Summary
Appropriate methylation of genomes is essential for gene regulation. We describe here the six-member ORTHRUS (ORTH) gene family of Arabidopsis thaliana that plays a role in DNA methylation in vivo. ORTH1-5 are predicted to encode proteins each containing one PHD, two RING domains, and one SRA domain whereas ORTHlike-1 encodes a protein with only one RING and SRA domain. cDNAs for ORTH1, 2, 5 and ORTHlike-1 were isolated, and when expressed as GST fusion proteins were capable of promoting ubiquitylation in vitro with the E2 AtUBC11. ORTH1 promotes ubiquitylation when paired with additional AtUBC8 family members. ORTH1 proteins with substitutions in metal ligand binding residues in each ORTH1 RING domain individually and ORTH1 truncation derivatives lacking one or both RING domains were tested for ability to catalyze ubiquitylation in vitro. In these assays, either ORTH1 RING domain is capable of promoting ubiquitylation. The PHD alone is not active as an E3 ligase, nor required for ligase activity. GFP-ORTH1 and GFP-ORTH2 are nuclear-localized in transgenic Arabidopsis plants. Overexpression of ORTH1 or 2 in Arabidopsis leads to an altered flowering time. Inspection of DNA methylation at FWA and Cen180 repeats revealed hypomethylation when ORTH proteins were over-expressed. Once initiated, a late-flowering phenotype persisted in the absence of the ORTH transgene, consistent with epigenetic effects at FWA. We conclude that ORTH proteins are E3 ligases mediating DNA methylation status in vivo.
Introduction
Cytosine methylation of genomic DNA is an important element in the epigenetic regulation of gene expression [for recent reviews see (Gehring and Henikoff, 2007; Henderson and Jacobsen, 2007)]. In plants, cytosine methylation can occur in CG, CHG, or CHH sequence context (where H = A, T, or C). In Arabidopsis, DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) is responsible for de novo methylation at all sites, while maintenance methylation requires multiple activities. The DNA methyltransferase MET1 is responsible primarily for maintenance of pre-existing CG methylation (Finnegan and Dennis, 1993; Ronemus et al., 1996), while maintenance of CHG methylation requires a plant-specific DNA methyltransferase, CHROMOMETHYLASE 3 (CMT3), and also a histone H3 lysine 9 methyltransferase KRYPTONITE/SUVH4 (KYP) (Henderson and Jacobsen, 2007). Small RNAs play a key role in targeting DNA methylation, especially in plants, in a process not completely understood (Wassenegger et al., 1994; Henderson and Jacobsen, 2007; Huettel et al., 2007; Matzke et al., 2007). Asymmetric methylation of CHH is maintained in a small RNA-dependent de novo methylation pathway performed by DRM2 (Henderson and Jacobsen, 2007).
Plants defective in either de novo methylation or maintenance methylation exhibit a wide variety of phenotypic alterations. In met1 mutants dramatic hypomethylation takes place genome-wide (Zhang et al., 2006). This leads to stochastic phenotypes caused by the aberrant expression of genes silenced by methylation, including a late-flowering phenotype derived from the ectopic expression of FWA, altered flower development, and decreased fertility (Finnegan et al., 1996; Soppe et al., 2000; Kankel et al., 2003).
Another family of proteins implicated in regulating DNA methylation in Arabidopsis that is much less understood, is the ORTHRUS/VARIANT IN METHYLATION (ORTH/VIM) proteins (Johnson et al., 2007; Liu et al., 2007; Woo et al., 2007). They share domains with and are likely orthologous to the mammalian UHRF (Ubiquitin-like containing, PHD, Ring Finger) proteins (Bronner et al., 2007), which are important for DNA methylation maintenance (Bostick et al., 2007; Sharif et al., 2007). ORTH/VIM/UHRF proteins are characterized by the presence of an SRA domain [SET (Su(var), Enhancer of Zeste, Trithorax) and RING (Really Interesting New Gene) Associated], a PHD, and a RING domain.
The SRA domain, also known as the YDG domain (for three highly conserved amino acids), as the name suggests, usually occurs in conjunction with two different types of proteins - SET domain proteins or the UHRF/ORTH/VIM proteins with one or two RING domains. The SET domain generally performs the catalytic histone methyltransferase activity of the Arabidopsis SRA-SET proteins, such as KYP (Baumbusch et al., 2001). The SRA-SET proteins are unique to the plant kingdom, while the SRA-RING proteins, UHRF/ORTH/VIM, are found in many species with cytosine methylation, such as mammals, zebrafish, honeybee, and plants. The SRA domain has been reported as a methylated DNA binding domain (Unoki et al., 2004; Johnson et al., 2007; Woo et al., 2007). Additionally, the UHRF1 SRA domain has a strong preference for hemimethylated CG (Bostick et al., 2007). However, histone binding has also been ascribed to the SRA domain. The SRA domain of the murine UHRF1 binds H1, H2B and H3 and ubiquitylates histones in vitro (Citterio et al., 2004). The human UHRF1 selectively binds to methylated H3K9 peptides, and this activity is dependent on both the PHD and SRA domains (Karagianni et al., 2008). ORTH2/VIM1 has also been shown to associate with histones H2B, H3, H4, and the centromere H3 histone variant HTR12 in vitro (Woo et al., 2007), although the domain responsible is not known.
The RING domain is an approximately 40–60 amino acid region with an octet of histidine/cysteine residues spaced to chelate two zinc atoms (Borden and Freemont, 1996). This structure binds the E2 carrying activated ubiquitin and facilitates ubiquitin transfer to substrate proteins (Lorick et al., 1999). Curiously, only a few Arabidopsis RING proteins, in addition to the ORTH/VIM proteins, contain multiple RING domains out of the more than 400 predicted (Stone et al., 2005). PRT1 (PROTEOLYSIS1) has two N-terminal RING domains and participates in the proteolysis of a subset of ubiquitin pathway substrates bearing specific N-terminal amino acids (Stary et al., 2003). It remains to be determined whether the two RING domains can function independently in supporting ubiquitylation. Multiple members of the Arabidopsis ARIADNE family of RING proteins, like their animal counterparts, contain two RING or RING-like domains separated by a conserved region, designated IBR for in between RING (Mladek et al., 2003). Mutations in either RING domain results in lethality in Drosophila (Aguilera et al., 2000); however, no clear evidence exists to suggest the second RING domain is independently active in ubiquitylation.
