Abstract
Ets factors play a critical role in oncogenic Ras- and growth factor-mediated regulation of the proximal rat prolactin (rPRL) promoter in pituitary cells. The rPRL promoter contains two key functional Ets binding sites (EBS): a composite EBS/Pit-1 element located at –212 and an EBS that co-localizes with the basal transcription element (BTE, or A-site) located at –96. Oncogenic Ras exclusively signals to the –212 site, which we have named the Ras response element (RRE); whereas the response of multiple growth factors (FGFs, EGF, IGF, insulin and TRH) maps to both EBSs. Although Ets-1 and GA binding protein (GABP) have been implicated in the Ras and insulin responses, respectively, the precise identity of the pituitary Ets factors that specifically bind to the RRE and BTE sites remains unknown. In order to identify the Ets factor(s) present in GH4 and GH3 nuclear extracts (GH4NE and GH3NE) that bind to the EBSs contained in the RRE and BTE, we used EBS-RRE and BTE oligonucleotides in electrophoretic mobility shift assays (EMSAs), antibody supershift assays, western blot analysis of partially purified fractions and UV-crosslinking studies. EMSAs, using either the BTE or EBS-RRE probes, identified a specific protein–DNA complex, designated complex A, which contains an Ets factor as determined by oligonucleotide competition studies. Using western blot analysis of GH3 nuclear proteins that bind to heparin–Sepharose, we have shown that Ets-1 and GABP, which are MAP kinase substrates, co-purify with complex A, and supershift analysis with specific antisera revealed that complex A contains Ets-1, GABPα and GABPβ1. In addition, we show that recombinant full-length Ets-1 binds equivalently to BTE and EBS-RRE probes, while recombinant GABPα/β preferentially binds to the BTE probe. Furthermore, comparing the DNA binding of GH4NE containing both Ets-1 and GABP and HeLa nuclear extracts devoid of Ets-1 but containing GABP, we were able to show that the EBS-RRE preferentially binds Ets-1, while the BTE binds both GABP and Ets-1. Finally, UV-crosslinking experiments with radiolabeled EBS-RRE and BTE oligonucleotides showed that these probes specifically bind to a protein of ∼64 kDa, which is consistent with binding to Ets-1 (54 kDa) and/or the DNA binding subunit of GABP, GABPα (57 kDa). These studies show that endogenous, pituitary-derived GABP and Ets-1 bind to the BTE, whereas Ets-1 preferentially binds to the EBS-RRE. Taken together, these data provide important insights into the mechanisms by which the combination of distinct Ets members and EBSs transduce differential growth factor responses.
INTRODUCTION
The Ets family of transcription factors comprises more than 30 members, which play important roles in a variety of biological responses, including cell proliferation, differentiation, development and apoptosis (1). Ets family members are characterized by an 85 amino acid, winged helix–turn–helix DNA binding domain that is highly conserved from Drosophila to humans (2,3). The conserved Ets domain recognizes a core 5′-GGA(A/T)-3′ DNA element and sequences flanking this core contribute to binding specificity (1,4). Further specificity occurs through tissue-specific expression of Ets genes and through interactions of Ets proteins with co-factors at adjacent DNA elements (1). However, due to overlapping DNA binding specificities and the ubiquitous expression of some Ets factors, it has been difficult to identify the precise function of individual Ets proteins.
Several members of the Ets family of transcription factors are important nuclear targets of various growth factor signaling pathways via MAP kinase. Elk-1 and the Drosophila Pointed P2 protein were among the first Ets factors shown to be targets of the MAP kinase pathway (5–8). Further studies have shown that members of at least six subfamilies of Ets factors contain this highly conserved Pointed domain and are MAP kinase targets (9). Phosphorylation by MAP kinase has been shown to enhance the transcriptional activity of certain Ets proteins and to increase their specificity (1,9).
Regulation of the rat prolactin (rPRL) promoter in GH4 rat pituitary cells provides a well-defined and physiologically relevant model system to study the regulation of tissue-specific gene expression. GH4 cells are a highly differentiated cell line which retain cell-specific functions, normal hormonal responses and express the phenotypic marker, PRL. The developmental and cell-type-specific expression of the PRL gene is under the control of the pituitary-specific POU homeodomain protein, Pit-1 (10). The rPRL promoter contains several consensus binding sites for the Ets family of transcription factors, and these sites have been shown to be the targets for multiple signaling pathways mediated by MAP kinase. The Ets binding site (EBS) at –212 is part of a previously identified composite Ets-1/Pit-1 Ras response element (RRE) (11–13) that is also targeted by fibroblast growth factor (FGF) (14) and thyrotropin-releasing hormone (TRH) (15). The more proximal EBS centered at –96 co-localizes with a previously identified basal transcription element (BTE) (16), which is a target of several growth factor pathways, including FGF-2 and -4 (14), epidermal growth factor (EGF) (17), insulin (18,19), insulin-like growth factor-1 (IGF-1) (20) and TRH (15).
The precise identities of the endogenous, pituitary-derived Ets factors that bind to the RRE and BTE Ets sites in a functionally relevant manner remain unknown. Based on our previous reports, it is clear that the EBS-RRE can bind to the DNA binding domain of recombinant Ets-2; that GH4 cells contain an Ets factor (or factors) which can bind to the RRE; that GH4 cells contain Ets-1; and that the Ras response via the RRE is enhanced selectively by chicken Ets-1 (12). While these data strongly support Ets-1 as the key nuclear effector of the Ras response via the RRE, it has not yet been shown that endogenous Ets-1 is the specific Ets member that binds to the RRE in vivo and transduces the Ras response. Similarly, the endogenous pituitary Ets factor that binds to the BTE has not been identified. Recombinant Ets proteins GA binding protein α (GABPα), Elk-1 and SAP-1 can bind to the BTE in vitro (18,19); however, transfected GABPα and its partner, the ankyrin repeat protein GABPβ, block the insulin response (19). Thus, the role of GABP at the BTE remains unclear. Indeed, expression of Ets-1 and Pit-1, which enhance the Ras response via the RRE (11–13), actually inhibit the FGF response (14), suggesting that the RRE and BTE utilize distinct Ets factors to elicit the Ras and FGF responses. Moreover, a rigorous approach characterizing the pituitary cell-derived Ets factors that bind to the RRE and the BTE has not been reported.
In this study, we have characterized the Ets factors derived from GH4 or GH3 pituitary cells that bind to the Ets sites of the BTE and RRE of the rPRL promoter. Using the EMSA, we show that both EBSs form a similar protein–DNA complex (complex A) with GH4 and GH3 nuclear extracts (GH4NE and GH3NE), and oligonucleotide competition studies indicate that complex A contains an Ets factor. Super-shift analysis with partially purified GH3NE and the BTE and EBS-RRE probes show that complex A contains GABP and Ets-1. Gel shifts with recombinant Ets-1 and GABPα/β show that Ets-1 binds equivalently to both the BTE and EBS-RRE probes while GABP preferentially binds to the BTE probe. Similarly, using HeLa nuclear extracts devoid of Ets-1 but containing GABP, we were able to show that the EBS-RRE preferentially binds Ets-1, while the BTE binds both GABP and Ets-1. UV-crosslinking studies show that both probes bind to a protein of ∼64 kDa, which, allowing for the additional mass of the cross-linked probe, is consistent with the reported masses for both Ets-1 and GABPα. Taken together, the data show that the BTE preferentially binds to GABP and the EBS-RRE preferentially binds to Ets-1. Thus, this system provides an ideal model to study the regulation of transcription by Ets factors and the mechanism by which Ets factors achieve specific transcriptional responses transduced by different signaling pathways.
