via Egr-1, nogalamycin, surprisingly, increased PDGF-B transcription by stimulating Sp1 phosphorylation through atypical protein kinase C-ζ (PKC-ζ). These findings demonstrate for the first time that the phosphorylation state of this nuclear protein regulates the inducible expression of PDGF.
MATERIALS AND METHODS
Cell culture
WKY12-22 pup rat aortic SMCs (21) were cultured in Waymouth’s MB752/1 medium (Life Technologies, Inc) supplemented with 10% fetal bovine serum, 30 µg/ml l-glutamine, 10 U/ml penicillin and 10 µg/ml streptomycin at 37°C and 5% CO2, and routinely passaged every 4–5 days in 75 cm2 flasks.
Transient cell transfection analysis
WKY12-22 cells were seeded in 100 mm tissue culture plates 48 h prior to transfection. When ∼60–70% confluent, the cells were transfected with 8 µg of the indicated promoter-reporter plasmids using FuGENE6 (Roche). A precipitate was formed using 3 µl of FuGENE6 per mg of transfected DNA, and the transfection mix made up to 1 ml with serum-free medium. After incubation at 22°C for 10 min, the DNA/FuGENE6 mixture was added to cells containing 4 ml of complete medium. Two days post-transfection cell lysates were prepared for assessment of CAT activity (14). The concentration of protein in the lysates was assessed using the BCA protein assay and used to correct CAT data. PDGF-B promoter-CAT constructs d26, d77 and d75 contain 153, 82 and 19 bp of promoter sequence upstream of TATA (10).
Oligonucleotide synthesis, purification and radiolabelling
Oligodeoxynucleotides were synthesised by Pacific Oligos (Lismore, Australia). Double-stranded oligonucleotides were radiolabelled with [γ-32P]dATP (Gene Works) using T4 polynucleotide kinase (NEB) and purified using Chromaspin-10 columns (Clontech).
Western blot analysis
Electrophoresis, transfer and immunodetection of Sp1, Sp3, Egr-1 and AP2 were performed using rabbit polyclonal antibodies obtained from Santa Cruz Biotechnology, as described previously (22).
Preparation of nuclear extracts
Cells in culture dishes were washed twice with PBS pH 7.4 at 4°C and detached by scraping. Cells were pelleted by centrifugation at 1200 r.p.m. for 10 min at 4°C. Pellets were resuspended in ice-cold PBS pH 7.4 and the resulting suspension transferred to Eppendorf tubes. Cells were repelleted in a microcentrifuge by spinning at 14 000 r.p.m. for 20 s at 4°C. The cells were lysed by resuspending the pellet in an ice-cold buffer A, which consists of 10 mM HEPES pH 8, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 200 mM sucrose, 0.5% Nonidet P-40, 1 mg/ml leupeptin and 1 mg/ml aprotinin. The samples were incubated on ice for 5 mins followed by recentrifugation. The nuclei were lysed in ice-cold buffer C, which consisted of 20 mM HEPES pH 8, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM PMSF, 1 mg/ml leupeptin and 1 mg/ml aprotinin. The cellular debris was removed by centrifugation, and the supernatant containing DNA binding proteins was combined with an equal volume of ice-cold buffer D (20 mM HEPES pH 8, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF, 1 mg/ml leupeptin and 1 mg/ml aprotinin). Nuclear extracts were stored at –80°C until use.
Electrophoretic mobility shift analysis
Gel shift reactions were performed in 20 µl of 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% glycerol, 1 µg salmon sperm DNA, 1 µg poly(dI-dC) poly(dI-dC), 32P-labelled oligonucleotide probe and 5 µg of nuclear extract (determined by BCA protein assay). The reaction was incubated for 35 min at 22°C. Bound complexes were separated from free probe by loading samples onto a 6% non-denaturing polyacrylamide gel and electrophoresed at 100 V for 3.5 h. The gels were vacuum dried at 80°C and subjected to autoradiography overnight at –80°C. For supershift analysis, the binding mixture was incubated with rabbit polyclonal antibodies to Sp1, Egr-1 or Sp3 (Santa Cruz Biotechnology) for 10 min prior to the addition of the probe. For competition analysis, the binding mixture was incubated with 100-fold molar excess of unlabelled oligonucleotide prior to the addition of the probe.
RT–PCR
Total RNA was prepared using the TRIzol reagent (Gibco BRL, Life Technologies) in accordance with manufacturer’s instructions. For the RT reaction 15 µg of RNA, 300 ng random primer (Promega) and 150 U of M-MLV reverse transcriptase (Stratagene) were combined in a total volume of 50 µl containing 50 mM Tris–HCl pH 8.3, 75 mM KCl, 3 mM MgCl2, 40 U RNasin and 0.5 mM of each dNTP. Reaction tubes were incubated at 65°C for 5 min and then tubes were cooled slowly to room temperature. PDGF-B amplification was performed as follows. One hundred picomoles of 5′ and 3′ PDGF-B primers were combined in a total volume of 100 µl containing 20 mM (NH4)2SO4, 75 mM Tris–HCl pH 9.0, 0.1% (w/v) Tween-20, 4.0 mM MgCl2 and 0.25 mM of each dNTP and the reverse transcribed cDNA. Samples were heated to 91°C for 4 min before the addition of 1 U Taq polymerase. The samples were cycled through 91°C for 1 min, 56°C for 1 min and 72°C for 1 min, followed by a further 3 min extension at 72°C to facilitate complete extension of the product. GAPDH amplification was performed to allow relative quantification of PCR products. GAPDH amplification was performed as follows: 100 pm of 5′ and 3′ primers were combined in a total volume of 20 µl containing 20 mM (NH4)2SO4, 75 mM Tris–HCl pH 9.0, 0.1% (w/v) Tween-20, 2.0 mM MgCl2 and 0.25 mM of each dNTP. The samples were cycled through 95°C for 10 s, 55°C for 30 s followed by 72°C for 1.5 min. Thirty cycles were followed by a further 4 min extension at 72°C. The PCR reaction was loaded onto 1% agarose gels, electrophoresed and stained with ethidium and photographed under ultraviolet illumination. Expected size products were GAPDH of 420 bp and PDGF-B of 317 bp.