ORTH/VIM proteins also contain a PHD, which consists of an octet of histidine/cysteine residues reminiscent of the RING domain (Schindler et al., 1993; Aasland et al., 1995; Borden and Freemont, 1996). Searches for physiological substrates for the PHD have demonstrated binding to substrates as diverse as phosphoinositols (Gozani et al., 2003) and the tri-methyl lysine 4 of histone H3 (Li et al., 2006; Pena, 2006; Shi et al., 2006; Wysocka et al., 2006). It has also been suggested that PHD can confer E3 ligase activity to proteins, although those proteins were either determined to be RING domains or the assays were not reproducible (Lu et al., 2002; Goto et al., 2003; Yonashiro et al., 2006).
Previous studies have yielded some insight into the biological function of ORTH/VIM proteins. A screen for natural variation in cytosine methylation in Arabidopsis revealed a role for ORTH2/VIM1 in centromeric repeat methylation (Liu et al., 2007; Woo et al., 2007). Additionally, the amino terminus of ORTH2/VIM1 containing the PHD binds to NtSET1 in a yeast two hybrid experiment (Liu et al., 2007). Over-expression of green fluorescent protein (GFP)-tagged VIM1 also led to a late-flowering phenotype with increased levels of FLC (Liu et al., 2007).
Substantial work on the role of the vertebrate ORTH/VIM homolog, UHRF1, has revealed roles in cell cycle regulation (Bonapace et al., 2002; Jeanblanc et al., 2005), cellular transformation (Mousli et al., 2003; Jenkins et al., 2005), DNA damage repair (Muto et al., 2002), replication of pericentromeric heterochromatin (Papait et al., 2007), liver regeneration (Sadler et al., 2007), and recruiting DNMT1 to replication foci (Bostick et al., 2007; Sharif et al., 2007). The mechanism of many of these processes has not been thoroughly investigated, and further studies of the domains of UHRF1 and its Arabidopsis counterparts are essential.
Herein we report additional characterization of the Arabidopsis ORTH/VIM family of proteins. We demonstrate that ORTH proteins possess E3 ligase activity requiring either, but not both, RING domains for E3 activity with AtUBC8 family members in vitro. The PHD did not contribute to in vitro E3 ligase activity. As reported previously (Liu et al., 2007), ORTH1/2 over-expressing lines flower later. We further explored the molecular mechanism for this alteration and demonstrate decreased methylation at FWA as the likely cause for delayed flowering. ORTH1/2 over-expression also reduces methylation at Cen180 repeats. These data support the role of the ORTH family as E3 ligases that play a role in controlling epigenetic mechanisms at both heterochromatic and euchromatic loci in plants.
Results
ORTH Domains and Phylogenetic Analyses
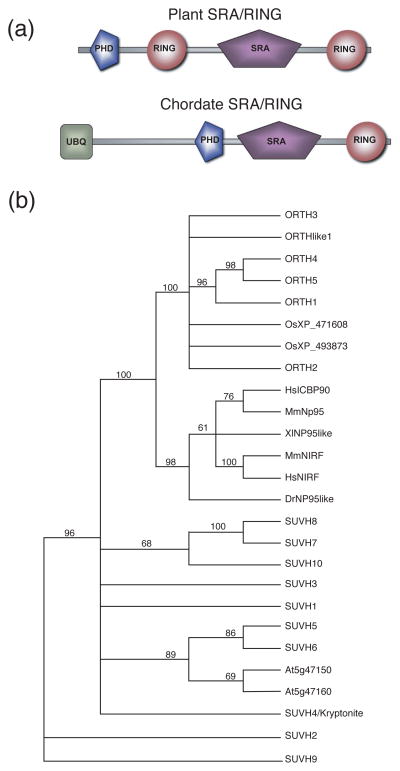
Inspection of the Arabidopsis predicted proteome for multiple RING domain-containing proteins identified five members of the ORTHRUS/VIM family and a related protein ORTH-LIKE1 (ORL1/VIM6). ORTH proteins contain an N-terminal PHD and two RING domains flanking an SRA domain (Figure 1a, upper). ORL1/VIM6 is missing the N-terminal PHD and C-terminal RING domain, but contains the SRA and N-terminal RING domains. Proteins with both SRA and RING domains are referred to here as SRA-RING proteins. Mammalian SRA-RING proteins have an altered domain organization compared to plant ORTH proteins, with an N-terminal ubiquitin-like domain and an SRA domain that is flanked N-terminally by a PHD and C-terminally by a RING domain (Figure 1a, lower).
Figure 1. ORTH domains and phylogenetic analysis.
(a). Domain organization of plant SRA-RING (top) and chordate SRA-RING (bottom) proteins.
(b). Phylogenetic tree of the SRA domain-containing proteins. The tree is based on the SRA domain only. Included in the analysis are all SRA domain-containing proteins of Arabidopsis and select SRA-RING proteins of other organisms. ORTH1/VIM3 (At5g39550), ORTH2/VIM1 (At1g57820), ORTH3/VIM5 (At1g57800), ORTH4/VIM4 (At1g66040), ORTH5/VIM2 (At1g66050), and ORL/VIM6 (At4g08590). Arabidopsis SRA-domain only proteins are listed by their AGI code (At5g47150, At5g47160). Arabidopsis SRA-SET domain-containing proteins are listed as previously designated with the abbreviation SUVH (Baumbusch et al., 2001). Rice SRA-RINGs are listed by NCBI locus code. NCBI locus codes for other organisms are as follows: HsICBP90 (AAF28469), MmNP95 (AAK55743), XlNP95like (AAH72079), MmNIRF (AAH60241), HsNIRF(NP_690856), DrNP95like (AAT68031). At (Arabidopsis thaliana), Os (Oryza sativa), Hs (Homo sapiens), Mm (Mus musculus), Xl (Xenopus laevis), Dr (Danio rerio).
Additional Arabidopsis proteins contain an SRA domain without a PHD or RING domain. These include ones with a SET domain (referred to here as SRA-SET proteins) as well as those without any other described protein-protein interaction domain (SRA-only). The SRA domains of all Arabidopsis SRA-RING family members were compared with selected SRA-RING proteins from other species and to the other Arabidopsis SRA-SET and SRA-only proteins (Figure 1b). Phylogenetic analysis using the entire protein results in the same conclusions (Supplemental Figure 1 and data not shown).