MATERIALS AND METHODS
Cell culture
Adherent GH4T2 rat pituitary tumor cells and HeLa human cervical carcinoma cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Grand Island, NY) supplemented with 12.5% horse serum and 2.5% fetal calf serum. Cells were maintained at 37°C in 5% CO2. GH3 rat pituitary cells were grown in spinner culture at 37°C in high glucose DMEM (Life Technologies) supplemented with 15% horse serum and 2.5% fetal bovine serum (Life Technologies).
Preparation of nuclear extracts
GH4T2 and HeLa cells were harvested at 75–80% confluency from tissue culture plates. GH3 cells were grown to a density of 8–10 × 105 cells/ml and collected by centrifugation at 1500 g. Nuclear extracts were prepared from cycling cells, as described by Dignam et al. (21). Briefly, cells were washed once in ice-cold phosphate-buffered saline (PBS) and harvested on ice with PBS containing 3 mM EDTA. The cells were resuspended in two packed cell volumes of buffer A [10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.15 mM spermine, 0.5 mM spermidine, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate and 1× complete protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN)], incubated for 10 min on ice and lysed with a glass Dounce homogenizer using 20 strokes. The lysate was centrifuged for 5 min at 2000 r.p.m., and the nuclear pellet was resuspended in an equal volume of buffer B (20 mM HEPES pH 7.9, 25% glycerol, 0.42 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 10 mM NaF, 1 mM Na3VO4, 25 mM β-glycerophosphate and 1× protease inhibitors), homogenized with 20 strokes, as above, and incubated at 4°C for 30 min. The cell lysate was centrifuged at 25 000 g for 20 min, and the supernatant was dialyzed against buffer C (20 mM HEPES pH 7.9, 100 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT and 17% glycerol) for 2 h or overnight. The dialysate was centrifuged at 25 000 g for 20 min and the pellet was discarded. Protein concentrations were determined using the Bio-Rad (Hercules, CA) protein assay.
Electrophoretic mobility shift assay
Double-stranded oligonucleotides were annealed and labeled using Klenow DNA polymerase and [α-32P]dCTP, as described previously (12). In the oligonucleotides shown below, only the message strand is depicted and the SalI linker is shown in lower case. The wild-type rPRL promoter BTE sequence (–107 to –88) used is 5′-tcgaCTTAATGACGGAAATAGATG-3′. The mutant BTE oligonucleotide is identical to the wild-type BTE oligonucleotide, except the core Ets site (underlined sequences) was changed to a XhoI site (5′-CTCGAG-3′). The EBS-RRE oligonucleotide sequence (–219 to –203) used is 5′-tcgaCTCATTTCCTTTTGCTG-3′. The mutant EBS-RRE oligonucleotide sequence used is (Bgl II linker in lower case): 5′-gatcCTCTCATCTCGACTTGCTG-3′. The MSV-EBS oligonucleotide sequence, from the murine sarcoma virus long terminal repeat (MSV-LTR), is 5′-tcgaCTCGGAGAGCGGAAGCGCGCA-3′ (22). The mutant MSV-EBS oligonucleotide is identical, except the underlined sequences (GCGG) were changed to 5′-CGCC-3′. Finally, the rPRL promoter FPI sequence (–69 to –41) used as a heterologous competitor is 5′-tcgaTGCCTGATTATATATATATTCATGAAGGT-3′.
For the gel shift assays, 1.5 µg of nuclear extract was incubated with 0.05 ng of the indicated probe (~20 000 c.p.m.) in binding buffer [10 mM HEPES, 50 mM KCl, 4% glycerol, 1 mM EDTA, 1 mM DTT and 250 ng poly(dI-dC) (Pharmacia Biotech, Piscataway, NJ)], in a final volume of 20 µl. Oligonucleotide competition experiments were performed with the indicated amount of unlabeled oligonucleotide. After a 30 min incubation at room temperature, 2 µl of loading dye (25% Ficoll, 0.01% bromophenol blue in 0.25× TBE) was added to each sample and the protein–DNA complexes were separated on pre-electrophoresed 6% non-denaturing polyacrylamide gels in 0.25× TBE buffer (22.5 mM Tris borate, 0.5 mM EDTA) at 200 V. After electrophoresis, gels were dried and protein–DNA complexes were visualized by autoradiography. For the supershift experiments, the Ets-1 antisera was included at a final dilution of 1:50, the GABP antisera was used at a final dilution of 1:25 and the control rabbit IgG sera (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:30. The antisera were added to each gel shift reaction for 15 min prior to the addition of probe. The Ets-1 antibody (4-123) was kindly provided by Dr James Hagman (National Jewish Medical and Research Center, Denver, CO) and the GABP antisera was generously provided by Dr Tom Brown (Pfizer, Groton, CT).
Ammonium sulfate fractionation and heparin–Sepharose chromatography
GH3 cells were used for large-scale nuclear extract preparation, since these cells are similar to GH4 cells (23,24) and are easily grown in spinner culture. GH3NE (262 mg) was partially purified by precipitation in 50% ammonium sulfate. Precipitated proteins were collected by centrifugation at 20 000 g for 20 min at 4°C and the pellet was resuspended in 20.5 ml of binding buffer (10 mM HEPES–KOH pH 7.9, 7 mM KCl, 4% glycerol, 1 mM EDTA, 1 mM DTT with 1× protease inhibitors and 1 mM Na3VO4). After dialysis, fractions were assayed by EMSA, and BTE and EBS-RRE binding activity was present in the 50% ammonium sulfate fraction.
The 50% ammonium sulfate fraction was further purified by heparin–Sepharose chromatography. Briefly, the 50% ammonium sulfate cut was divided into five equal samples and loaded sequentially onto a 5 ml Hi-Trap heparin–Sepharose column (Pharmacia Biotech) equilibrated with binding buffer. The extract was re-circulated using a peristaltic pump for 20 min at 2 ml/min at 4°C and the column was washed with 10 ml of binding buffer. Bound proteins were eluted with a 50 ml 70–800 mM KCl linear gradient with a flow rate of 1 ml/min and 1 ml fractions were collected for each column, using a Pharmacia FPLC. An equal volume (3 µl) of each fraction was assayed for BTE binding activity by EMSA. Peak fractions 1 (∼350 mM KCl) and 2 (∼600 mM KCl) were pooled separately and dialyzed against binding buffer containing 7 mM KCl. The pooled peak fractions from each of the ammonium sulfate samples were concentrated by reloading on the 5 ml heparin–Sepharose column and step-eluted with 550 mM KCl for peak 1 and 1 M KCl for peak 2. Peak fractions 1 and 2 were dialyzed against binding buffer as described above, and equal volumes of each fraction were assayed for binding activity in gel shift assays. Protein concentrations were determined by the Bio-Rad protein assay.
Preparation of recombinant proteins
The pGEX c-Ets-1 bacterial expression vector, which expresses the full-length chicken Ets-1 protein, was kindly provided by B.Wasylyk (Institut de Genetique et de Biologie Moleculaire et Cellulaire, CNRS, Illkirch, France). GST–Ets-1 was grown in the Escherichia coli BL21 cells at 30°C to an optical density of 0.8 at 600 nm, and expression was induced with 2 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 25°C. Cells were collected by centrifugation at 2000 g and washed three times with ice-cold 1× PBS. Cells were lysed in GST-lysis buffer (1% Triton-X, 1% NP-40, 40 mM DTT and 0.4 mM PMSF in 1× PBS), along with 1× complete protease inhibitors (Boehringer Mannheim), and the lysed cells were incubated at 4°C for 30 min with rotation, and subsequently sonicated four times for 15 s each at 4°C. The cell lysate was centrifuged at 10 000 g for 5 min at 4°C to remove cellular debris, and the supernatant containing GST–Ets-1 was added to a 250 µl slurry of glutathione–Sepharose (Boehringer Mannheim). After a 2 h incubation at 4°C, the beads were washed three times with ice-cold lysis buffer and GST–Ets-1 was eluted from the glutathione beads with 5 mM glutathione in 50 mM Tris, pH 8, overnight at 4°C. Protein concentrations were determined by the Bio-Rad protein assay or by silver-staining on an SDS–polyacrylamide gel with known protein standards.