RESULTS AND DISCUSSION
Nogalamycin stimulates PDGF-B expression
Egr-1 is a positive regulator of PDGF-B transcription in vascular endothelial cells (10) and SMCs (23). Based on the capacity of nogalamycin to block Egr-1 binding to DNA in vitro (20) we evaluated the effect of nogalamycin on the temporal expression of PDGF-B mRNA. Nogalamycin (10 µM) was incubated with WKY12-22 smooth muscle cells (14) for various times prior to extraction of total RNA and semi-quantitative RT–PCR analysis. PDGF-B mRNA was inducibly expressed within 5 h of exposure to nogalamycin and continued to increase over the next 21 h (Fig. 1A). In contrast, levels of GAPDH mRNA were unchanged over this time course (Fig. 1A).
Figure 1.
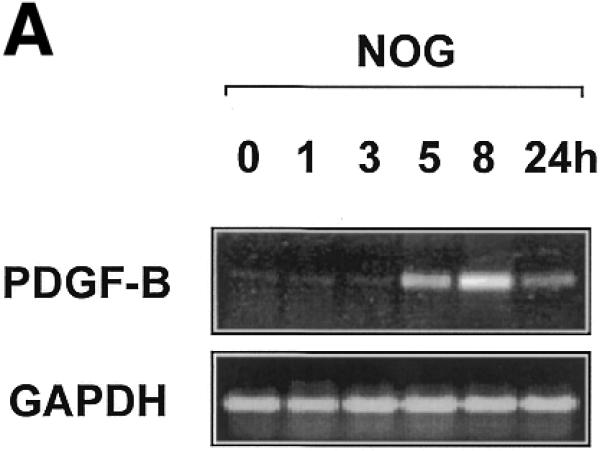
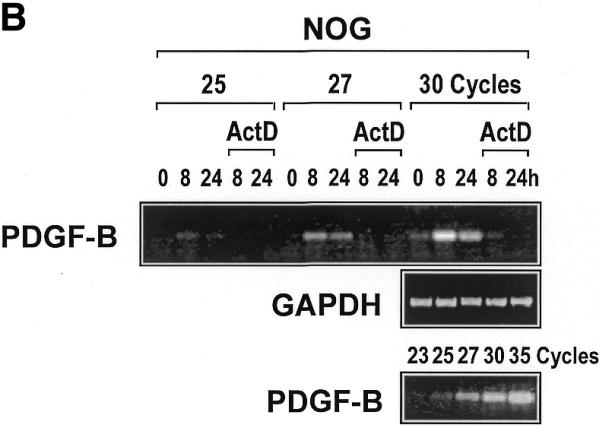
PDGF-B mRNA expression is induced in WKY12-22 cells exposed to nogalamycin. WKY12-22 cells were exposed to 10 µM nogalamycin for various times, and total RNA was prepared using TRIzol reagent according to the manufacturer’s instructions. The RNA was reverse-transcribed and the cDNA was used as the template for PCR. (A) Time-course of nogalamycin-inducible PDGF-B expression. (B) Actinomycin D pretreatment (10 µg/ml for 2 h) blocks PDGF-B expression inducible by nogalamycin. Cycle-based PDGF-B PCR (bottom panel) was performed on 8 h samples without actinomycin D preincubation. The data are representative of two independent determinations.
To demonstrate that nogalamycin-inducible PDGF-B mRNA expression is mediated at the level of transcription, we incubated the cells with the inhibitor of RNA polymerase II, actinomycin D, prior to exposure to nogalamycin. This prevented any increase in PDGF-B expression without affecting GAPDH expression (Fig. 1B), therefore indicating that nogalamycin stimulates new PDGF-B transcription.
Nogalamycin activates the PDGF-B promoter
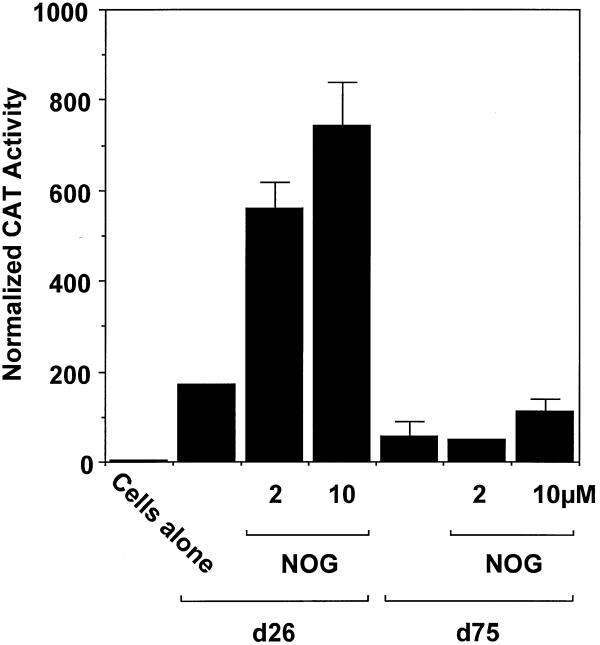
To confirm these observations and begin dissecting the mechanism of nogalamycin-inducible PDGF-B expression, WKY12-22 cells were transfected with d26 and d75, PDGF-B promoter–CAT reporter constructs containing 153 and 19 bp of PDGF-B promoter sequence relative to the TATA box, respectively (10). Basal CAT activity detectable in SMCs transfected with construct d26 (Fig. 2) exceeded reporter expression driven by d75, which lacks binding sites for Sp1, Sp3 and Egr-1 (Fig. 2). Incubation with nogalamycin (2 or 10 µM) increased CAT activity in a dose-dependent manner (Fig. 2). In contrast, the agonist had no significant effect on CAT activity driven by construct d75 (Fig. 2).