Within the AtORTH family, ORTH1, 4 and 5 form a clade (Figure 1a). ORTH1 has 95% similarity to ORTH4 and 91% amino acid identity to ORTH5 across the entire protein and ORTH4 and 5 have overall 99% amino acid and 99% nucleotide identity. In contrast, ORTH1 is 70% and 62% identical to ORTH2 and ORTH3, respectively, and ORTH2 and 3 have 73% amino acid identity. Interestingly, ORTH2 and 3 (At1g57820 and At1g57800, respectively) are separated by only one predicted pseudogene, and ORTH4 and 5 (At1g66040 and At1g66050, respectively) are similarly one predicted open reading frame apart, suggesting that duplication events gave rise to the ORTH2/3 and ORTH4/5 pairs.
Comparisons of the SRA domains reveal that the SRA-RING proteins of Arabidopsis have greater similarity to SRA-RING proteins from chordates than they do to Arabidopsis SRA-SET proteins. This implies that SRA-RING proteins were present in the common ancestor of plants and animals. In contrast, the plant-specific Arabidopsis SRA-SET proteins are present in a clade with the SRA-only proteins, suggesting that they are more closely related to each other than to the SRA-RING proteins.
To determine which genes are expressed, effort was expended to isolated cDNAs for all six members. cDNAs were isolated for ORTH1, 2, 5 and ORL1 using RT-PCR. Attempts to isolate cDNAs for ORTH3 and 4 failed using TAIR (The Arabidopsis Information Resource, www.arabidopsis.org) annotations for their predicted open reading frames. Thus, the expression of ORTH3 and ORTH4 is uncertain. The 99% nucleotide identity between ORTH4 and 5 coding regions makes EST/cDNA gene assignments challenging. Two ORTH4 ESTs/cDNAs are listed on TAIR (1/08), but their sequences (GenBank EL177826 and EL265513) are 100% identical to both ORTH4 and ORTH5 and the clones are not available for further sequencing, so their assignment to a single locus cannot be made. The cDNA sequences that are ascribed to ORTH5 have the few ORTH5-specific nucleotides, indicating that these are annotated correctly and that there is no ORTH4 cDNA in that set. Only a single EST is listed for ORTH3, and this assignment appears to be correct. Thus, there is no evidence for expression of ORTH4 and scant evidence for ORTH3 expression. These data suggest that only a subset of ORTHs are significant contributors to ORTH activity in vivo.
ORTH Family Members are Active as E3 Ubiquitin Ligases
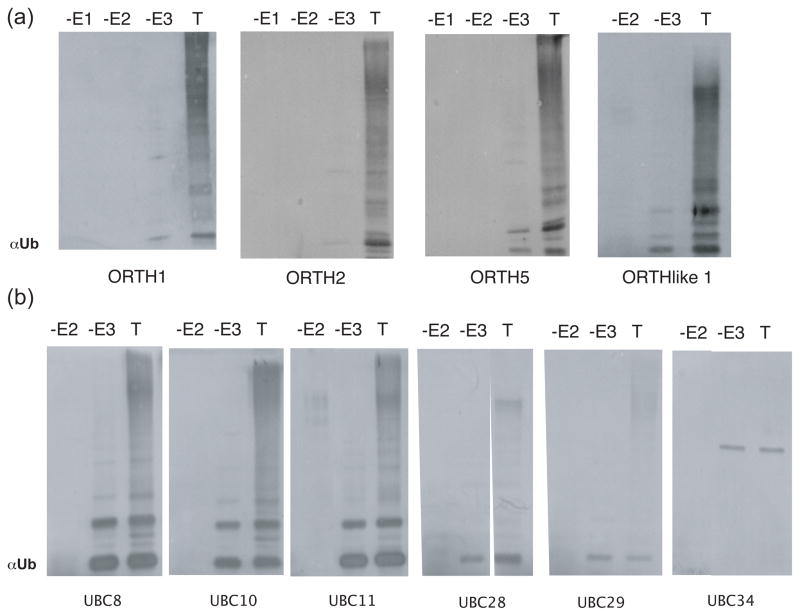
The RING domain is associated with the ability to function in ubiquitin transfer (Lorick et al., 1999; Borden, 2000; Kraft et al., 2005; Stone et al., 2005), although not all RING proteins demonstrate in vitro E3 ligase activity. The murine SRA-RING domain-containing protein mUHRF1 is active in in vitro ubiquitylation assays with the E2 UBCH5B (Citterio et al., 2004). To determine if ORTH proteins function as E3 ligases, full-length ORTH1, 2, 5 and ORL proteins were expressed in E. coli as glutathione-S-transferase (GST) fusions, purified on glutathione beads and tested for the ability to promote ubiquitylation in vitro as previously described (Kraft et al., 2005; Stone et al., 2005). All three ORTH proteins were active with the Arabidopsis E2 UBC11 (Figure 2a) and as expected, omission of E1, E2 or ORTH protein eliminates attachment of ubiquitin to higher molecular weight proteins. Similarly, ORL1, which lacks the C-terminal RING and N-terminal PHD, was also active (Figure 2a).
Figure 2. ORTH Family Members Possess E3 Ligase Activity.
(a-b) In vitro ubiquitylation assays. The complete reaction (T) contains E1, 6HIS-E2 and GST-tagged E3, ubiqutin and ATP. The reaction was fractionated by SDS-PAGE, the proteins transferred to a membrane, and the products visualized with anti-ubiquitin antibodies. Free ubiquitin migrates at 5.5 kDa and has migrated off of the bottom of the gel. Anti-ubiquitin bands visualized in the -E3 lane correspond to E2-Ub adducts. Activity was determined by comparing -E3 reactions to complete reactions for the presence of high molecular weight proteins immunoreactive with anti-ubiquitin antibody (Stone et al., 2005, Kraft et al., 2005). The experiments were repeated at least two times with similar results.
(a) GST-ORTH proteins were tested with the E2 UBC11.
(b) GST-ORTH1 was tested with different E2s to determine E2-E3 specificity. The E2 used is listed below its corresponding western blot.
We asked whether ORTH proteins require a specific E2 or E2s for activity. UBC11 is a member of the UBC8 family of Arabidopsis E2s (Kraft et al., 2005). ORTH1 was tested with additional E2s of the UBC8 family (UBC8, 10, 28, 29, 30), as well as with members of three other E2 subgroups (UBC27, 36, 32 and 34) (Kraft et al., 2005). ORTH1 shows robust activity with UBC8 family members UBC8, 10 and 11 (Figure 2b). ORTH1 also shows activity, albeit diminished, with other more divergent UBC8 family members UBC28 and 29 (Figure 2b). ORTH1 was not active with the other E2s tested; UBC34 (Figure 2b) and UBC27, 30, 32 and 36 (data not shown). Thus, recombinant GST-ORTH1 is most active in vitro with E2s from the UBC8 subfamily.