Recombinant mouse GABPα contains amino acids 311–430, which includes the ETS DNA binding domain, and mouse GABPβ includes residues 1–157 which are required for the interaction with GABPα (25). These recombinant proteins are the same proteins used for the crystallization of GABPα/β, which were previously co-expressed as precipitates in E.coli and purified by anion- and cation-exchange chromatography (25), and were kindly provided by Dr Cynthia Wolberger (Johns Hopkins University School of Medicine, Baltimore, MD). Where indicated, 50 ng of recombinant GABPα/β1 was included in the gel shift reaction in place of nuclear extract.
Western blot analysis
Equal amounts (25 µg) of the peak heparin–Sepharose fractions, along with 25 or 50 µg of GH3NE, where indicated, were resolved on 10% SDS–polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Bedford, MA), according to the manufacturer’s protocol. Membranes were probed with primary antibodies to GABPα and β (Tom Brown, Pfizer, Groton, CT) or Ets-1 (Santa Cruz Biotechnology), diluted 1:1000 in blocking buffer (5% non-fat dried milk in TBST) (20 mM Tris pH 7.4, 138 mM NaCl, 0.1% Tween), for 1 h at room temperature. Membranes were washed in TBST three times for 5 min each and probed with a goat anti-rabbit horseradish peroxidase enzyme antibody (Life Technologies) diluted 1:8000 in blocking buffer. Protein was detected using an enhanced chemiluminescence (ECL) method (Pierce, Rockford, IL), according to the manufacturer’s protocol. Membranes were stripped for 30 min at 50°C, according to the manufacturer’s protocol, where indicated.
UV-crosslinking studies
The BTE probe used for UV-crosslinking experiments was identical to the gel shift probe, except two thymidine residues opposite the core EBS on the complementary strand were substituted with two bromodeoxyuridine residues (U): 5′-tcgaCATCTATUUCCGTCATTAAG-3′ (Macromolecular Resources, Fort Collins, CO). The double-stranded BTE was annealed and radioactively labeled as described above, and the standard gel shift reaction was scaled up 2-fold and incubated with the indicated probe in a final volume of 40 µl for 20 min at room temperature. Cold, competitor oligonucleotide was added at 400-fold molar excess, where indicated. The reactions were transferred to a 96-well plate, and irradiated at a distance of ∼1.5 cm using a 254 nm hand-held UV light for 30 min at 4°C. To digest free probe, the reactions were incubated with 1 U of DNase (Promega, Madison, WI) supplemented with 8 mM CaCl2 for 10 min at 37°C. After the addition of 2× sample buffer (0.25 M Tris pH 6.8, 2% SDS, 30% glycerol and 0.2 M DTT) to the crosslinking reactions, the samples were boiled for 5 min and separated on 10% SDS–polyacrylamide gels. Proteins crosslinked to the radiolabeled probe were visualized by autoradiography.
RESULTS
Complex A binds specifically to the Ets sites within the BTE, RRE and MSV-LTR
In order to characterize the endogenous Ets factors present in GH4 pituitary cells capable of binding to the BTE and EBS-RRE of the rPRL promoter, we used gel shift assays with probes encompassing the EBSs of the BTE and EBS-RRE. Figure 1 shows that incubation of GH4NE with the BTE probe leads to the formation of several protein–DNA complexes, denoted as A, B and NS (Fig. 1, lane 2). Addition of a 10-fold molar excess of unlabeled wild-type BTE oligonucleotide (Fig. 1, lane 3) reduced all three complexes. Further increasing the amount of cold BTE oligonucleotide to 100× molar excess completely abolished complex A (Fig. 1, lane 4), whereas complex B is not completely abolished until competition with 1000× BTE (Fig. 1, lane 5). Competition with a mutant BTE oligonucleotide, where the EBS has been mutated, does not affect complexes A and B, but reduced complex NS (lanes 6–8). Competition with a 100× excess of an oligonucleotide encompassing the EBS of the rPRL promoter RRE (EBS-RRE) abolished band A, did not affect band B at any concentration of oligonucleotide competitor and had variable effects on complex NS (Fig. 1A, lanes 9–11). Competition with a mutant EBS-RRE oligonucleotide, where the Ets site has been mutated (mEBS-RRE), does not compete complexes A, B or NS at any level of competitor input (Fig. 1, lanes 12–14). Based on these data, complex A binds to both the BTE and the EBS-RRE via the EBS contained in these oligonucleotides, whereas complex B appears to bind specifically to the BTE. The faster migrating NS band did display some variability in its competition by wild-type cold competitor oligonucleotides. Nevertheless, it appears to be non-specific since it is affected by both wild-type and mutant BTE oligonucleotides, and more importantly, this NS complex was detected using a radiolabeled mutant BTE probe (data not shown). Thus, based on the criteria of binding to a radiolabeled mutant BTE probe and competition by wild-type BTE probe, we labeled this band as NS.
Figure 1.
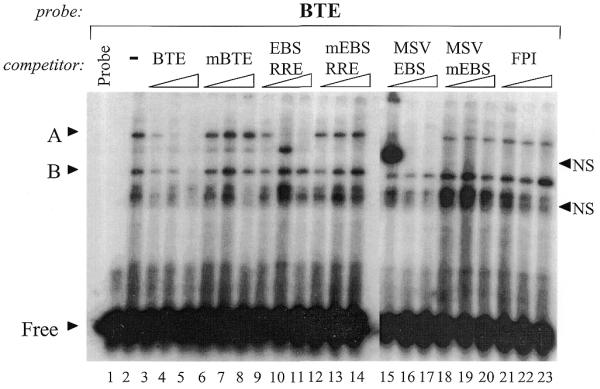
Characterization of GH4 nuclear factors binding to the BTE probe. GH4NE (1.5 µg) was incubated with a 32P-labeled oligonucleotide corresponding to the BTE (–106 to –87) of the rPRL promoter. Protein–DNA complexes were competed with increasing amounts (10×, 100× or 1000×) of cold, unlabeled oligonucleotides encompassing the intact or mutant EBSs of the rPRL promoter BTE or EBS-RRE or the MSV-LTR promoter EBS, as indicated. The FPI oligonucleotide corresponds to the proximal Pit-1 binding site of the rPRL promoter. Protein–DNA complexes were resolved by non-denaturing gel electrophoresis, as described in the Materials and Methods. Lanes 1–14 were run on one gel, and lanes 15–23 were run on a second. Specific protein–DNA complexes are indicated by ‘A’ and non-specific complexes are denoted with ‘NS’. ‘B’ indicates a protein–DNA complex that is competed with high amounts of the wild-type but not mutant unlabeled oligonucleotides. ‘Free’ indicates free probe.