Figure 2.
Nogalamycin increases PDGF-B promoter-dependent gene transcription. WKY12-22 cells were transiently transfected with 8 µg of PDGF-B promoter–CAT construct d26 or d75 and CAT activity in the cell lysates was determined after 48 h. CAT activity was normalised to the concentration of protein in the lysates. Nogalamycin was added (2 and 10 µM final concentration) to the transfectants 24 h prior to harvesting. The data are representative of two independent determinations.
Zinc finger binding site in the PDGF-B promoter is crucial for inducible transcription upon exposure to nogalamycin
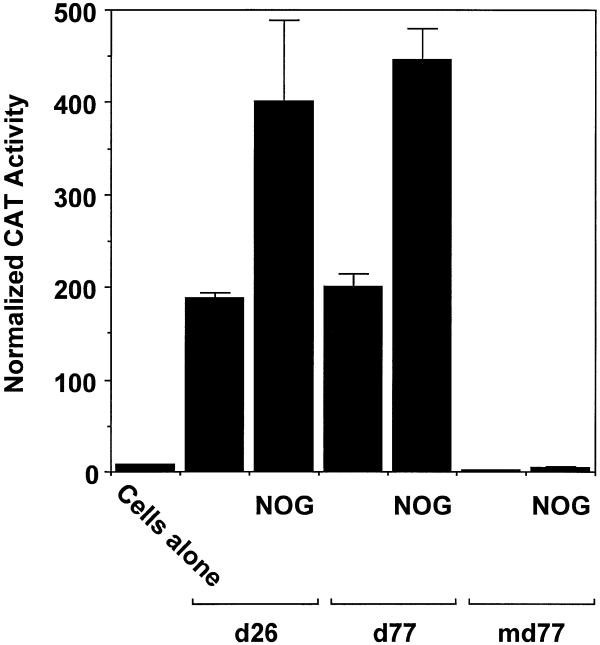
The preceding data demonstrated the capacity of 153 bp of proximal PDGF-B promoter sequence to mediate inducible PDGF-B transcription upon exposure to nogalamycin. To demonstrate the importance of the zinc finger binding site in nogalamycin-induced PDGF-B transcription, we transiently transfected WKY12-22 cells with a CAT reporter vector (md77) driven by a fragment of the PDGF-B promoter containing a mutation of the consensus binding elements recognised by Sp1, Sp3 and Egr-1. Basal CAT activity generated in cells transfected with construct d77, containing 82 bp of wild-type PDGF-B promoter sequence, was induced by nogalamycin (Fig. 3). In contrast, mutation of the zinc finger binding site in this fragment of the PDGF-B promoter abrogated both basal and nogalamycin-inducible CAT activity (Fig. 3). These data demonstrate the requirement of the proximal region of the PDGF-B promoter in nogalamycin-inducible PDGF-B transcription.
Figure 3.
The zinc finger binding site in the proximal PDGF-B promoter mediates nogalamycin-inducible PDGF-B promoter-dependent gene expression. WKY12-22 cells were transiently transfected with construct d26, d77 or md77. After 24 h, the transfectants were incubated with 10 µM of nogalamycin for a further 24 h prior to harvest. The cells were lysed and CAT activity was normalised to the concentration of protein. The data are representative of two independent determinations.
Nogalamycin increases Sp1 DNA binding
The ability of nogalamycin to increase transcription driven by d77, but not md77, prompted us to explore whether nogalamycin could increase DNA binding activity of Sp1, Egr-1 or Sp3. A 32P-labelled double-stranded oligonucleotide (Oligo A) bearing high-affinity consensus binding sites for Sp1, Sp3 and Egr-1 was incubated with nuclear extracts of WKY12-22 cells exposed to nogalamycin at various concentrations for 5 h. EMSA and supershift analysis revealed the formation of specific Sp1, Egr-1 and Sp3 DNA binding complexes (Fig. 4A), as previously observed (14). Nogalamycin stimulated the capacity of Sp1 to bind DNA at concentrations as low as 2 µM (Fig. 4A and B), whereas the intensity of the Egr-1 and Sp3 binding complexes were only slightly affected (Fig. 4A and B). Western blot analysis using these nuclear extracts showed that levels of an unrelated transcription factor AP2 were not affected by exposure to nogalamycin (Fig. 4C).
Figure 4.