A single RING domain is sufficient for robust in vitro ubiquitylation activity
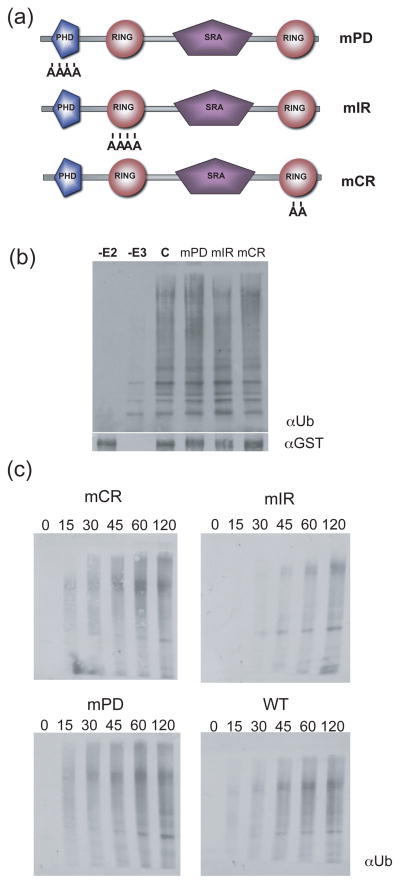
To determine the domain associated with the E3 ligase activity of ORTH1, codons for conserved metal ligand residues in the PHD and each RING domain were individually targeted for mutagenesis. These residues have been shown for many RING proteins to be essential for ligase activity (Lorick et al., 1999; Stone et al., 2005). Codons for the PHD metal-ligand binding positions 3–6 or for the N-terminal RING metal ligand binding position 4 and 5 of the N-terminal or C-terminal RINGs were mutagenized to encode alanine, creating proteins called mPD for mutant PHD, mNR for mutant N-terminal RING and mCR for mutant C-terminal RING (Figure 3a). When these GST-ORTH1 fusion proteins were tested for ubiquitylation activity with UBC8, all three retained robust activity in end-point assays (Figure 3b). Time course experiments to detect differences in ubiquitylation rates showed that activity of each mutant protein was largely unaffected relative to wild-type GST-ORTH1 (Figure 3c), indicating that alteration of one RING domain or the PHD does not dramatically interfere with the in vitro ubiquitylation activity of ORTH1.
Figure 3. Substitutions in a single zinc-chelating domain do not affect in vitro ubiquitylation activity of full length ORTH1.
(a). Schematic of the ORTH1 protein and amino acid changes introduced. Codons for the internal CHCC of the ORTH1 proteins mPD or mNR or the internal HC of mCR were altered to encode for alanine.
(b). Single domain mutations have no effect on ubiquitylation in endpoint assays. GST-tagged ORTH1 and mutant proteins were tested for ubiquitylation activity as described in Figure 1 to determine the contribution of each domain. Ubiquitylated proteins were visualized by anti-ubiquitin western. Anti-GST western (below) illustrate equal input of the E3 ORTH1 at time zero.
(c). Time course of ubiquitylation activity of wild-type ORTH1 (WT, bottom left) and mutant ORTH1 proteins (designated as in (a)). Aliquots of assays were removed at times indicated (in minutes) and stopped with SDS.
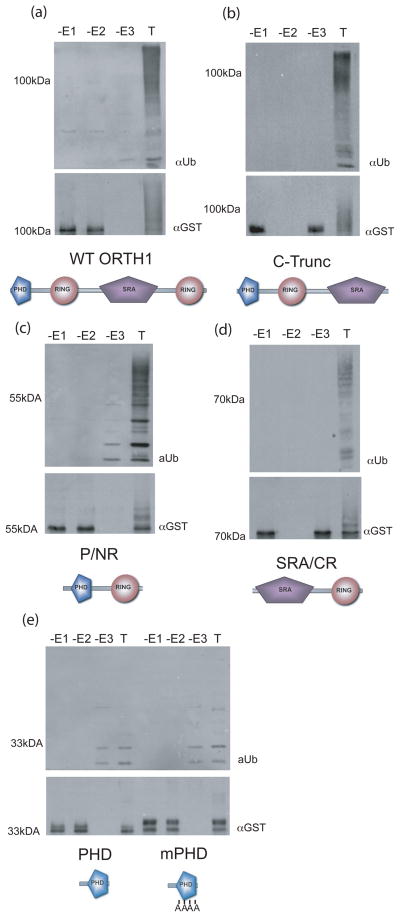
To further characterize the RING domain responsible for E3 ligase activity of ORTH1, the open reading frame was truncated to express GST-ORTH1 proteins lacking one or more domains; lacking either the C-terminal RING only (C-Trunc, Figure 4b), the C-terminal RING and SRA domains (P/NR, Figure 4c), or the PHD and N-terminal RING domains (SRA/CR, Figure 4d). These truncations were assayed with UBC11 in in vitro ubiquitylation assays and compared to the activity of the full-length protein (Figure 4a). All truncated proteins exhibited E3 ligase activity with UBC11, demonstrating that in vitro either RING domain can provide E3 ligase activity. The autoubiquitylation of these proteins can be seen in the anti-GST blots, visualized as slowed migration in the complete reactions only, resulting from covalent addition of ubiquitin to the GST-ORTH fusion protein (Figure 4, below each activity blot).
Figure 4. ORTH1 proteins with either RING domain, but not the PHD, have E3 ligase activity.
Truncations of ORTH1 (name and schematic form under their respective western blots) were expressed as GST fusions and tested for in vitro ubiquitylation activity.
(a-e) Top, anti-ubiquitin immunoblot to detect ubiquitylated products; bottom, anti-GST to visualize E3 ligase. Reactions lacking either E1, E2 or E3 (-E1, -E2, -E3, respectively) are compared to comparable reaction with all components (T).
(e) mPHD has the CCHC of the core metal ligands, positions 3-6, all changed to alanine. Full length ORTH1 assays (a) were included for comparison.
The contribution of the PHD to ubiquitylation activity was additionally tested by comparing the reactions of a truncated ORTH1 protein with only the PHD to that of the identical protein except with substitutions in four of the eight metal ligand binding residues in the PHD (mPHD, Figure 4e). In these reactions, there were faint higher molecular weight anti-ubiquitin immunoreactive bands, but the profile was identical between reactions with or without the ORTH-PHD (-E3 lanes) and reactions containing either ORTH-PHD or ORTH-mPHD (T lanes). There does not appear to be any ubiquitylation activity associated with the closely related PHD, at least in these in vitro assays.