To further determine whether complex A contains an Ets factor, we used an oligonucleotide containing a canonical EBS from the MSV-LTR promoter (MSV-EBS) (22). Competition with increasing amounts of the MSV-EBS oligonucleotide specifically and completely eliminated complex A at 10-fold excess dose level (Fig. 1, lanes 15–17), whereas complexes B and NS are minimally affected. In contrast, the mutant MSV-EBS oligonucleotide did not compete complexes A, B or NS (Fig. 1, lanes 18–20). Finally, competition with an unrelated oligonucleotide encompassing FPI from the rPRL promoter, which does not contain an EBS, did not affect complexes A or B, and minimally reduced complex NS (Fig. 1, lanes 21–23). These data suggest that protein A is an Ets-like factor, since complex A is specifically competed by the EBSs of the BTE, RRE and MSV-LTR. In contrast, protein B appears to bind specifically to the BTE oligonucleotide. These data suggest that complex B does not contain an Ets protein. Indeed, gel shift analysis with a mutant BTE probe, where the core EBS was mutated, also binds to band B, but not band A, indicating that band A is specific to the EBS within the BTE probe and appears to be an Ets factor, while complex B binds to both the wild-type and mutant BTE probes, suggesting that protein B is binding to the BTE probe non-specifically (data not shown). The appearance of the slower migrating NS complex present in Figure 1, lane 15, was highly variable, and also bound to a radiolabeled, mutant BTE probe (data not shown).
In order to characterize endogenous GH4 pituitary nuclear factors that bind to the EBS-RRE probe, an EMSA was performed using GH4NE and a radiolabeled EBS-RRE probe (Fig. 2, lanes 1 and 2). The results revealed multiple complexes, denoted as A, B and NS1-3, of which A, B and NS3 co-migrated with complexes A, B and NS when bound to the BTE probe (data not shown). Oligonucleotide competition studies were carried out using a 400× excess of the cold competitor oligonucleotides used previously (Fig. 2, lanes 2–9). Addition of excess wild-type EBS-RRE oligonucleotide competed away complex A and reduced complexes B, NS1 and NS2 (Fig. 2, lane 3). Competition with the mutant EBS-RRE oligonucleotide did not affect binding of any of these complexes (Fig. 2, lane 4). Addition of excess BTE oligonucleotide competed for complexes A, B and NS1, whereas the mBTE does not compete away any complex (Fig. 2, lanes 5 and 6). Competition with the MSV-EBS only eliminated complex A, whereas the mutant MSV-EBS did not affect complex A (Fig. 2, lanes 7 and 8). Finally, competition with the FPI oligonucleotide did not affect the binding of complex A, but reduced complexes B, NS1 and NS3 (Fig. 2, lane 9). These data indicate that complex A is also specific to the EBS-RRE, and that the binding of GH4 nuclear factors to the BTE and EBS-RRE is very similar. In addition, these oligonucleotide competition studies further support the hypothesis that the protein present in complex A is an Ets factor.
Figure 2.
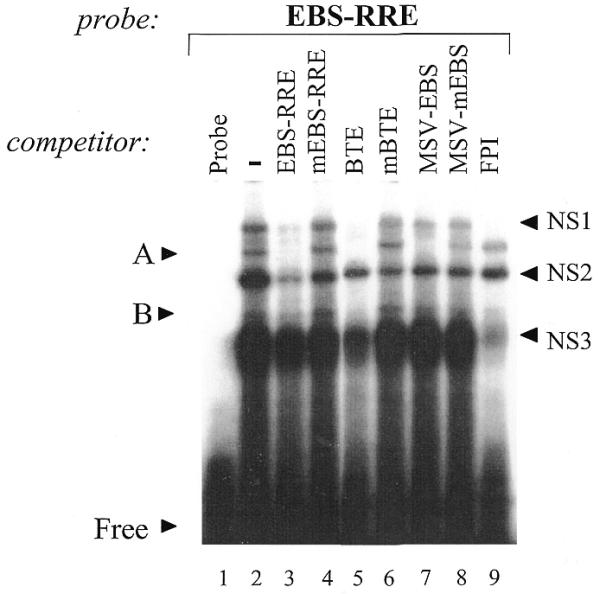
Characterization of GH4 nuclear factors binding to the EBS-RRE probe. GH4NE (1.5 µg) was incubated with a radiolabeled oligonucleotide corresponding to EBS of the RRE (–220 to –205) of the rPRL promoter. Protein–DNA complexes were competed with 400-fold molar excess of cold, unlabeled oligonucleotides as described in Figure 1. Specific protein–DNA complexes are indicated by ‘A’ and non-specific complexes are denoted with ‘NS’. ‘Free’ indicates free probe.
Complex A contains Ets-1 and GABP
To further characterize the Ets factor present in complex A, GH3NEs were partially purified by a 50% ammonium sulfate precipitation followed by heparin–Sepharose chromatography. The 50% ammonium sulfate fraction containing band A activity was loaded onto a 5 ml HiTrap heparin–Sepharose column. Proteins were eluted with a 70–800 mM KCl linear gradient, and 1 ml fractions were collected using an FPLC. Band A activity that bound to the BTE probe eluted in two peaks, one at ∼300 mM KCl (peak 1) and the second at ∼600 mM KCl (peak 2) (data not shown). Peaks 1 and 2 were pooled separately and assayed for binding activity with radiolabeled BTE and EBS-RRE probes. Interestingly, band A binding activity for the BTE probe was detected in peaks 1 and 2, whereas only peak 1 contained band A binding activity for the EBS-RRE probe (data not shown).
In order to identify the Ets factors present in the peak heparin–Sepharose fractions, western blot analysis using antibodies to various Ets factors was done. Figure 3 shows that the anti-GABPα antibody detected a band migrating at ∼59 kDa in GH3NE (25 µg), consistent with the size of GABPα. The anti-GABPα antibody detected a faster-migrating band (∼57 kDa) in the heparin–Sepharose column input (50% ammonium sulfate cut; 25 µg). Both forms, in particular the faster-migrating form of GABPα, were enriched in peak 1, but neither was detected in the flow-through fraction or peak 2 (Fig. 3). GABPβ1 was not detected in GH3NE (25 µg), the heparin–Sepharose input (50% ammonium sulfate; 25 µg), flow-through (25 µg) or peak 2 (25 µg), but a band of ∼47 kDa, which is consistent with the size of GABPβ, was enriched in peak 1 (25 µg) (Fig. 3). Western blot analysis with an antibody to Ets-1 showed that Ets-1 (∼54 kDa) was detected in GH3NE (50 µg), was enriched in the 50% ammonium sulfate cut (25 µg) and was further enriched in peak 1 (25 µg) (Fig. 3). However, Ets-1 was not detected in the flow-through (25 µg) (Fig. 3). The presence of GABPα and β1, and Ets-1 in peak 1, suggests that these factors may be present in complex A which binds to the BTE and EBS-RRE probes.
Figure 3.
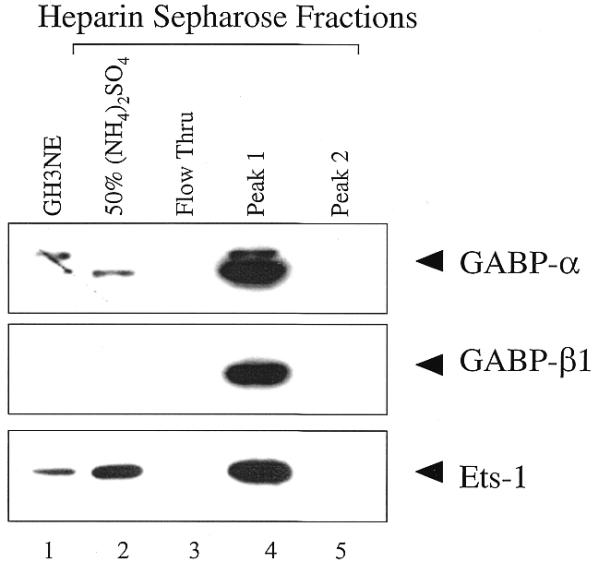
Analysis of heparin–Sepharose purified GH3NEs by western blotting. Equal amounts (25 µg) of each heparin–Sepharose fraction, along with 25 µg of GH3NE for the GABP westerns, and 50 µg of GH3NE for the Ets-1 and Ets-2 westerns, were resolved on 10% SDS–polyacrylamide gels and transferred to Immobilon-P membranes. Blots were probed with antibodies against GABPα, GABPβ or Ets-1, where indicated. Protein was detected using ECL, as described in the Materials and Methods.