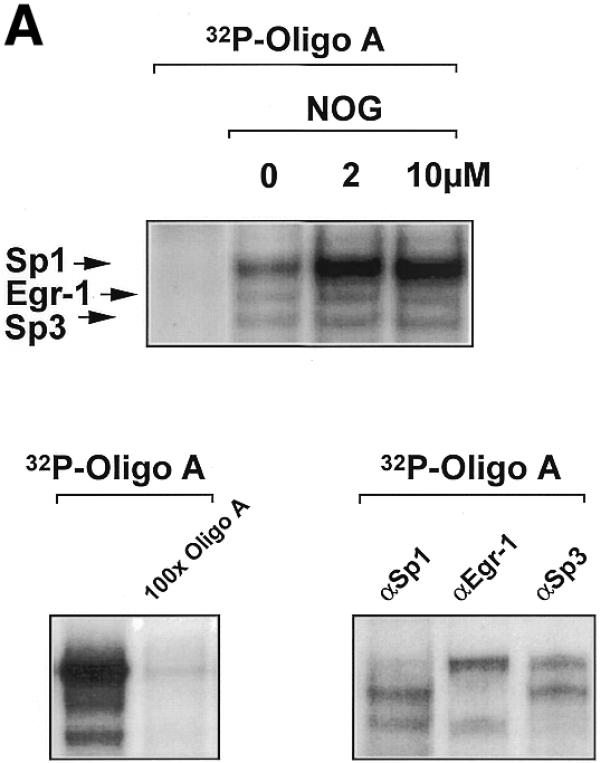
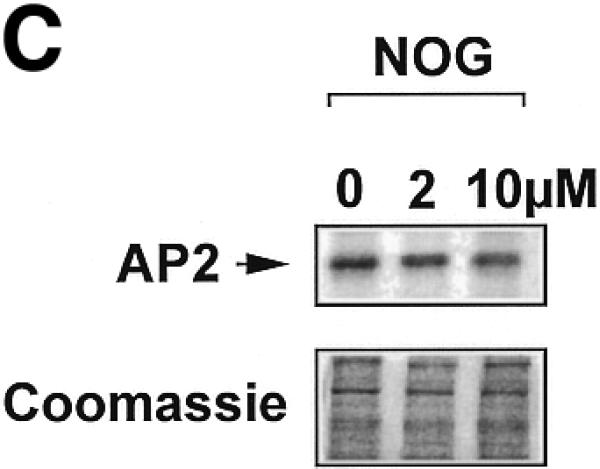
Nogalamycin increases Sp1 binding to a GC-rich oligonucleotide probe. 32P-labelled oligonucleotide bearing consensus elements for Sp1, Egr-1 and Sp3 (32P-Oligo A, 5′-GGG GGG GGC GGG GGC GGG GGC GGG GGA GG-3′, two overlapping consensus Egr-1 binding sites italicised). (A) EMSA was performed as described in Materials and Methods. Supershift analysis shows a distinct pattern of nucleoprotein complexes (Sp1, Egr-1, Sp3), whereas competition analysis demonstrates the specificity of these complexes. (B) Densitometric quantitation of band intensity from EMSA. (C) Western blot analysis for AP2 using the same extracts as those used for the EMSA. The data are representative of two independent determinations.
Enhanced PDGF-B transcription by nogalamycin is Sp1 dependent
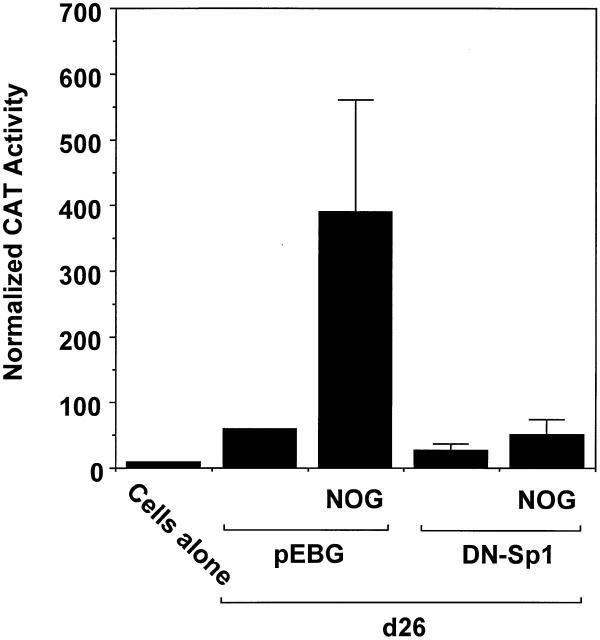
The preceding data demonstrate that Sp1 (but not Sp3 or Egr-1) DNA binding increased upon exposure to nogalamycin. To demonstrate the functional involvement of Sp1 in nogalamycin-induced PDGF-B transcription, we overexpressed a dominant-negative (DN) mutant form of Sp1 (pEBG-Sp1), containing the DNA binding domain of human Sp1 (24), in a transient transfection setting with construct d26. CAT activity generated by construct d26 cotransfected with the empty expression vector increased several-fold in cells exposed to nogalamycin (Fig. 5). Overexpression of mutant Sp1 blocked nogalamycin-induced PDGF-B promoter-dependent gene expression (Fig. 5). These results thus demonstrate the requirement of Sp1 for nogalamycin-induced PDGF-B transcription.
Figure 5.
PDGF-B transcription inducible by nogalamycin is Sp1-dependent. WKY12-22 cells were cotransfected with 8 µg of d26 and 5 µg of either pEBG or pEBG-DN Sp1. After 24 h, 10 µM nogalamycin was added to transfectants. CAT activity in the lysates was determined 48 h after transfection and normalised to the concentration of protein. The data are representative of two independent determinations.
Nogalamycin modulates Sp1 DNA binding to the PDGF-B promoter
We next tested whether nogalamycin modulates the direct interaction of Sp1 with the PDGF-B promoter. A 32P-labelled double-stranded oligonucleotide spanning the –30/–13 region of the PDGF-B promoter was incubated with nuclear extracts of WKY12-22 cells exposed to nogalamycin (10 µM) for various times up to 24 h. EMSA revealed the inducible formation of a distinct Sp1 DNA binding complex (14) within 3 h which was still apparent after 24 h (Fig. 6A). The temporal pattern of increased Sp1 binding precedes the earliest detection of inducible PDGF-B expression (Fig. 1). Further experiments revealed that Sp1 binding is induced by nogalamycin in a dose-dependent manner at concentrations as low as 2 µM (Fig. 6B). We next investigated whether the increased capacity of Sp1 to bind the PDGF-B promoter was due to elevated levels of nuclear Sp1 or chemical modification of the transcription factor.
Figure 6.