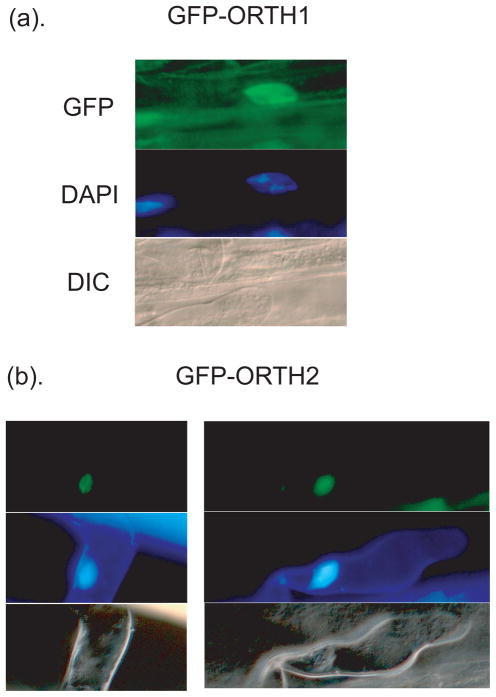
GFP-ORTH1 and GFP-ORTH2 are nuclear-localized
To determine the intracellular localization of ORTH proteins, constructs for expression of ORTH1 and ORTH2 in fusion with GFP were made and stably transformed into Arabidopsis. GFP was visualized by fluorescence microscopy and compared to DAPI nuclear staining. Both GFP-ORTH2 and GFP-ORTH1 co-localized with DAPI-staining nuclei (Figure 5a and b, respectively). Thus ORTH1 and ORTH2 are located primarily in nuclei.
Figure 5. ORTH1 and 2 are Nuclear-localized.
Confocal analysis shows GFP-ORTH1 (a) and GFP-ORTH2 (b) are localized to nuclei (top panels). Corresponding DAPI (middle panels) and DIC images (bottom panels) are included to show position of nuclei and cellular organization/outline.
Overexpression of ORTH1 or ORTH2 affects DNA methylation-mediated gene silencing
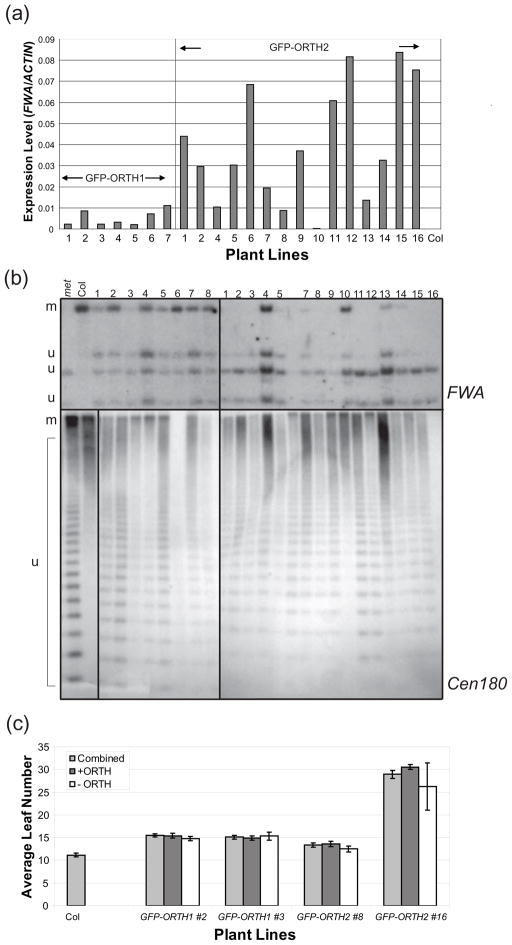
While producing plants for intracellular localization studies, we observed that GFP-ORTH1 and GFP-ORTH2 expressing transgenic plants exhibited delayed flowering, while GFP-ORL plants did not (data not shown and Supplemental Figure 2). Similarly, Liu et al (2007) recently reported that expression of GFP-ORTH2/VIM1 under the control of a constitutive promoter leads to a mild to moderate late flowering phenotype, while expression of GFP-ORL/VIM6 does not. To further explore the molecular basis of this effect, multiple T1 plants were analyzed for FWA expression. FWA is a homeodomain transcription factor completely silenced by DNA methylation in all cells of wild-type plants, except in the developing endosperm. Hypomethylation of the FWA promoter leads to ectopic expression causing a late flowering phenotype (Soppe et al., 2000). Using RNA extracted from leaf tissue, no expression of FWA is observed in wild-type plants, while varying levels of expression are seen in almost all independent T1s transformed with either GFP-ORTH1 or GFP-ORTH2 by real time PCR (polymerase chain reaction) of cDNA relative to ACTIN (Figure 6a). Further analysis by Southern blotting confirmed hypomethylation at the FWA promoter. Increased digestion of genomic DNA by the methylation-sensitive enzyme CfoI was present in all GFP-ORTH1- and GFP-ORTH2-expressing lines, compared to the negative control, untransformed Col ecotype (Figure 6b, top panel).
Figure 6. The late-flowering phenotype of GFP-ORTH1 and GFP-ORTH2 plants is caused by decreased DNA methylation.
(a) Relative FWA levels from seven GFP-ORTH1 and sixteen GFP-ORTH2 independent T1 plants, compared to Col.
(b) Southern blots for FWA (upper) and Cen180 bp repeats (lower) with DNA from eight GFP-ORTH1 and fifteen GFP-ORTH2 independent T1 plants. Col (Col) DNA is fully methylated (m), while met1-3 (met) DNA is severely unmethylated (u).
(c) Flowering time analysis for the T2 generation of two independent GFP-ORTH1 (#2, #3) and GFP-ORTH2 (#8, #16) lines. The number of leaves produced before the transition from vegetative tissue was counted in at least 26 plants from each line. The plants were then genotyped for the presence (+ ORTH) or absence (-ORTH) of the transgene.
A correlation is observed between FWA expression levels and FWA methylation levels throughout the independent T1s for both GFP-ORTH1 and GFP-ORTH2. Higher FWA expression is generally coincident with decreased methylation (GFP-ORTH2 #16), and lower FWA expression coincident with a more wild-type level of methylation (GFP-ORTH2 #10). Noticeable late-flowering phenotypes are only observed in lines with the highest expression of FWA (GFP-ORTH2, #12, 15, and 16, data not shown).