To further characterize the Ets factors present in complex A, super-shift experiments with antisera raised against Ets-1, GABPα, GABPβ or control IgG, were included in the gel shift reactions with the BTE (Fig. 4, lanes 1–6) or EBS-RRE probes (Fig. 4, lanes 7–12). Figure 4 shows the appearance of complex A with the BTE probe and the heparin–Sepharose fraction peak 1 (lane 2). Pre-incubation with an antibody to Ets-1 super-shifted ~50% of complex A (A*, lane 3). In contrast, incubation with antisera to GABPα super-shifted nearly the entire complex to a slower migrating band A* (lane 4), whereas antisera to GABPβ1 resulted in a complete loss of complex A and a minimally detectable super-shifted band A* (lane 5). The control rabbit IgG did not affect complex A formation (lane 6), substantiating the specificity of these super-shift reactions. Of note, protein A binding to the BTE was not affected by addition of an antibody to YY1 (data not shown), which has been shown to bind Ets sites (26). Super-shift experiments with the EBS-RRE probe yielded similar results (Fig. 4). Addition of heparin fraction peak 1 resulted in the formation of complex A (lane 8), which is the lower band of the duplex, as shown by specific EBS-RRE competition and by gel shifts with mutant EBS-RRE probe (data not shown). Pre-incubation with the Ets-1 antisera resulted in the strong appearance of super-shift band A* (lane 9). Similarly, pre-incubation with antisera to GABPα led to a similar strong super-shift, while antisera to GABPβ1 resulted in a reduced formation of super-shift complex A* (lanes 10 and 11). Again, the rabbit IgG did not affect the formation of complex A (lane 12). These data show that the Ets factors Ets-1 and GABP can bind to both the BTE and EBS-RRE probes to form complex A. However, there also appears to be qualitative differences in the ability of these factors to bind to these two probes, since the intensity of the super-shifted band A* and of the remaining original complex A is different for the BTE and EBS-RRE super-shifts (Fig. 4).
Figure 4.

Supershift analysis of GH3 nuclear factors that bind to the BTE and EBS-RRE probes. Gel shifts were performed as in Figure 1, except that the heparin–Sepharose peak 1 was pre-incubated with antibodies to Ets-1, GABPα, GABPβ or rabbit IgG, where indicated, for 15 min prior to the addition of the BTE (left) or EBS-RRE (right) probes. Protein–DNA complexes were separated by non-denaturing electrophoresis, as described in Figure 1. ‘A’ indicates the specific protein–DNA complex, and ‘A*’ indicates the supershifted complex. Non-specific bands are indicated by ‘NS’, and ‘Free’ indicates free probe.
In order to determine whether Ets-1 and GABP can bind to the BTE and EBS-RRE probes, gel shifts were performed with recombinant Ets-1 or GABP (Fig. 5). It has previously been demonstrated that recombinant GABP can bind to the BTE probe (19); however, the binding of GABP to the EBS-RRE probe and the binding of Ets-1 to either probe has not been shown. As shown in Figure 5, recombinant Ets-1 and GABPα/β1 bind equally to the BTE probe. Recombinant Ets-1 and GABPα/β1 also bind to the EBS-RRE probe; however, the level of binding of GABPα/β1 to the EBS-RRE probe is lower than that observed for the BTE probe, whereas Ets-1 binds equivalently to each probe (Fig. 5).
Figure 5.

Recombinant Ets-1 and GABP can bind to the BTE and EBS-RRE probes. Recombinant Ets-1 (100 ng) or GABPα and β (50 ng total) were incubated with the BTE or EBS-RRE probes in a gel shift assay, as described in the Materials and Methods.
To determine whether Ets-1 or GABP preferentially bind to the BTE and/or EBS-RRE probes to generate complex A, gel shift analysis with HeLa nuclear extracts were performed (Fig. 6). HeLa cells lack Ets-1 (12), but contain GABP (27), whereas GH4 cells contain both factors (12). Thus, if Ets-1 preferentially binds to either probe, complex A should be seen only in GH4, but not HeLa nuclear extracts. Figure 6 shows that the addition of the BTE probe to GH4 or HeLa nuclear extract led to the formation of several complexes, with the intensity of complex A formed by GH4NE being stronger than that formed by HeLa nuclear extract (Fig. 6, lanes 2–7). As before, complex A was competed by wild-type, but not mutant, BTE oligonucleotides, and complex C was specific for the BTE probe only in HeLa nuclear extract (Fig. 6, lanes 1–7). The addition of the EBS-RRE probe to GH4 and HeLa nuclear extract led to a distinct pattern of binding (Fig. 6, lanes 8–14), in that complex A was evident in GH4NE, but not in HeLa nuclear extract (Fig. 6, lanes 8–14). Complex C formed by the EBS-RRE probe does not appear to be specific since it fails to be competed by the wild-type probe (Fig. 6, lanes 12–14). These data show that the BTE probe binds to an Ets factor that is present in both GH4 and HeLa nuclear extracts, and that the EBS-RRE probe specifically binds to an Ets factor that is only expressed in GH4 pituitary cells. Taken together, the data support a model whereby the BTE site binds to GABP and Ets-1, whereas the EBS-RRE site preferentially binds to Ets-1.
Figure 6.

Gel shift analysis of GH4 and HeLa nuclear extracts with the BTE and EBS-RRE probes. GH4 or HeLa nuclear extract (1.5 µg) was incubated with the BTE or EBS-RRE probes (as indicated) as described above. ‘A’ indicates a specific protein–DNA complex, and ‘C’ indicates a HeLa-specific protein–DNA. Non-specific complexes are denoted by ‘NS’ and ‘B’. ‘Free’ indicates free probe.
UV-crosslinking reveals complex A as a protein–DNA complex of ∼64 kDa
In order to determine the size of the protein(s) present in complex A, UV-crosslinking experiments were done with the heparin–Sepharose input and peak 1 fractions, and the BTE or EBS-RRE probes. The 50% ammonium sulfate cut (used as the heparin–Sepharose column input) and peak 1 from the heparin–Sepharose column, were incubated with the BTE or EBS-RRE probes, exposed to UV light and separated by SDS–PAGE, as described in the Materials and Methods. Figure 7A, lane 1, shows that UV-crosslinking of the 50% ammonium sulfate fraction with the BTE probe leads to a smear of bands of ∼50 kDa and a distinct band at 64 kDa. The ∼50 and 64 kDa bands are enriched in peak 1 (Fig. 7A, lane 2). However, the ∼50 kDa bands are competed by both wild-type and mutant BTE oligonucleotides, whereas the 64 kDa band is specifically competed by the wild-type, but not mutant BTE oligonucleotide (Fig. 7A, lanes 3 and 4). Additionally, the ∼50 kDa band binds to a radiolabeled mutant BTE probe, whereas the 64 kDa protein only binds to a wild-type BTE probe (data not shown). Based on these two criteria, we have labeled the 50 kDa band as ‘non-specific’.
Figure 7.