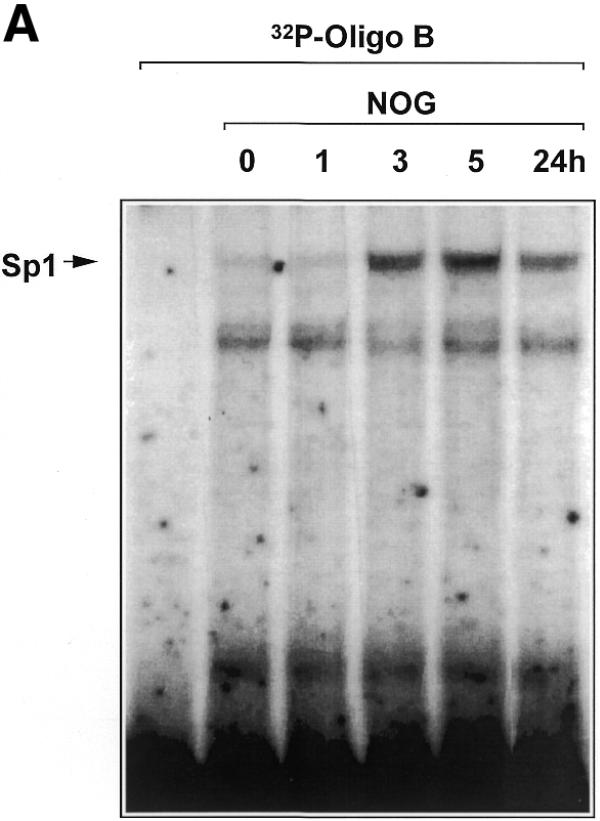

Nogalamycin increases Sp1 DNA binding to the proximal PDGF-B promoter. (A) EMSA was performed using nuclear extracts of WKY12-22 cells exposed to 10 µM nogalamycin for the indicated times and 32P-labelled Oligo B. Binding reactions and non-denaturing gel electrophoresis were performed as described in Materials and Methods. (B) Nogalamycin increases Sp1 DNA binding in a dose-dependent manner. Nuclear extracts from WKY12-22 cells exposed to various concentrations of nogalamycin for 5 h were incubated with 32P-labelled Oligo B (5′-GCT GTC TCC ACC CAC CTC TCG CAC TCT-3′). Binding reactions and non-denaturing gel electrophoresis were performed as described in Materials and Methods. Nucleoprotein complexes were visualised by autoradiography. The data are representative of two independent determinations.
Nogalamycin stimulates Sp1 phosphorylation
WKY12-22 cells were exposed to nogalamycin for various times and nuclear extracts were analysed for Sp1 immunoreactivity by western blot analysis. Sp1 migrates on an SDS–polyacrylamide gel as two polypeptide species with apparent molecular masses of 95 and 105 kDa, which result from differential post-translational modification of the polypeptide (25,26); the 105 kDa species represents the phosphorylated form of Sp1 (27). If the agonist modulates the phosphorylation state of Sp1, the relative intensity of the 105 kDa species should increase. If, on the other hand, nogalamycin increased total levels of Sp1, the overall intensity of immunoreactive Sp1 should increase. Time-course studies indicate that while total Sp1 levels remained relatively unchanged, there was a clear transition in the phosphorylation state of Sp1 upon nogalamycin treatment as early as 3 h (Fig. 7). To further demonstrate that the 105 kDa band represents the phosphorylated form of Sp1, nuclear extracts were exposed to calf intestinal phosphatase (CIP) for 30 min prior to western blot analysis. The upper band was completely abrogated by CIP treatment (Fig. 7).
Figure 7.
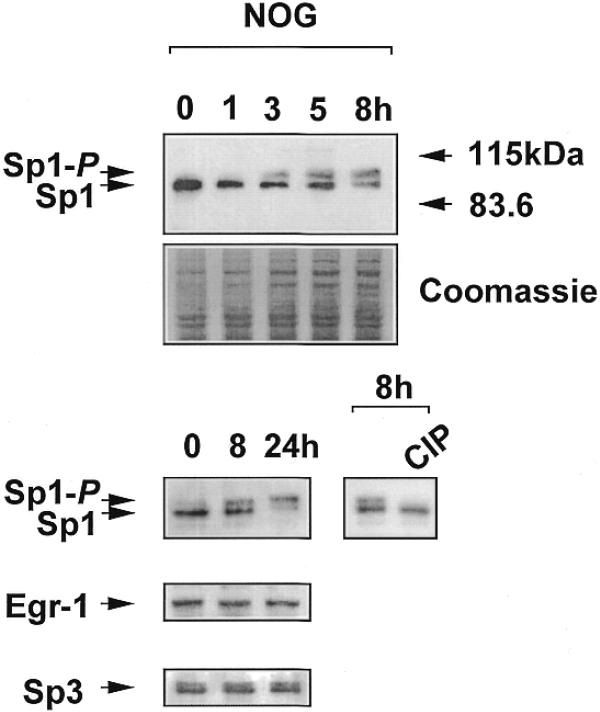
Nogalamycin phosphorylates endogenous Sp1 but not Egr-1 or Sp3. Nuclear extracts from WKY12-22 cells exposed to 10 µM nogalamycin for various times were resolved by electrophoresis prior to transfer to PVDF membranes and exposure to polyclonal antibodies to Sp1, Sp3 or Egr-1. Where indicated, the 8 h sample was incubated with CIP prior to western blot analysis for Sp1. The data are representative of two independent determinations.
Since Sp3 and Egr-1 also bind to the –30/–13 region of the PDGF-B promoter we next explored the effect of nogalamycin on the electrophoretic migration of these transcription factors by western blot analysis using the same extracts and appropriate antibodies. Unlike Sp1, the phosphorylation state of Sp3 and Egr-1 was unaltered by incubation with nogalamycin (Fig. 7). Furthermore, total levels of Sp3 and Egr-1 protein were unchanged by the agonist (Fig. 7).