To test if the late-flowering phenotype was the function of a dominant negative form of ORTH protein caused by the GFP tag, an expression construct for untagged-ORTH1 was also introduced into Col under control of the 35S promoter. Flowering time, expressed as the leaves present prior to transition to flowering, was measured in segregating T2 seedlings derived from independent T1s, and five of the ten lines had an average leaf number higher than wild type (Supp. Figure 3a). This late-flowering phenotype was also associated with decreased methylation at the FWA locus (Supp. Figure 3b, top panel). Therefore, plants over-expressing either ORTH1/VIM3 or ORTH2/VIM1 exhibit a late-flowering phenotype likely caused by the ectopic expression of a hypomethylated FWA locus.
Loss of ORTH2/VIM1 leads to decreased methylation at the heterochromatic Cen180 repeats (Woo et al., 2007). To test if overexpression of ORTH family members affects methylation at the Cen180 repeats in addition to the FWA locus, Cen180 Southern blots were performed on similarly digested genomic DNA from wild-type Columbia and transgenic lines. Decreased Cen180 DNA methylation was observed in GFP-ORTH1, GFP-ORTH2, and untagged-ORTH1 lines (Figure 6b, Supplemental Figure 3b; bottom panels).
Finally, to test if FWA hypomethylation was stable in subsequent generations, T2 plants from two GFP-ORTH1 (#2, #3) and GFP-ORTH2 (#8, #16) lines were planted. Flowering time was measured, and plants were genotyped for the presence of the ORTH transgene. As expected, the average flowering time of all T2 lines delayed relative to wild type Columbia (Figure 6c). In addition, the presence or absence of the GFP-ORTH expressing transgene did not affect flowering time (Figure 6c). This shows that ORTH over-expression in the T1 generation leads to stable and heritable late-flowering phenotype.
Discussion
The Arabidopsis ORTH/VIM family contains six family members; however, ORL1/VIM6 lacks the N-terminal PHD and C-terminal RING domain present in the other five family members. In addition, ORL1 over-expression does not have the same effect as over-expression of ORTH1 or ORTH2, suggesting that ORL1 may have a unique function [this work and (Liu et al., 2007)]. Of the ORTH1-5 proteins, only three may contribute significant activity in vivo because ORTH3/VIM5 and ORTH4/VIM4 appear to have very low or limited expression. Information on their expression cannot be determined from available microarray studies. ORTH3/VIM5 sequences are not on the most commonly used expression array (ATH1), and ORTH4 and ORTH5 are 99% identical in the coding region and cannot be distinguished on the current arrays. Among the expressed ORTH/VIM proteins, there are differences in expression patterns. Using in situ hybridization, Liu et al (2007) detected RNA in maturing or mature embryos from ORTH2/VIM1 and ORL1/VIM6, but not ORTH5/VIM2. Thus, only a subset of the ORTH proteins is likely to be present at significant levels in vivo at any given time.
Previous work by many laboratories has demonstrated that the presence of a RING domain confers E3 ligase activity (Lorick et al., 1999; Kraft et al., 2005; Stone et al., 2005). Consistent with these reports, we show here that ORTH1, 2, 5, and ORL1 show a clear ability to function as E3 ligases in vitro. The related mammalian SRA-RING proteins, mouse UHRF1/Np95 and human UHRF2/NIRF, were active in vitro with the human E2s, UbcH5b (Citterio et al., 2004) and UbcH5a, respectively, and UHRF2 was not active with UbcH7 (Mori et al., 2004). The UBC8 family of Arabidopsis is more similar to the human UbcH5 E2s than any other Arabidopsis E2 family (Kraft et al., 2005), and thus the E2-E3 pairing of UHRFs and AtORTH family members appears to be shared.
ORTH1-5 are distinguished from most other Arabidopsis RING proteins and from the chordate SRA-RING proteins in containing two RING domains. Activity assays with truncated ORTH1 proteins lacking one or more domains, or with full-length ORTH1 proteins with amino acid substitutions in one RING domain showed that either RING domain alone is functional in the transfer of ubiquitin. However, it still is possible that both RING domains function synergistically in the transfer of ubiquitin to the endogenous target(s) or that one RING domain has a novel function in vivo.
mUHRF1/Np95 interacts with and ubiquitylates core histones in vitro and ex vivo with a preference for H3 (Citterio et al., 2004). UHRF2/NIRF interacts with and ubiquitylates PCNP (PEST-containing nuclear protein), a protein of unknown function, in vitro, and enhances its ubiquitylation in vivo (Mori et al., 2004). Arabidopsis ORTH2/VIM1 expressed in plants interacted with recombinant histones H2B, H3, H4 and a centromere-localized H3, HTR12 (Woo et al., 2007), although a direct interaction was not demonstrated, as another protein in complex with ORTH2/VIM1 could be binding to the histones. In our hands, GST-ORTH1 was able to ubiquitylate histone H3 in vitro; however, another Arabidopsis non-ORTH RING E3 also shared this ability (unpublished data). Thus, we could not determine whether ORTH proteins specifically ubiquitylate histone H3 in our assays. ORTH2/VIM1 interacts with tobacco SET1, but whether SET1 is a ORTH substrate was not tested (Liu et al., 2007). Clearly, further analysis to determine additional interacting proteins or characterize ORTH-containing complexes will allow testing for ORTH-mediated ubiquitylation.
The SRA domain of SRA-RING and SRA-SET proteins has been shown to be a methylated DNA binding domain (Unoki et al., 2004; Johnson et al., 2007; Woo et al., 2007). The presence of this domain in a proteome correlates with the presence of methylated DNA in a genome. For example, SRA domain-containing proteins are not predicted from the annotated genomes of Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe and Saccharomyces cerevisiae by BLAST and SMART searches, and CpG methylation has not been reliably detected in their DNA. The SET domain associated with the SRA domain is recognized to be required for protein methyltransferase activity, most notably but not exclusively onto histone lysyl ε-amino groups (Trievel et al., 2002; Marmorstein, 2003; Ng et al., 2007). Those organisms lacking SRA domains contain SET domain proteins as well as methylated histones. C. elegans protein Maternal-effect sterile protein 4 (MES-4) and D. melanogaster protein Mes4, have either/both a PHD and/or RING domain(s), and the N-terminal MES-4 PHD is required for chromosome association (Bender et al., 2006). A SET domain in association with a RING and/or PHD without an SRA domain is also observed in human and mouse proteins, but not in plants. Thus, it appears the SRA domain is specific to organisms with cytosine methylation, while SET domains are more widespread.