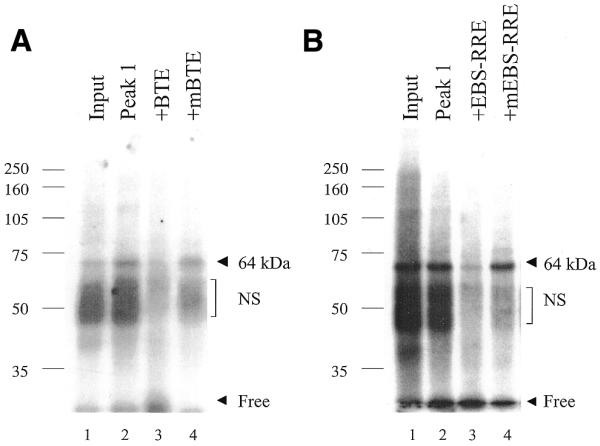
UV-crosslinking of GH3 nuclear factors that bind to the BTE and EBS-RRE probes. (A) The peak 1 heparin–Sepharose fraction (2 µl) and input (1 µl) were incubated with the BTE probe, containing two bromodeoxyuridine substitutions, in a standard EMSA reaction, as described in the Materials and Methods. The reaction was incubated for 20 min at room temperature, exposed to UV light (254 nm) for 30 min at 4°C and analyzed by SDS–PAGE. (B) The peak 1 heparin–Sepharose fraction (2 µl) and input (1 µl) were incubated with the EBS-RRE probe in a standard EMSA reaction, as described in (A). Molecular weight markers are indicated to the left.
A similar UV-crosslinking pattern is seen with the EBS-RRE probe (Fig. 7B). UV-crosslinking of the 50% ammonium sulfate fraction (input) with the EBS-RRE also shows a smear of proteins of ∼50 kDa and a distinct band of 64 kDa (Fig. 7B, lanes 1). The 64 and ∼50 kDa bands are enriched in the heparin–Sepharose peak 1; however, only the 64 kDa protein is specifically competed by the wild-type, but not mutant EBS-RRE oligonucleotides (Fig. 7B, lanes 2–4). These data indicate that the 64 kDa band is specific to both the BTE and EBS-RRE probes. Since the 64 kDa band size reflects the size of the bound factor and residual single- or double-stranded BTE or EBS-RRE probes (7–15 kDa), it is likely that protein A is in the 49–57 kDa size range. This size range is consistent with either Ets-1 (54 kDa) or GABPα (57 kDa), the DNA binding component of GABP.
DISCUSSION
The EBS-RRE and the BTE sites play critical roles in oncogenic Ras- and growth factor-mediated regulation of the proximal rPRL promoter, respectively, with oncogenic Ras acting exclusively via the EBS and certain growth factors targeting both sites (14,17,20,28–31). In this study, we have used gel shift analysis, antibody supershift studies, UV-crosslinking methods and cell extracts devoid of certain Ets factors, to show that pituitary Ets-1 preferentially binds to the EBS-RRE, while both Ets-1 and GABP bind to the BTE. These data are consistent with previous functional studies that have implicated Ets-1 as the sole Ets family member capable of mediating the Ras response via the EBS-RRE (13), and that have suggested that GABP is the nuclear effector of the insulin response via the BTE (19). Thus, the data reported here provide biochemical evidence supporting the idea that distinct Ets family members that have very similar DNA binding specificities, act at different sites on the rPRL promoter to mediate different signaling responses.
The EBS-RRE preferentially binds Ets-1
We have previously shown that the EBS-RRE at position –212 is the only Ets site required to mediate the oncogenic Ras and dominant-active Raf responses of the proximal rPRL promoter (30). Furthermore, co-transfection studies with several Ets family members has revealed that only Ets-1 is capable of enhancing the Ras response in GH4 rat pituitary cells (12). Moreover, using a HeLa reconstitution assay, we have shown that a precise molecular code of interacting partners, involving Pit-1 and Ets-1 acting via the EBS-RRE, is required for optimal rPRL promoter activation (13). A variety of other Ets factors, including Ets-2, Net, Elk and Ehf fail to enhance the Ras response in GH4 pituitary cells (12 and R.E.Schweppe and A.Gutierrez-Hartmann, unpublished observations). A potential biochemical basis for these observations became evident when we showed that Pit-1 and Ets-1 interact directly, and that the physical interaction of Pit-1 with Ets-1 maps to the Ets-1-unique domain (13). Additionally, we have shown that the EBS-RRE can bind to a GST–Ets fusion and that it can compete for the binding of a GH4-derived Ets factor to a canonical Ets-1 binding site from the MSV-LTR (12). While these functional and biochemical studies underscore the importance of Ets-1 on the EBS-RRE, the identity of the pituitary-derived Ets factor capable of binding to this site remains unknown (11–13). Here we present several different lines of evidence supporting the notion that the EBS-RRE preferentially binds to Ets-1 derived from GH4 and GH3 pituitary nuclear extracts. First, complex A formed by the EBS-RRE and GH4NEs contains Ets factor(s), based on specific competition by oligonucleotides containing consensus Ets sites (Fig. 2). Second, antibody supershift studies show that EBS-RRE complex A contains an Ets factor or factors that share an epitope with Ets-1 and GABPα (Fig. 4). Third, recombinant Ets-1 and GABP bind to the EBS-RRE (Fig. 5). Finally, HeLa nuclear extracts that are devoid of Ets-1, but contain GABP (13,27), fail to form complex A with the EBS-RRE (Fig. 6).
The BTE binds both Ets-1 and GABP
The region of the proximal rPRL promoter centered at –96 has been implicated in regulating basal promoter activity (16,32,33) and in mediating multiple hormone and growth factor responses, including TRH, cAMP, insulin, IGF-1, EGF, FGF-2 and FGF-4 (14,15,17,20,28,31). Our group first defined this region in the context of its role as a basal transcription element, and denoted it as the BTE (16), while other groups defined it as a modulator of the protein kinase A response and named it the A-site (34,35). Although the –99 to –92 region contains a CRE-like sequence (TGACGGAA), and CREB can mediate multiple hormonal responses (36), several groups reported that recombinant CREB was unable to footprint the proximal rPRL promoter, including the –99 to –92 region (28,35). Nevertheless, GC and GH3 pituitary nuclear extracts generated specific gel-shift activity using BTE DNA probes (16,32,35), showing that a nuclear factor was capable of binding to this site. Peers et al. (32) performed southwestern blot studies using a radiolabeled DNA oligonucleotide spanning human PRL promoter sequences from –115 to –85 to probe HeLa and GC nuclear extracts, and identified a protein band of ∼100 kDa. Moreover, this ∼100 kDa band was detected only with a wild-type probe, and not with a probe mutated in the core EBS (32). However, the identity of this ∼100 kDa protein remains unknown, and its size is not consistent with any candidate transcription factors mediating growth factor, cAMP and hormonal responses (36).
More recently, the 43 kDa B-Zip protein C/EBPα has been shown to bind to a DNA sequence (ATGACGGAAA) overlapping the BTE-EBS, and to slightly enhance the insulin and EGF responses, but not the cAMP response of the rPRL promoter (37). While exogenously transfected C/EBPα functionally activated the rPRL promoter in GH4 cells using the BTE-C/EBPα site, and recombinant C/EBPα bound to this site, the binding of endogenous GH4-derived C/EBPα to the BTE is weak (37). The oligonucleotide competition data presented here show that an EBS derived from the MSV-LTR and the EBS-RRE, which are both devoid of a C/EBPα binding site, completely compete for GH4 nuclear factor binding to the BTE probe (Fig. 1). Additionally, UV-crosslinking data indicate that the factor binding to the BTE is in the 47–57 kDa size range, which would not be consistent with the 43 kDa size of C/EBPα. However, the formal possibility remains that both C/EBPα and GABP (or Ets-1) simultaneously bind to the BTE site, although this has not been demonstrated.