PKC-ζ modulates PDGF-B transcription
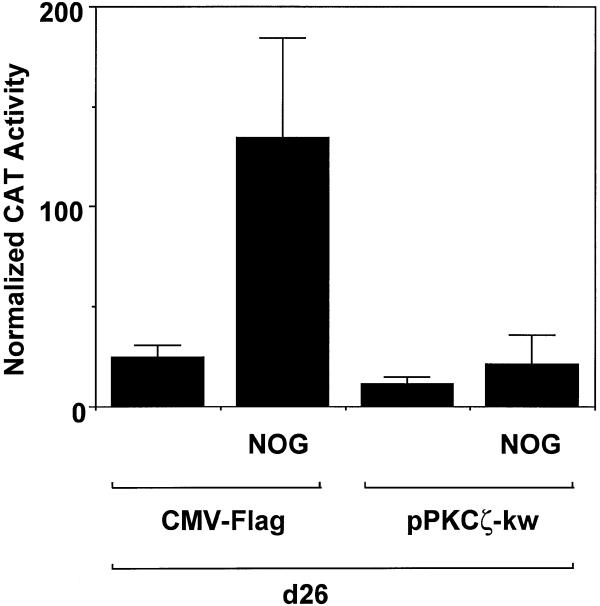
PKC-ζ is an atypical member of the PKC superfamily (28) recently found to interact directly and specifically with Sp1 and phosphorylate this transcription factor (29). To determine whether this protein kinase is involved in inducible PDGF-B promoter-dependent expression, we forced the expression of a kinase inactive mutant form of PKC-ζ (PKC-ζ-kw, lysine275 to tryptophan substitution) together with construct d26. Nogalamycin, as expected, induced reporter activity in cells transfected with the empty expression vector (Fig. 8). In contrast, the DN PKC-ζ virtually abolished PDGF-B promoter activation by the agonist (Fig. 8).
Figure 8.
Nogalamycin activation of the PDGF-B promoter is PKC-ζ-dependent. WKY12-22 cells were cotransfected with 8 µg of d26 and 5 µg of either CMV-Flag or pPKC-ζ-kw. Nogalamycin (10 µM) was added to the transfectants 24 h post-transfection prior to harvest 24 h later. CAT activity was normalised to the concentration of protein in the extracts. The data are representative of two independent determinations.
To provide direct evidence for the involvement of PKC-ζ in Sp1 phosphorylation following exposure to nogalamycin, we overexpressed pPKC-ζ-kw or its backbone control in WKY12-22 cells to demonstrate whether pPKC-ζ-kw could block Sp1 phosphorylation. Sp1 phosphorylation by an anthracycline has hitherto not been reported. Findings from immunoblot analysis revealed that pPKC-ζ-kw abrogated the phosphorylated form of Sp1 in cells incubated with nogalamycin. In contrast, Sp1 was still phosphorylated in backbone transfectants incubated with nogalamycin (Fig. 9). To demonstrate PKC-ζ activation by nogalamycin, we performed a western blot analysis using antibodies specifically recognising the activated form (phospho-Thr410) of PKC-ζ (30,31). Nogalamycin stimulated PKC-ζ phosphorylation in a transient manner (Fig. 10); PKC-ζ activation was apparent within 15 min but was no longer detectable at 3 h (Fig. 10). These data, taken together, demonstrate for the first time the involvement of PKC-ζ in inducible PDGF-B transcription and establish Sp1 phosphorylation as a positive regulatory event in the increased expression of this gene.
Figure 9.

DN PKC-ζ ablates Sp1 phosphorylation in WKY12-22 cells. Cells were transfected with 30 µg of CMV-Flag or pPKC-ζ-kw 24 h prior to the addition of nogalamycin (2 µM). Nuclear extracts were prepared 24 h later. Nuclear extracts were resolved by electrophoresis prior to transfer to PVDF membranes and exposure to polyclonal Sp1 antibodies. The data are representative of two independent determinations.
Figure 10.
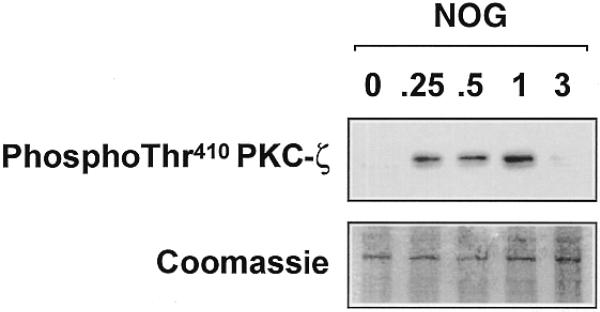
Nogalamycin stimulates PKC-ζ (Thr410) phosphorylation in WKY12-22 cells. Cells were exposed to 10 µM of nogalamycin for various times prior to resolution of the cytoplasmic extracts by electrophoresis and western blot analysis using antibodies to the Thr410 phosphorylated form of PKC-ζ. The data are representative of two independent determinations.
In this paper we report the positive influence of Sp1 phosphorylation on PDGF-B expression and the involvement of PKC-ζ in this process. RT–PCR and transient transfection analysis revealed that an agent capable of phosphorylating Sp1 in intact cells increased PDGF-B transcription and mRNA expression. Nogalamycin activation of the PDGF-B promoter was abrogated by mutation of the Sp1 binding site in the PDGF-B promoter or by overexpression of DN mutants of Sp1 or PKC-ζ. These studies demonstrate that Sp1 phosphorylation is a key regulatory process mediating inducible PDGF-B expression.
Sp1 phosphorylation has been linked to the constitutive and inducible expression of a diverse set of mammalian genes. For example, Lin et al. (32) correlated increased Sp1 hyperphosphorylation and transcriptional activity with inducible tissue factor gene expression. Also, Alroy et al. (33) determined that epidermal growth factor-like Neu differentiation factors stimulate Sp1 phosphorylation and activity and the inducible transcription of genes encoding specific subunits of the acetylcholine receptor. Finally, Ray et al. (34) found that serum amyloid A gene expression involves phosphorylation of both Sp1 and amyloid A-activating sequence binding factor. Chun et al. (35) reported that Sp1 and HIV type 1 Tat form a protein–protein complex mediating Sp1 phosphorylation by DNA-dependent protein kinase (35,36). Our present demonstration that PDGF-B transcription is stimulated through Sp1 phosphorylation is the first report of the inducible expression of PDGF as a function of the phosphorylation state of a transcriptional regulator.