The ORTH PHD fits the PHD consensus (SMART, e=1.77e−05), including the spacing limitations, nature of the metal ligands and the characteristic tryptophan residue one residue N-terminal to the seventh metal ligand. Here we present in vitro data to support the hypothesis that the PHD does not function as an E3 ligase. However, it remains to be determined if the PHD can function as an E3 in vivo, or possesses E3 ligase activity with an untested E2. Recently, Liu et al (2007) reported that ORTH2/VIM1 PHD was required and sufficient for interaction with NtSET in yeast two-hybrid assays. Thus, the ORTH/VIM PHD may be a protein interaction domain.
It is interesting that both loss of ORTH2/VIM1 (Woo et al., 2007) and expression of 35S::ORTH1 or 35S::GFP-ORTH2 lead to loss of DNA methylation. A similar phenomenon was seen with GL2 (GLABRA2) expression, where 35S::GL2 had the same phenotype as a loss-of-function line (Ohashi et al., 2002). The mechanism responsible is not understood. One possibility is that there are decreased ORTH mRNA levels rather than increased, due to co-suppression in the 35S expression lines. However, endogenous ORTH1 mRNA levels were not substantially changed in GFP-ORTH1 lines tested (data not shown), eliminating this as a mechanism. Another possibility is that over-expression leads to a dominant negative effect, for example, by sequestering other proteins. The large GFP tag is not responsible for the dominant negative effect, because expression of untagged ORTH1 also results in a late flowering phenotype, but over-expression of untagged and tagged ORTH proteins could reduce the free levels of other proteins. Finally, expression of hUHRF1 is regulated through the cell cycle, with highest expression just before S phase (Miura et al., 2001). It is possible that the phenotype is derived from 35S driven expression throughout the cell cycle.
In this study, over-expression of either ORTH1 or ORTH2 phenocopy the vim1 mutant at the Cen180 repeats, implicating both proteins in the same pathway. However, the 35S::ORTH lines have a more severe phenotype than vim1, since these lines lose CpG methylation additionally at FWA, as well as at the heterochromatic Cen180 repeats. This result implicates the ORTH family members in activities outside of centromeric DNA methylation maintenance. In addition to affecting flowering time, over-expression of ORTH2/VIM1 affects root growth (Liu, et al., 2007).
While the target for ubiquitylation by the ORTH/VIM family of proteins remains uncertain, it is clear that this family has an important role in DNA methylation and subsequent gene regulation. While single insertion lines have no obvious macroscopic phenotype [unpublished data and (Liu et al., 2007; Woo et al., 2007))], T-DNA insertions in ORTH2/VIM1 have decreased methylation at centromeric repeats and the vim1-2 allele exhibited increased centromeric decondensation by FISH hybridization (Woo et al., 2007). Generation of multiple knockout lines in ORTH family members will be required to further elucidate ORTH in vivo function. Additional work to understand the intricacies of how ORTHs regulate DNA modifications will not only prove valuable in plants but aid in the understanding of the orthologous chordate proteins as well.
Experimental Procedures
Identification of Arabidopsis ORTH proteins
The RING domain of Arabidopsis CIP8 was used in BLAST searches against the complete non-redundant Arabidopsis genome (TAIR, The Arabidopsis Information Resource, April 16. 2003, http://www.arabidopsis.org; see also (Stone et al., 2005)). BLAST searches identified five proteins each with two RING domains, a PHD, and an SRA domain as well as one protein with only a single RING and SRA domain. The SMART database was used to analyze retrieved sequences (Simple Modular Architecture Research Tool, version 4.0 05/28/2004, http://smart.embl-heidelberg.de/) followed by manual inspection to confirm the presence of the complete PHD/RING/SRA domains.
Phylogenetic Analysis
The ClustalX program was used to generate an alignment of the ORTH protein sequence. The alignment was generated using a PAM350 protein matrix, with gap opening and gap extension penalty parameters of 35.0 and 0.75, respectively, in pairwise alignment and 15.0 and 0.3, respectively, in the multiple alignments (Thompson et al., 1997). MacClade sequence editor (Sinauer Associates, Inc., Sunderland MA) was used to manually edit the alignment. The phylogenetic trees were created by PAUP* (Phylogenetic Analysis Using Parsimony version 4.0, Sinauer Associates, Inc.) using the neighbor-joining method with 1000 bootstrap replicates.
Cloning and Mutagenesis
Arabidopsis ORTH cDNAs were cloned by reverse transcription (RT) reactions followed by polymerase chain reactions (PCR) to amplify the predicted open reading frame (ORF) and recombined into Gateway pDONR vector (Invitrogen, Carlsbad, CA). RNA was isolated from either Arabidopsis thaliana ecotype Col-0 10 day-old seedlings or floral tissue from 6 to 7-week-old plants using Qiagen RNeasy plant RNA extraction kit (Qiagen, Valencia, CA) per manufacturer instructions. DNA sequence was identical to the predicted ORF (http://www.arabidopsis.org). ORTHs were expressed in bacteria as glutathione-S-transferase fusions from pDEST15-based plasmids (Invitrogen, Carlsbad, CA). Quik-Change site-directed mutagenesis (Stratagene, La Jolla, CA) was used to make a series of point mutations in RING domain metal ligand codons. Truncations were made with gene-specific primers with in-frame stop codons using cDNA clones as template and the sequences were verified. Positions of truncations (nucleotide relative to ATG) are as follows: PHD only, nt 198; PHD and N-terminal RING nt 675; no C-terminal RING nt 1476. The protein with the SRA and C-terminal RING domains contains nucleotides 676 to stop. ORTH1 and ORTH2 coding regions were recombined into the pGWB6 binary transformation vector for localization studies (Nakagawa et al., 2007). Over-expression of tagless ORTH1 was achieved by recombination into pYL TAP C-t but with a stop codon present (Rubio et al., 2005).
Protein Expression and Purification
GST-ORTH fusions were expressed in E. coli strain BL21 AI or BL21-pLysS. Transformed cells were grown at 37 °C for 2 to 3 hours or to an OD600 of 0.4 – 0.6 before induction with 0.2% arabinose or 0.5mM IPTG, respectively, for 2 to 3 hours at 25°C. Cells were harvested by centrifugation and lysed in a buffer containing 25 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% Triton X-100. For purification, glutathione agarose (Sigma, St. Louis, MO) was added to cleared lysates and incubated for one hour at 4 °C. Beads were then washed four times with wash buffer containing 25 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.1% Triton X-100. GST fusion proteins were eluted with elution buffer containing 25 mM Tris-HCl pH 8.1, 50 mM Glutathione, and 0.1% Triton X-100. Glycerol was added to the eluted protein to a final concentration of 40%. Proteins were stored at −80 °C until needed.