The importance of Ets factors acting via the BTE became apparent when the core GGAA Ets binding sequence within the BTE was identified as the insulin response element of the proximal rPRL promoter (18). Furthermore, the insulin response was abrogated by a dominant-negative Ets construct, and recombinant SAP-1 and Elk-1 were shown to bind to the BTE (18). Subsequently, GABPα, an Ets family member, was shown to bind to the BTE and was implicated in mediating the insulin response of the rPRL promoter (19). Of note, the ability of Ets-1 to bind to the BTE has not been shown previously. In this study, we use several different approaches to show that GABP and Ets-1 bind to the BTE. First, the BTE binds an Ets factor, based on competition studies using well-characterized EBSs (Fig. 1). Second, anti-Ets-1 and anti-GABP antibodies supershift the BTE complexes (Fig. 4). Third, recombinant Ets-1 and GABP bind to the BTE (Fig. 5). Finally, HeLa nuclear extracts devoid of Ets-1 but containing GABP result in efficient formation of complex A (Fig. 6).
UV-crosslinking of GH3NE with the BTE and EBS-RRE probes in this study identifies a protein band of 64 kDa (Fig. 7). Once the contribution of the cross-linked probe is taken into account (∼15 kDa for double-stranded and ∼8 kDa for single-stranded probe), these data imply that the GH3 pituitary nuclear factor(s) that binds to these probes has a mass of 49–57 kDa (Fig. 7). This size estimate is clearly different from the ∼100 kDa protein previously identified as binding to the BTE using a southwestern blot approach (32). Of note, we also have identified an ∼100 kDa protein band by southwestern analysis of GH4NEs; however, a mutated probe also binds to this band (R.E.Schweppe and A.Gutierrez-Hartmann, unpublished observations). The 49–57 kDa size range identified here is consistent with either Ets-1 (54 kDa) or GABPα (57 kDa); thus, the UV-crosslinking studies support our conclusions that Ets-1 and GABP bind to these two sites, but cannot distinguish which specific factor binds to which specific probe.
Distinct Ets factors with similar DNA binding specificities bind to the rPRL promoter
Members of the Ets family of transcription factors contain a highly conserved ∼85 amino acid DNA binding domain, called the ETS domain. The ETS domain is sufficient for DNA binding and all members of the family bind to a core 5′-GGAA/T-3′ motif. Thus, the problem of binding specificity arises. DNA binding site selection studies have been completed for several members of the Ets family and indicate that subtle differences in sequences flanking the GGAA/T core contribute to binding specificity. Sequences selected by various Ets factors 5′ of the GGAA/T core vary significantly and are therefore thought to contribute to specificity (1). Sequences selected 3′ of the GGAA/T core are highly conserved and these sequences are believed to be critical for the binding of all Ets factors (1). The core sequence of the rPRL promoter BTE EBS (5′-GACGGAAATA-3′) and of the EBS-RRE (5′-AAAGGAAATG-3′) closely resemble the DNA elements selected by either GABPα [5′-(G/a)(C/g)(C/a)GGA(A/t)(G/a)(T/c)] (38) or by Ets-1 [5′-(A/g)C(C/a)GGA(A/T)(G/a)(C/T)-3′)] (39), where the lower case letters represent nucleotides selected less often. However, there is a slight bias of the BTE site for the GABPα consensus sequence and of the EBS-RRE for the Ets-1 consensus sequence, based on nucleotide preference. Moreover, the Pit-1 site adjacent to the EBS in the RRE appears to confer selectivity for Ets-1 via protein–protein interactions, whereby Pit-1 physically associates selectively with Ets-1 via an Ets-1-unique domain (13). In contrast, as noted above, the BTE-EBS site adjacent to the FPII element and is overlapped by a putative C/EBPα binding site (ATGACGGAAA) (16,37). Although both GABP (and Ets-1) and C/EBPα can individually bind to this site, the ability of both factors to co-occupy the BTE site has not been shown. Nevertheless, the possibility remains that C/EBPα confers selectivity to the BTE-EBS, and thus contributes to the preferred recruitment of GABP to this site. For example, GABP functionally interacts with C/EBPα to activate the liver-specific gene Factor IX (40) and the neutrophil elastase gene (41). In addition, the unidentified factor that binds to the adjacent FPII site may also confer binding specificity by interacting with GABP at the BTE.
The data presented here demonstrate that, despite similar DNA sequences of the EBSs contained in the EBS-RRE and the BTE and of near overlapping sequence selectivity of Ets-1 and GABP (38,39), these two Ets factors bind preferentially to the EBS-RRE and BTE, respectively. In order to unequivocally identify the Ets and other nuclear factors that bind to these sites, DNA-affinity purification of these proteins using large-scale preparative techniques followed by protein sequencing methods is required.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Andrew Bradford, Dawn Duval, David Gordan and James Hagman for discussion and reading of the manuscript. We thank Kelley Fantle Brodsky for technical assistance, Drs James Hagman and Tom Brown for the Ets-1 and GABP antibodies, respectively, and Dr Cynthia Wolberger for recombinant GABPα/β. This study utilized the Tissue Culture Core Facility of the University of Colorado Comprehensive Cancer Center, supported by NIH grant NCI P30 CA46934. This research was supported by NIH grant DK46868.