Currently it is controversial whether Sp1 phosphorylation increases or decreases the capacity of Sp1 to bind DNA. Armstrong et al. (37), for example, showed that casein kinase II-mediated phosphorylation of the C-terminus of Sp1 decreases its DNA binding activity. Conversely, phosphorylation of Sp1 by ERK-2 stimulates DNA binding (38). Kumar and Butler (39) demonstrated a correlation between phosphorylation of Sp1 and increased DNA binding activity. Additionally, Merchant et al. (38) showed that ERK-2-induced Sp1 phosphorylation enhancing its capacity to bind DNA and induce gastrin promoter activity. These latter findings support the present data showing increased DNA binding and transcription as a consequence of Sp1 phosphorylation, a process mediated by PKC-ζ and a distinct element in the proximal PDGF-B promoter.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Alex Toker and Debabrata Mukhopadhyay (Beth Israel Deaconess Medical Center, Boston) for PKC-ζ phospho-Thr410 antibody and DN PKC-ζ plasmid, respectively, Dr Tucker Collins (Brigham and Women’s Hospital, Boston) for PDGF-B–CAT constructs, Dr Gerald Thiel (Institute for Genetics, University of Cologne) for the DN Sp1 plasmid, and Dr Trevor Biden (Garvan Institute, Sydney) for helpful advice. This work was supported by grants from the National Health and Medical Research Council of Australia and NSW Department of Health. L.A.R. was supported by an Australian Postgraduate Research Award. L.M.K. is a Research Fellow of the National Health and Medical Research Council of Australia.
References
- 1.Antoniades H.N., Scher,C.D. and Stiles,C.D. (1979) Purification of platelet-derived growth factor. J. Biol. Chem., 76, 1809–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deuel T.F., Huang,J.S., Proffit,R.T., Baenzinger,J.U., Chang,D. and Kennedy,B.B. (1981) Human platelet-derived growth factor: purification and resolution into two active protein fractions. J. Biol. Chem., 256, 8896–8899. [PubMed] [Google Scholar]
- 3.Soma Y., Takehara,K. and Ishibashi,Y. (1994) Alteration of the chemotactic response of human skin fibroblasts to PDGF by growth factors. Exp. Cell Res., 212, 274–277. [DOI] [PubMed] [Google Scholar]
- 4.Rydziel S., Ladd,C., McCarthy,T.L., Centrella,M. and Canalis,E. (1992) Determination and expression of platelet-derived growth factor-AA in bone cell cultures. Endocrinology, 130, 1916–1922. [DOI] [PubMed] [Google Scholar]
- 5.Thommen R., Humar,R., Misevic,G., Pepper,M.S., Hahn,A.W., John,M. and Battegay,E.J. (1997) PDGF-BB increases endothelial migration on cord movements during angiogenesis in vitro. J. Cell. Biochem., 64, 403–413. [PubMed] [Google Scholar]
- 6.Okura T., Igase,M., Kitami,Y., Fukuoka,T., Maguchi,M., Kohara,K. and Hiwada,K. (1998) Platelet-derived growth factor induces apoptosis in vascular smooth muscle cells: roles of the Bcl-2 family. Biochim. Biophys. Acta, 1403, 245–253. [DOI] [PubMed] [Google Scholar]
- 7.Raines E.W., Bowen-Pope,D.F. and Ross,R. (1990) In Sporn,M.B. and Roberts,A.B. (eds), Handbook of Experimental Pharmacology: Peptide Growth Factors and their Receptors. Springer-Verlag, Berlin, pp. 173–262.
- 8.Ross R. (1993) The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature, 362, 801–809. [DOI] [PubMed] [Google Scholar]
- 9.Pech M., Rao,C.D., Robbins,K.C. and Aaronson,S.A. (1989) Functional identification of regulatory elements within the promoter region of platelet-derived growth factor 2. Mol. Cell. Biol., 9, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khachigian L.M., Fries,J.W.U., Benz,M.W., Bonthron,D.T. and Collins,T. (1994) Novel cis-acting elements in the human platelet-derived growth factor B-chain core promoter that mediate gene expression in cultured vascular endothelial cells. J. Biol. Chem., 269, 22647–22656. [PubMed] [Google Scholar]
- 11.Jin H.-M., Robinson,D.F., Liang,Y. and Fahl,W.E. (1994) SIS/PDGF-B promoter isolation and characterization of regulatory elements necessary for basal expression of the SIS/PDGF-B gene in U2-OS osteosarcoma cells. J. Biol. Chem., 269, 28648–28654. [PubMed] [Google Scholar]
- 12.Khachigian L.M. and Collins,T. (1997) Inducible expression of Egr-1-dependent genes: a paradigm of transcriptional activation in vascular endothelium. Circ. Res., 81, 457–461. [DOI] [PubMed] [Google Scholar]
- 13.Liang Y., Robinson,D.F., Dennig,J., Suske,G. and Fahl,W.E. (1996) Transcriptional regulation of the SIS/PDGF-B gene in human osteosarcoma cells by the Sp family of transcription factors. J. Biol. Chem., 271, 11792–11797. [DOI] [PubMed] [Google Scholar]
- 14.Rafty L.A. and Khachigian,L.M. (1998) Zinc finger transcription factors mediate high constitutive PDGF-B expression in smooth muscle cells derived from aortae of newborn rats. J. Biol. Chem., 273, 5758–5764. [DOI] [PubMed] [Google Scholar]
- 15.Fok J. and Waring,M. (1972) Breakdown of pulse-labeled ribonucleic acid in Bacillus megaterium, revealed by exposure to the antibiotics mithramycin, chromomycin and nogalamycin. Mol. Pharmacol., 8, 65–74. [PubMed] [Google Scholar]