In Vitro Ubiquitylation Assay
Ubiquitylation reactions (30 μL) containing 50 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 0.05 mM ZnCl2; 1 mM ATP; 0.2 mM dithiothreitol; 10 mM phosphocreatine; 0.1 unit creatine kinase (Sigma, St. Louis, MO); 50 ng yeast E1 (Boston Biochem, Cambridge, MA); 250 ng of recombinant Arabidopsis E2 (Kraft et al., 2005); 500 ng of GST-ORTH; and 2 μg ubiquitin (Sigma, St. Louis, MO) were incubated at 30°C for 2 h. For histone ubiquitylation assays, 2 μg of crude bovine nucleosomes (Sigma, St. Louis, MO) were added to the ubiquitylation reaction. Reactions were stopped by adding 6 μL of 5x SDS-PAGE sample buffer (125mM Tris-HCl, pH 6.8, 20% [v/v] glycerin, 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) and analyzed by SDS-PAGE followed by western blotting using ubiquitin and/or GST antibodies as described previously (Stone et al., 2005).
Plant Growth Conditions and GFP visualization
All transgenes were expressed in ecotype Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col). Seed was surface sterilized with 30% (v/v) bleach and 0.1% (v/v) Triton X-100 and grown on GM containing 1% (w/v) agar with 1X Murashige and Skoog salts (Sigma Chemical Co) under continuous light. Seedling roots were examined for GFP expression using a Nikon Optiphot 2 fluorescent microscope with a Nikon Super High Pressure Mercury Lamp power supply model HB-10101AF (Nikon, Melville, NY). Images were captured by a QImaging Retiga 1300 camera and QCapture 2.68.2 software (Quantitative Imaging Corporation, Burnaby, British Columbia).
FWA Expression
Six micrograms of RNA extracted from adult leaf tissue (Trizol, Invitrogen, Carlsbad, CA) was converted to cDNA (First Strand, Invitrogen, Carlsbad, CA). FWA transcript levels were analyzed by semi-quantitative real-time PCR, using primers JP3130 (CCCACCAAGATCTGAAGTCC) and JP3133 (CAGGTGCAATGGTGGTGTAT) and internal primer M12 (CCTTCGGGATTTTCGATAGTGCCA). ACTIN was used to standardize cDNA levels and was quantified with primers JP2452 (TCGTGGTGGTGAGTTTGTTAC) and JP2453 (CAGCATCATCACAAGCATCC) and visualized with Brillant SYBR Green QPCR Master Mix (Statagene, La Jolla, CA).
Southern Blot Analysis
DNA was extracted from leaves or flowers of recently bolted plants using a slightly modified CTAB/sarkosyl method (Bernatzky and Tanksley, 1986). For Cen180 Southern blots, one half to one microgram of DNA was digested with HpaII overnight and run for 4 hr on a 1 % agarose gel, and transferred to membrane. The membrane was incubated overnight with radiolabeled probe for Cen180 bp repeats (Vongs et al., 1993). After washing, the membrane was exposed to film for four hours. FWA Southern blots were performed as previously described (Chan et al., 2004).
Supplementary Material
Acknowledgments
We thank Drs X-W. Deng (Yale University) and T. Nakagawa (Shimane University) for plant transformation plasmids. Research in the Callis laboratory supported in part by the National Science Foundation (2010 Program MCB-0519970) and the Department of Energy (DE-FG02-03ER15416). E.B. was partially funded by National Institutes of Health Predoctoral Training Grant in MCB (GM-0007377-27). M.B. was partially funded by the American Heart Association Postdoctoral Fellowship 0625014Y and the National Institutes of Health National Research Service Award 5F32CA126302. Work in the Jacobsen lab is funded by NIH grant GM060398. S.E.J. is an Investigator of the Howard Hughes Medical Institute. JC is the Paul K. and Ruth R. Stumpf Professor of Plant Biochemistry.
Contributor Information
Edward Kraft, Email: ealkraft@gmail.com.
Magnolia Bostick, Email: mbostick@ucla.edu.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Aguilera M, Oliveros M, Martinez-Padron M, Barbas JA, Ferrus A. Ariadne-1: a vital Drosophila gene is required in development and defines a new conserved family of ring-finger proteins. Genetics. 2000;155:1231–1244. doi: 10.1093/genetics/155.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4433. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD. Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics. 1986;112:887–898. doi: 10.1093/genetics/112.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace IM, Latella L, Papait R. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SWL, Zilberman D, Xie Z, Johansen LK, Carrington JC, SEJ RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim Biophys Acta. 2007;1769:276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, Hotta H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem. 2003;278:14657–14668. doi: 10.1074/jbc.M211285200. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Jeanblanc M, Mousli M, Hopfner R. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- Jenkins Y, Markovtsov V, Lang W. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. Genome Analysis and Functional Characterization of the E2 and RING domain E3 ligase Ubiquitination Enzymes of Arabidopsis thaliana. Plant Physiol. 2005;139:1597–1611. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu Y, Ruan Y, Meyer D, Wolff M, Xu L, Wang N, Steinmetz A, Shen WH. The SET- and RING-associated domain proteins in heterochromatinization. Plant J. 2007;5:914–926. doi: 10.1111/j.1365-313X.2007.03286.x. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Miura M, Watanabe H, Sasaki T, Tatsumi K, Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- Mladek C, Guger K, Hauser MT. Identification and characterization of the ARIADNE gene family in Arabidopsis. A group of putative E3 ligases. Plant Physiol. 2003;131:27–40. doi: 10.1104/pp.012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Li Y, Hata H, Kochi H. NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 2004;557:209–214. doi: 10.1016/s0014-5793(03)01495-9. [DOI] [PubMed] [Google Scholar]
- Mousli M, Hopfner R, Abbady AQ. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Brit J Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta. 2007;1769:316–329. doi: 10.1016/j.bbaexp.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Ruberti I, Morelli G, Aoyama T. Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 2002;29:359–369. doi: 10.1046/j.0960-7412.2001.01214.x. [DOI] [PubMed] [Google Scholar]
- Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol Biol Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci USA. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Stary S, Yin XJ, Potuschak T, Schlogelhofer P, Nizhynska V, Bachmair A. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 2003;133:1360–1366. doi: 10.1104/pp.103.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nishidate T, Nakamura Y. CBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA Methylation Mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura- Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.