References
- 1.Graves B. and Petersen,J. (1998) Specificity within the ets family of transcription factors. Adv. Cancer Res., 75, 1–55. [DOI] [PubMed] [Google Scholar]
- 2.Watson D., McWilliams,M., Lapis,P., Lautenberger,J., Schweinfest,C. and Papas,T. (1988) Mammalian Ets-1 and Ets-2 genes encode highly conserved proteins. Proc. Natl Acad. Sci. USA, 85, 7862–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth A., Ascione,R., Fisher,R., Mavrothalassitis,G., Bhat,N. and Papas,T. (1992) The Ets gene family. Cell Growth Differ., 3, 327–334. [PubMed] [Google Scholar]
- 4.Wasylyk B., Hahn,S.H. and Giovane,A. (1993) The Ets family of transcription factors. Eur. J. Biochem., 211, 7–18. [DOI] [PubMed] [Google Scholar]
- 5.O’Neill E., Rebay,I., Tijan,R. and Rubin,G. (1994) The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell, 78, 137–147. [DOI] [PubMed] [Google Scholar]
- 6.Gille H., Sharrocks,A.D. and Shaw,P.E. (1992) Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at the c-fos promoter. Nature, 358, 414–417. [DOI] [PubMed] [Google Scholar]
- 7.Hill C.S., Marais,R., John,S., Wynne,J., Dalton,S. and Treisman,R. (1993) Functional analysis of a growth-factor responsive transcription complex. Cell, 73, 395–406. [DOI] [PubMed] [Google Scholar]
- 8.Marais R., Wynne,J. and Treisman,R. (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell, 73, 381–393. [DOI] [PubMed] [Google Scholar]
- 9.Wasylyk B., Hagman,J. and Gutierrez-Hartmann,A. (1998) Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci., 23, 213–216. [DOI] [PubMed] [Google Scholar]
- 10.Ryan A. and Rosenfeld,M. (1997) POU domain family values: flexibility, partnerships and developmental codes. Genes Dev., 11, 1207–1225. [DOI] [PubMed] [Google Scholar]
- 11.Bradford A.P., Conrad,K.E., Wasylyk,C., Wasylyk,B. and Gutierrez-Hartmann,A. (1995) Functional interaction of c-Ets-1 and GHF-1/Pit-1 mediates Ras activation of pituitary specific gene expression: Mapping of essential c-Ets-1 domain. Mol. Cell. Biol., 15, 2849–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford A., Conrad,K., Tran,P., Ostrowski,M. and Gutierrrez-Hartmann,A. (1996) GHF-1/Pit-1 functions as a cell specific integrator of Ras signaling by targeting the Ras pathway to a composite Ets-1/GHF-1 response element. J. Biol. Chem., 271, 24639–24648. [DOI] [PubMed] [Google Scholar]
- 13.Bradford A., Wasylyk,C., Wasylyk,B. and Gutierrez-Hartmann,A. (1997) Interaction of Ets-1 and the POU-homeodomain GHF-1/Pit-1 reconstitutes pituitary specific gene expression. Mol. Cell. Biol., 17, 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweppe R., Frazer-Abel,A., Gutierrez-Hartmann,A. and Bradford,A. (1997) Functional components of FGF signal transduction in pituitary cells: identification of FGF response elements in the prolactin gene. J. Biol. Chem., 272, 30852–30859. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y.-H. and Maurer,R. (1999) A role for the mitogen-activated protein kinase in mediating the ability of thyrotropin-releasing hormone to stimulate the prolactin promoter. Mol. Endocrinol., 13, 1094–1104. [DOI] [PubMed] [Google Scholar]
- 16.Jackson S.M., Keech,C.A., Williamson,D.J. and Gutierrez–Hartmann,A. (1992) Interaction of positive and negative transcription elements controls repression of the proximal rat prolactin promoter in nonpituitary cells. Mol. Cell. Biol., 12, 2708–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob K., Wininger,E., DiMinni,K. and Stanley,F. (1999) The EGF response element in the prolactin promoter. Mol. Cell. Endocrinol., 152, 137–145. [DOI] [PubMed] [Google Scholar]
- 18.Jacob K., Ouyang,L. and Stanley,F. (1995) A consensus insulin response element is activated by an ETS-related transcription factor. J. Biol. Chem., 270, 27773–27779. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang L., Jacob,K. and Stanley,F. (1996) GABP mediates insulin-increased prolactin gene expression. J. Biol. Chem., 271, 10425–10428. [DOI] [PubMed] [Google Scholar]
- 20.Castillo A., Tolon,R. and Aranda,A. (1998) Insulin-like growth factor-1 stimulates rat prolactin gene expression by a Ras, ETS and phosphatidylinositol 3-kinase dependent mechanism. Oncogene, 16, 1981–1991. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J., Lebovitz,R. and Roeder,R. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasylyk C., Gutman,A., Nicholson,R. and Wasylyk,B. (1991) The c-ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J., 10, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tashjian A.H.,Jr (1979) Clonal strains of hormone-producing pituitary cells. Methods Enzymol., 58, 527–535. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez-Hartmann A., Siddiqui,S. and Loukin,S. (1987) Selective transcription and DNase I protection of the rat prolactin gene by GH3 pituitary cell-free extracts. Proc. Natl Acad. Sci. USA, 84, 5211–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batchelor A., Piper,D., de la Brousse,F., McKnight,S. and Wolberger,C. (1998) The structure of GABPα/β: an ETS domain–ankyrin repeat heterodimer bound to DNA. Science, 279, 1037–1041. [DOI] [PubMed] [Google Scholar]
- 26.Liu S., Peng,B., Ma,J., Liu,Y. and Ng,S. (1995) Serum response element associated transcription factors in mouse embryos: serum response factor, YYI and PEA3 factor. Dev. Genet., 16, 229–240. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe H., Imai,T., Sharp,P. and Handa,H. (1988) Identification of two transcription factors that bind to specific elements in the promoter of the adenovirus early-region 4. Mol. Cell. Biol., 8, 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keech C.A., Jackson,S.M., Siddiqui,S.K., Ocran,K.W. and Gutierrez–Hartmann,A. (1992) Cyclic AMP activation of the rat prolactin promoter is restricted to the pituitary-specific cell type. Mol. Endocrinol., 6, 2059–2070. [DOI] [PubMed] [Google Scholar]
- 29.Berwaer M., Peers,B., Nalda,A.M., Monget,P., Davis,J.R.E., Belayew,A. and Martial,J.A. (1993) Thyrotropin-releasing hormone and epidermal growth factor induce human prolactin expression via identical multiple cis elements. Mol. Cell. Endocrinol., 92, 1–7. [DOI] [PubMed] [Google Scholar]
- 30.Conrad K.E., Oberwetter,J.M., Vallaincourt,R., Johnson,G.L. and Gutierrez-Hartmann,A. (1994) Identification of the functional components of the Ras signaling pathway regulating pituitary cell-specific gene expression. Mol. Cell. Biol., 14, 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob K. and Stanley,F. (1994) The insulin and cAMP response elements of the prolactin gene are overlapping sequences. J. Biol. Chem., 269, 25515–25520. [PubMed] [Google Scholar]
- 32.Peers B., Nalda,A.M., Monget,P., Voz,M.L., Belayew,A. and Martial,J.A. (1992) Binding of a 100-kDa ubiquitous factor to the human prolactin promoter is required for its basal and hormone regulated activity. Eur. J. Biochem., 210, 53–58. [DOI] [PubMed] [Google Scholar]
- 33.Iverson R.A., Day,K.H., d’Emden,M., Day,R.N. and Maurer,R.A. (1990) Clustered point mutation analysis of the rat prolactin promoter. Mol. Endocrinol., 4, 1564–1571. [DOI] [PubMed] [Google Scholar]
- 34.Peers B., Monget,P., Nalda,M.A., Voz,M.L., Berwaer,M., Belayew,A. and Martial,J.A. (1991) Transcriptional induction of the human prolactin gene by cAMP requires two cis-acting elements and at least the pituitary-specific factor Pit-1. J. Biol. Chem., 266, 18127–18134. [PubMed] [Google Scholar]
- 35.Liang J., Kim,K.E., Schoderbeck,W.E. and Maurer,R.A. (1992) Characterization of a nontissue-specific, 3′,5′-cyclic adenosine monophosphate-responsive element of the proximal region of the rat prolactin gene. Mol. Endocrinol., 6, 885–892. [DOI] [PubMed] [Google Scholar]
- 36.Shaywitz A. and Greenberg,M. (1999) CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem., 68, 821–861. [DOI] [PubMed] [Google Scholar]
- 37.Jacob K. and Stanley,F. (1999) CCAAT/enhancer binding protein a is a physiological regulator of prolactin gene expression. Endocrinology, 140, 4542–4550. [DOI] [PubMed] [Google Scholar]
- 38.Brown T. and McKnight,S. (1992) Specificities of protein–protein and protein–DNA interaction of GABPα and two newly defined ets-related proteins. Genes Dev., 6, 2502–2512. [DOI] [PubMed] [Google Scholar]
- 39.Nye J.A., Petersen,J.M., Gunther,C.V., Jonsen,M.D. and Graves,B.J. (1992) Interaction of murine Ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev., 6, 975–990. [DOI] [PubMed] [Google Scholar]
- 40.Boccia L., Lillicrap,D., Newcombe,K. and Mueller,C. (1996) Binding of the Ets factor GA-binding protein to an upstream site in the factor IX promoter is a critical event in transactivation. Mol. Cell. Biol., 16, 1929–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nuchprayoon I., Simkevich,C., Luo,M., Friedman,A. and Rosmarin,A. (1997) GABP cooperates with c-Myb and C/EBP to activate the neutrophil elastase promoter. Blood, 89, 4546–4554. [PubMed] [Google Scholar]