- 16.Williams L.D., Egli,M., Qi,G., Bash,P., van der Marel,G.A., van Boom,J.H., Rich,A. and Frederick,C.A. (1990) Structure of nogalamycin bound to a DNA hexamer. Proc. Natl Acad. Sci. USA, 87, 2225–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das G.C., Dasgupta,S. and Das Gupta,N.N. (1974) Interaction of nogalamycin with DNA. Biochim. Biophys. Acta, 353, 274–282. [DOI] [PubMed] [Google Scholar]
- 18.Fox K.R., Brassett,C. and Waring,M.J. (1985) Kinetics of dissociation of nogalamycin from DNA: comparison with other anthracycline antibiotics. Biochim. Biophys. Acta, 840, 383–392. [DOI] [PubMed] [Google Scholar]
- 19.Wassermann K. and Steiness,E. (1986) Doxorubicin binds in a cooperative manner to myocardial cells: two binding sites. Cancer Chemother. Pharmacol., 17, 241–246. [DOI] [PubMed] [Google Scholar]
- 20.Chiang S.-Y., Welch,J.J., Rauscher,F.J.,III and Beerman,T.A. (1996) Effect of DNA-binding drugs on early growth response factor-1 and TATA box-binding protein complex formation with the Herpes simplex virus latency promoter. J. Biol. Chem., 271, 23999–24004. [DOI] [PubMed] [Google Scholar]
- 21.Lemire J.M., Covin,C.W., White,S., Giachelli,C.M. and Schwartz,S.M. (1994) Characterization of cloned aortic smooth muscle cells from young rats. Am. J. Pathol., 144, 1068–1081. [PMC free article] [PubMed] [Google Scholar]
- 22.Khachigian L.M., Santiago,F.S., Rafty,L.A., Chan,O.L.W., Delbridge,G.J., Bobik,A., Collins,T. and Johnson,A.C. (1999) GC factor 2 represses platelet-derived growth factor A-chain transcription and is itself induced by arterial injury. Circ. Res., 84, 1258–1267. [DOI] [PubMed] [Google Scholar]
- 23.Silverman E.S., Khachigian,L.M., Lindner,V., Williams,A.J. and Collins,T. (1997) Inducible PDGF A-chain transcription in vascular smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am. J. Physiol., 42, H1415–H1426. [DOI] [PubMed] [Google Scholar]
- 24.Petersohn D. and Thiel,G. (1996) Role of zinc-finger proteins Sp1 and zif268/egr-1 in transcriptional regulation of the human synaptobrevin II gene. Eur. J. Biochem., 239, 827–834. [DOI] [PubMed] [Google Scholar]
- 25.Kadonaga J.T., Carner,K.R., Masiarz,F.R. and Tjian,R. (1987) Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell, 51, 1079–1090. [DOI] [PubMed] [Google Scholar]
- 26.Jackson S.P. and Tjian,R. (1988) O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell, 55, 125–133. [DOI] [PubMed] [Google Scholar]
- 27.Jackson S.P., MacDonald,J.J., Lees-Miller,S. and Tjian,R. (1990) GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell, 5, 155–165. [DOI] [PubMed] [Google Scholar]
- 28.Hug H. and Sarre,T.F. (1993) Protein kinase C isoenzymes: divergence in signal transduction? Biochem. J., 291, 329–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal S., Claffey,K.P., Cohen,H.T. and Mukhopadhyay,D. (1998) Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C zeta. J. Biol. Chem., 273, 26277–26280. [DOI] [PubMed] [Google Scholar]
- 30.Standaert M.L., Bandyopadhyay,G., Perez,L., Price,L., Galloway,L., Poklepovic,A., Sajan,M., Cenni,V., Sirri,A., Moscat,J., Toker,A. and Farese,R.V. (1999) Insulin activates protein kinases C by an autophosphorylation-dependent mechanism and stimulates their translocation to GLUT4 vesicles and other membrane fractions in rat adipocytes. J. Biol. Chem., 274, 25308–25316. [DOI] [PubMed] [Google Scholar]
- 31.Kanoh Y., Bandyopadhyay,G., Sajan,M.P., Stanaert,M.L. and Farese,R.V. (2000) Thiazolidinedione treatment enhances insulin effects on protein kinase C activation and glucose transport in adipocytes of nondiabetic and Goto-Kikizaki type II diabetic rats. J. Biol. Chem., 275, 16690–16696. [DOI] [PubMed] [Google Scholar]
- 32.Lin M.-C., Almus-Jacobs,F., Chen,H.-H., Parry,G.C.N., Mackmann,N., Shyy,J.Y. and Chen,S. (1997) Shear stress induction of the tissue factor gene. J. Clin. Invest., 99, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alroy I., Soussan,L., Seger,R. and Yarden,Y. (1999) Neu differentiation factor stimulates phosphorylation and activation of the Sp1 transcription factor. Mol. Cell. Biol., 19, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray A., Schatten,H. and Ray,B.K. (1999) Activation of Sp1 and its functional co-operation with serum amyloid A-activating sequence binding factor in synoviocyte cells trigger synergistic action of interleukin-1 and interleukin-6 in serum amyloid A gene expression. J. Biol. Chem., 274, 4300–4308. [DOI] [PubMed] [Google Scholar]
- 35.Chun R.F., Semmes,O.J., Neuveut,C. and Jeang,K.T. (1998) Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol., 72, 2615–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottlieb T.M. and Jackson,P. (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 37.Armstrong S.A., Barry,D.A., Leggett,R.W. and Mueller,C.R. (1997) Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem., 272, 13489–13495. [DOI] [PubMed] [Google Scholar]
- 38.Merchant J.L., Du,M. and Todisco,A. (1999) Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem. Biophys. Res. Commun., 254, 454–461. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A.P. and Butler,A.P. (1998) Serum responsive gene expression mediated by Sp1. Biochem. Biophys. Res. Commun., 252, 517–523. [DOI] [PubMed] [Google Scholar]